Published online Mar 9, 2024. doi: 10.5409/wjcp.v13.i1.90755
Peer-review started: December 12, 2023
First decision: December 19, 2023
Revised: January 1, 2024
Accepted: February 6, 2024
Article in press: February 6, 2024
Published online: March 9, 2024
Processing time: 85 Days and 8.7 Hours
Ulcerative colitis (UC) is an immune-mediated chronic inflammatory condition with a worldwide distribution. Although the etiology of this disease is still unknown, the understanding of the role of the microbiota is becoming increas
To investigate the predictive power of the gut microbiota for the diagnosis of UC in a cohort of newly diagnosed treatment-naïve Saudi children with UC.
The study population included 20 children with a confirmed diagnosis of UC and 20 healthy controls. Microbial DNA was extracted and sequenced, and shotgun metagenomic analysis was performed for bacteria and bacteriophages. Biostatistics and bioinformatics demonstrated significant dysbiosis in the form of reduced alpha diversity, beta diversity, and significant difference of abundance of taxa between children with UC and control groups. The receiver operating characteristic curve, a probability curve, was used to determine the difference between the UC and control groups. The area under the curve (AUC) represents the degree of separability between the UC group and the control group. The AUC was calculated for all identified bacterial species and for bacterial species identified by the random forest classification algorithm as important potential biomarkers of UC. A similar method of AUC calculation for all bacteriophages and important species was used.
The median age and range were 14 (0.5-21) and 12.9 (6.8-16.3) years for children with UC and controls, respectively, and 40% and 35% were male for children with UC and controls, respectively. The AUC for all identified bacterial species was 89.5%. However, when using the bacterial species identified as important by random forest classification algorithm analysis, the accuracy increased to 97.6%. Similarly, the AUC for all the identified bacteriophages was 87.4%, but this value increased to 94.5% when the important bacteriophage biomarkers were used.
The very high to excellent AUCs of fecal bacterial and viral species suggest the potential use of noninvasive microbiota-based tests for the diagnosis of unusual cases of UC in children. In addition, the identification of important bacteria and bacteriophages whose abundance is reduced in children with UC suggests the potential of preventive and adjuvant microbial therapy for UC.
Core Tip: This study reports the predictive power of fecal microbiota, bacteria and bacteriophages, in predicting the diagnosis of ulcerative colitis in children. This was demonstrated by the calculation of the area under the receiver operating characteristic curve (AUC). High values of the AUC up to 97.6% and 94.5% for bacteria and bacteriophage, respectively, indicate excellent predictive power in differentiating children with ulcerative colitis (UC) from controls. This finding may lead to the development of noninvasive microbiota-based test for the diagnosis of unusual cases of UC in children.
- Citation: El Mouzan M, Al Sarkhy A, Assiri A. Gut microbiota predicts the diagnosis of ulcerative colitis in Saudi children. World J Clin Pediatr 2024; 13(1): 90755
- URL: https://www.wjgnet.com/2219-2808/full/v13/i1/90755.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v13.i1.90755
Ulcerative colitis (UC) is an immune-mediated inflammatory bowel disease. Although the incidence of this disease is highest in Western populations, it is increasing globally[1-3]. The etiology of UC is unknown; however, multifactorial factors involving interactions between genetics, host immunity, the mucosal barrier, and the gut microbiome are highly suspected[4-6]. The role of the microbiota has been extensively reported mainly in Western populations, with strong evidence of an association with UC.
In Saudi Arabia, a developing country in transition, the incidence and clinical patterns of UC have been reported[7-10]. In addition, the microbiota profile of Saudi children with Crohn’s disease (CD) has been reported to be significantly associated with not only the presence of bacteria but also the high area under curve (AUC) for bacteria in fecal samples, suggesting high accuracy in predicting the diagnosis of CD[11,12]. However, there are no reports on the predictive power of the microbiota for the diagnosis of UC. The objective of this study was to evaluate the role of the microbiota in predicting the diagnosis of UC in Saudi children.
Children with a confirmed diagnosis of UC were enrolled in the study. The children were recruited from multiple hospitals in Riyadh, Kingdom of Saudi Arabia. The inclusion criteria included new-onset and untreated disease, as well as no antibiotic exposure for at least 6 months before stool collection. Fecal samples from the children with UC were collected before bowl preparation. Healthy school children were randomly selected as controls. Stool samples from children with UC and controls were collected in cryovials without fixatives or stabilizers and immediately stored at −80°C until analysis.
