Copyright
©The Author(s) 2016.
World J Obstet Gynecol. Feb 10, 2016; 5(1): 39-49
Published online Feb 10, 2016. doi: 10.5317/wjog.v5.i1.39
Published online Feb 10, 2016. doi: 10.5317/wjog.v5.i1.39