Bacterial and viral DNA from fecal samples was isolated using the QIAGEN DNeasy PowerSoil Pro Kit according to the manufacturer’s protocol. DNA libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina) and IDT Unique Dual Indexes with a total DNA input of 1ng. Library were subsequently sequenced on an Illumina NovaSeq S4 platform.
Shannon alpha diversity metrics were calculated in R using the R package “vegan”. Wilcoxon rank-sum tests were performed between groups using the R package ggsignif[13,14]. Bray-Curtis dissimilarity was calculated in R using the vegan package with the function vegdist, and PCoA tables were generated using the ape function pcoa. PERMANOVA tests for each distance matrix were generated using the vegan’s6 function adonis2, and beta dispersion was calculated and compared using the ANOVA method for the betadispering function from vegan[15]. DESeq2 was used to estimate differential abundance between cohorts based on count data[16]. The random forest classification algorithm was applied to the relative abundance data to predict bacterial and viral species biomarkers that might improve prediction[17].
The receiver operating characteristic (ROC) curve was used to determine the difference between the UC and control groups. The area under the curve (AUC) represents the degree of separability between the UC group and the control group. The AUC was calculated for all identified bacterial and bacteriophage species in this study and for bacterial and bacteriophage species identified by the random forest classification algorithm as important potential biomarkers of UC[18].
Ethical aspects: The study was approved by the Institutional Board Review of the College of Medicine, King Saud University in Riyadh, Kingdom of Saudi Arabia [No: 10/2647/IRB,26/6/2010]. Guardians and/or children signed informed consent and/or assent before enrollment in the study.
The median age and range were 14 (0.5-21) and 12.9 (6.8-16.3) years for children with UC and controls, respectively, and 40% and 35% were male for children with UC and controls, respectively. A high number of significant bacterial and bacteriophage dysbiosis events were found (unpublished data). Among these, 11 bacterial species biomarkers were identified. These included the Bifidobacterium angulatum, Alistipes putredinis, Bacteroides caccae, and Bifidobacterium adolescentis (Table 1). Similarly, among the high number of bacteriophages, four were identified as biomarkers. These included the Salmonella phage SEN4, Streptococcus phage YMC-2011, and uncultured crAssphage (Table 2).
| S. No. | Bacterial species | Mean | Median | Minimum | Maximum | Decision |
| 1 | Alistipes communis | 3.199 | 3.222 | 1.528 | 5.055 | Confirmed |
| 2 | Alistipes putredinis | 6.748 | 7.094 | 3.605 | 8.565 | Confirmed |
| 3 | Bacteroides caccae | 5.914 | 6.28 | 2.717 | 7.552 | Confirmed |
| 4 | Bifidobacterium adolescentis | 5.843 | 6.123 | 3.073 | 7.578 | Confirmed |
| 5 | Bifidobacterium angulatum | 8.89 | 9.47 | 4.265 | 10.827 | Confirmed |
| 6 | Bifidobacterium bifidum | 4.138 | 4.293 | 1.512 | 5.794 | Confirmed |
| 7 | Bifidobacterium catenulatum | 5.544 | 5.823 | 2.246 | 7.352 | Confirmed |
| 8 | Dialister succinatiphilus | 3.418 | 3.594 | -0.47 | 4.86 | Confirmed |
| 9 | Peptostreptococcus stomatis | 3.367 | 3.411 | 1.358 | 4.983 | Confirmed |
| 10 | Prevotella copri | 3.826 | 3.812 | 1.463 | 5.595 | Confirmed |
| 11 | Streptococcus_u_s | 3.987 | 3.93 | 1.595 | 6.232 | Confirmed |
| S. No. | Bacteriophage | Mean | Median | Minimum | Maximum | Decision |
| 1 | Salmonella phage SEN4 | 5.311 | 5.474 | 2.349 | 8.294 | Confirmed |
| 2 | Siphoviridae_u_s | 7.224 | 7.591 | 3.16 | 10.1 | Confirmed |
| 3 | Streptococcus phage YMC-2011 | 7.989 | 8.611 | 3.409 | 11.18 | Confirmed |
| 4 | uncultured crAssphage | 18.35 | 20.11 | 6.433 | 23.25 | Confirmed |
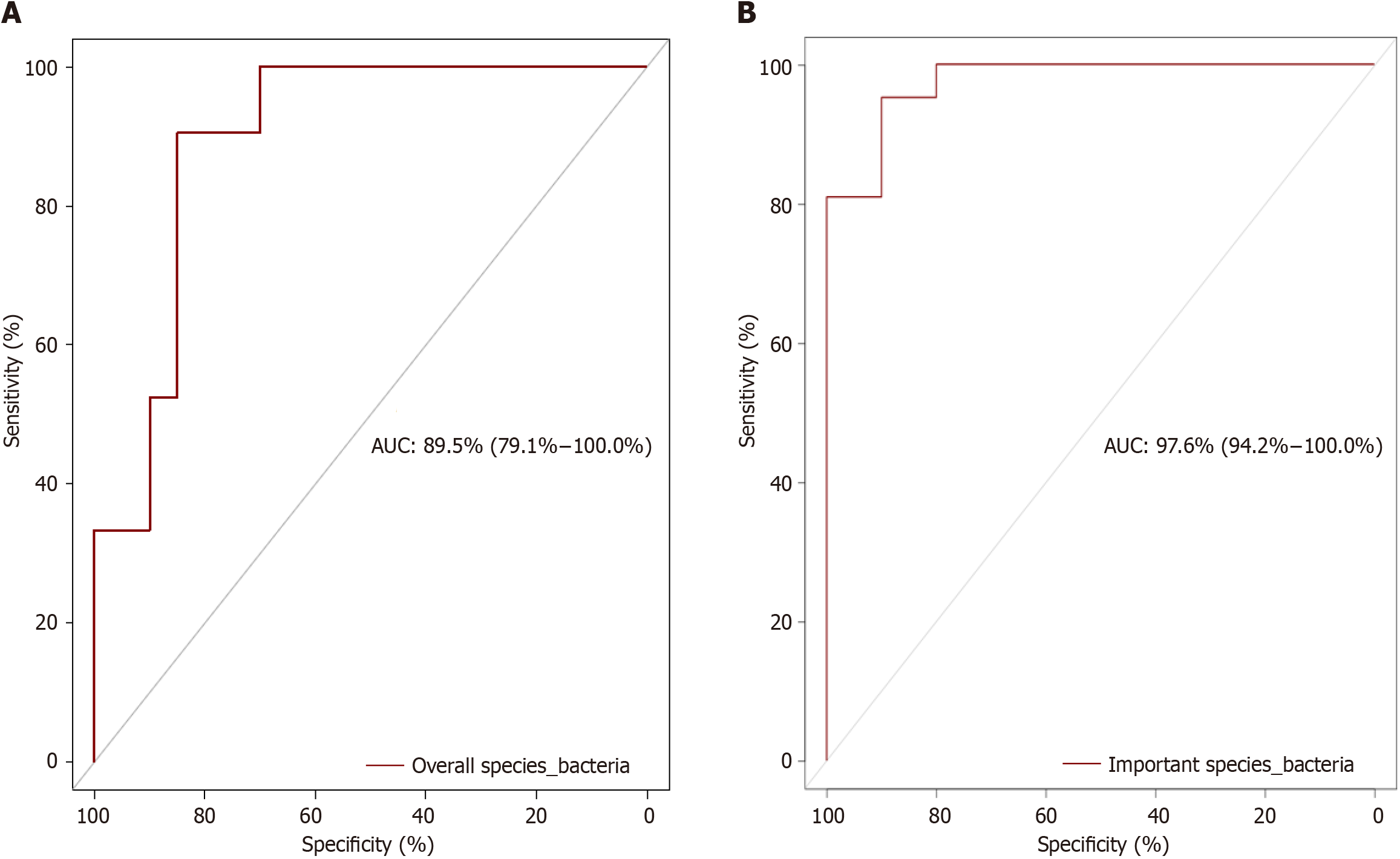
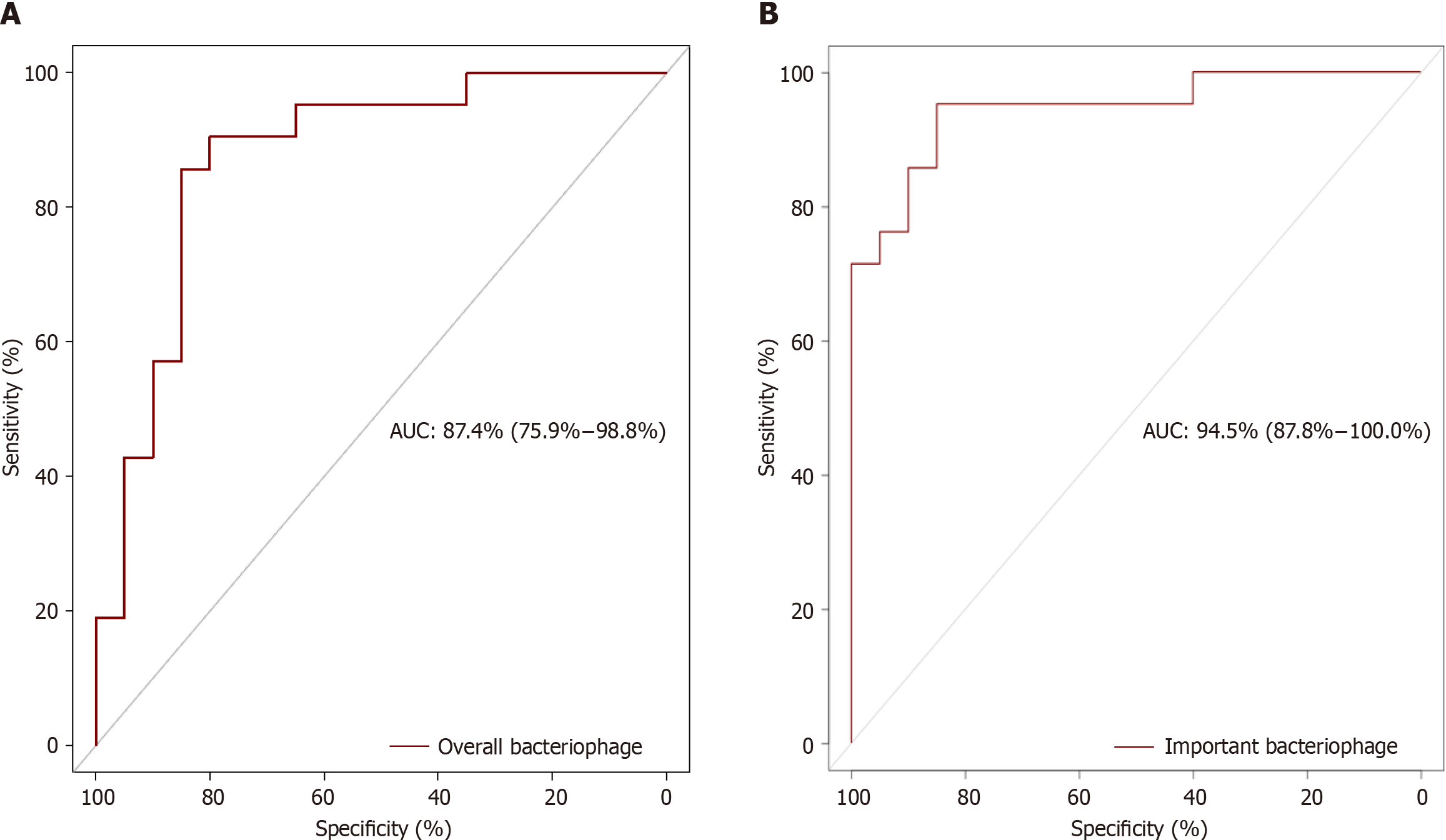
The AUC for all identified bacterial species was 89.5% (79.1%-100.0%), but when based on the biomarkers, the accuracy increased to 97.6% (94.2%-100.0%) indicating very good to excellent predictive power (Figure 1). Similarly, the AUC for all the identified bacteriophages was 87.4% (75.9%-98.8%), but the AUC increased to 94.5 % (87.8%-100%), when the identified important species were used, indicating very good to excellent predictive power (Figure 2).
Shotgun metagenomic analysis of bacterial and viral bacteriophage species in fecal samples of children with new-onset untreated UC revealed significant differential abundances between the UC group and the control group, indicating significant dysbiosis (unpublished data). The AUC of the ROC curve represents the degree of separability between the UC group and the control group, indicating the predictive power of the ROC curve for UC diagnosis.
In this study, we calculated the AUC based not only on the entire bacterial species and bacteriophages but also on important species identified by the random forest classification algorithm. The calculated AUC based on the abundance of all the bacterial species increased from 89.5% to 97.6% when only 11 bacterial species biomarkers were considered, indicating increased predictive power of the important bacterial species biomarkers. Similarly, the AUC calculated based on the bacteriophages increased from 87.4% to 94.5% when only four biomarkers were considered, indicating that the use of these bacteriophage biomarkers has greater predictive power for distinguishing UC patients from controls. The excellent predictive power of these biomarkers indicates the potential for the development of microbiota-based diagnostic tests. Among the bacteria and bacteriophages, Bifidobacterium angulatum and uncultured crAssphage had the highest median importance scores. Bifidobacterium angulatum is a species that belongs to the Bifidobacterium genus that is known to modulate the immune system and may be considered protective against UC[19,20]. Uncultured crAssphage is the most abundant human-associated virus and is found in the gut virome in approximately 50% of humans. This virus infects species of Bacteroides with mostly beneficial effects on health. Accordingly, Bifidobacterium angulatum and uncultured crAssphage could constitute the basis of prophylactic or therapeutic options[21-24].
The excellent predictive diagnostic power for UC in this report is slightly greater but consistent with the 93% accuracy for UC diagnosis reported within a multiclass disease in an adult study in Hong Kong[25] and the 91% accuracy in a group of children with UC in which shotgun metagenomic bacterial species-level abundance was used[26]. Finally, the 84.4% to 95% predictive power of the bacteriophage species identified in this study has not been reported thus far and deserves further study.
Study limitations: This study had a relatively small sample size, but it may be acceptable for this is the first study to use metagenomic analysis in a non-Western childhood population to determine the accuracy of the microbiota in predicting the diagnosis of UC.
The very high to excellent AUCs of fecal bacterial and viral species indicate the potential for the development of noninvasive microbiota-based tests for the diagnosis of UC and for preventive and adjuvant microbial therapy for UC. In addition, the identification of important bacteria and bacteriophages whose abundance is reduced in children with UC suggests the potential of preventive and adjuvant microbial therapy.
Microbiota dysbiosis has been reported in patients with ulcerative colitis (UC).
The role of the microbiota in predicting UC has rarely been reported.
To evaluate the predictive power of fecal bacteria and bacteriophages for diagnosing UC in children.
Metagenomic analysis of bacterial and bacteriophage DNA in the stool of children with newly diagnosed UC. The area under the curve (AUC) was calculated to evaluate the predictive power of the total bacteria and bacteriophages, and random forest analysis was used to identify important microbes for distinguishing UC patients from controls.
The discriminatory power of the entire bacterial species (AUC: 89.5%) and bacteriophages (AUC: 87.4%) was very high. The random forest classification algorithm analysis revealed the excellent predictive power of important bacterial species (AUC: 97.6%) and bacteriophages (AUC: 94.5%).
The very high to excellent AUCs of fecal bacterial and viral species indicate the potential for the development of noninvasive microbiota-based tests for the diagnosis of UC in children. In addition, the identification of important bacteria and bacteriophages whose abundance is reduced in children with UC suggests the potential of preventive and adjuvant microbial therapy for UC.
Future research in this area with larger sample sizes is needed to clarify the role of the microbiota in the diagnosis, prevention, and treatment of UC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ding X, China S-Editor: Liu JH L-Editor: A P-Editor: Zhao YQ
| 1. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3789] [Cited by in RCA: 3526] [Article Influence: 271.2] [Reference Citation Analysis (5)] |
| 2. | Malaty HM, Fan X, Opekun AR, Thibodeaux C, Ferry GD. Rising incidence of inflammatory bowel disease among children: a 12-year study. J Pediatr Gastroenterol Nutr. 2010;50:27-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 160] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 3. | Khan R, Kuenzig ME, Benchimol EI. Epidemiology of Pediatric Inflammatory Bowel Disease. Gastroenterol Clin North Am. 2023;52:483-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 4. | Turpin W, Goethel A, Bedrani L, Croitoru Mdcm K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm Bowel Dis. 2018;24:1133-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 5. | Kayama H, Okumura R, Takeda K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu Rev Immunol. 2020;38:23-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 439] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 6. | Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 1009] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 7. | El Mouzan MI, Saadah O, Al-Saleem K, Al Edreesi M, Hasosah M, Alanazi A, Al Mofarreh M, Asery A, Al Qourain A, Nouli K, Al Hussaini A, Telmesani A, AlReheili K, Alghamdi S, Alrobiaa N, Alzaben A, Mehmadi A, Al Hebbi H, Al Sarkhy A, Al Mehaidib A, Al Saleem B, Assiri A, Wali S. Incidence of pediatric inflammatory bowel disease in Saudi Arabia: a multicenter national study. Inflamm Bowel Dis. 2014;20:1085-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Saadah OI, El Mouzan M, Al Mofarreh M, Al Mehaidib A, Al Edreesi M, Hasosah M, Al-Hussaini A, AlSaleem K. Characteristics of Pediatric Crohn's Disease in Saudi Children: A Multicenter National Study. Gastroenterol Res Pract. 2016;2016:7403129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | AlSaleem K, El Mouzan MI, Saadah OI, AlSaleem B, Al-Hussaini A, Hassosa M, Ali AM, Banemai MO, Halaby H, El Edreesi M. Characteristics of pediatric ulcerative colitis in Saudi Arabia: a multicenter national study. Ann Saudi Med. 2015;35:19-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Al-Hussaini A, El Mouzan M, Hasosah M, Al-Mehaidib A, ALSaleem K, Saadah OI, Al-Edreesi M. Clinical Pattern of Early-Onset Inflammatory Bowel Disease in Saudi Arabia: A Multicenter National Study. Inflamm Bowel Dis. 2016;22:1961-1970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | El Mouzan MI, Winter HS, Assiri AA, Korolev KS, Al Sarkhy AA, Dowd SE, Al Mofarreh MA, Menon R. Microbiota profile in new-onset pediatric Crohn's disease: data from a non-Western population. Gut Pathog. 2018;10:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | El Mouzan MI, Winter HS, Al Sarkhy AA, Korolev K, Menon R, Assiri AA. Bacterial dysbiosis predicts the diagnosis of Crohn's disease in Saudi children. Saudi J Gastroenterol. 2021;27:144-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. vegan: Community Ecology Package 2019. R package version 2.5-6. Accessed May 15, 2023. Available from: https://CRAN.R-project.org/package=vegan. |
| 14. | Ahlmann-Eltze C. ggsignif: Significance Brackets for ‘ggplot2’ 2019. R package version 0.6.0. Accessed May 15, 2023. Available from: https://CRAN.R-project.org/package=ggsignif. |
| 15. | Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016. |
| 16. | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34752] [Cited by in RCA: 57280] [Article Influence: 5728.0] [Reference Citation Analysis (0)] |
| 17. | Kursa MB, Rudnicki WR. “Feature Selection with the Boruta Package. J Stat Softw. 2010;36:1-13. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1713] [Cited by in RCA: 1731] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 18. | Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6456] [Cited by in RCA: 8372] [Article Influence: 598.0] [Reference Citation Analysis (0)] |
| 19. | Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, Olin A, Wang J, Mikes J, Tan Z, Chen Y, Ehrlich AM, Bernhardsson AK, Mugabo CH, Ambrosiani Y, Gustafsson A, Chew S, Brown HK, Prambs J, Bohlin K, Mitchell RD, Underwood MA, Smilowitz JT, German JB, Frese SA, Brodin P. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884-3898.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 440] [Article Influence: 110.0] [Reference Citation Analysis (0)] |
| 20. | de Vrese M, Winkler P, Rautenberg P, Harder T, Noah C, Laue C, Ott S, Hampe J, Schreiber S, Heller K, Schrezenmeir J. Effect of Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, B. bifidum MF 20/5 on common cold episodes: a double blind, randomized, controlled trial. Clin Nutr. 2005;24:481-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 164] [Article Influence: 8.2] [Reference Citation Analysis (1)] |
| 21. | Aldars-García L, Chaparro M, Gisbert JP. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 115] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 22. | Wiredu Ocansey DK, Hang S, Yuan X, Qian H, Zhou M, Valerie Olovo C, Zhang X, Mao F. The diagnostic and prognostic potential of gut bacteria in inflammatory bowel disease. Gut Microbes. 2023;15:2176118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 23. | Manandhar I, Alimadadi A, Aryal S, Munroe PB, Joe B, Cheng X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am J Physiol Gastrointest Liver Physiol. 2021;320:G328-G337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 24. | Koonin EV, Yutin N. The crAss-like Phage Group: How Metagenomics Reshaped the Human Virome. Trends Microbiol. 2020;28:349-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 25. | Su Q, Liu Q, Lau RI, Zhang J, Xu Z, Yeoh YK, Leung TWH, Tang W, Zhang L, Liang JQY, Yau YK, Zheng J, Liu C, Zhang M, Cheung CP, Ching JYL, Tun HM, Yu J, Chan FKL, Ng SC. Faecal microbiome-based machine learning for multi-class disease diagnosis. Nat Commun. 2022;13:6818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 26. | Zuo W, Wang B, Bai X, Luan Y, Fan Y, Michail S, Sun F. 16S rRNA and metagenomic shotgun sequencing data revealed consistent patterns of gut microbiome signature in pediatric ulcerative colitis. Sci Rep. 2022;12:6421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |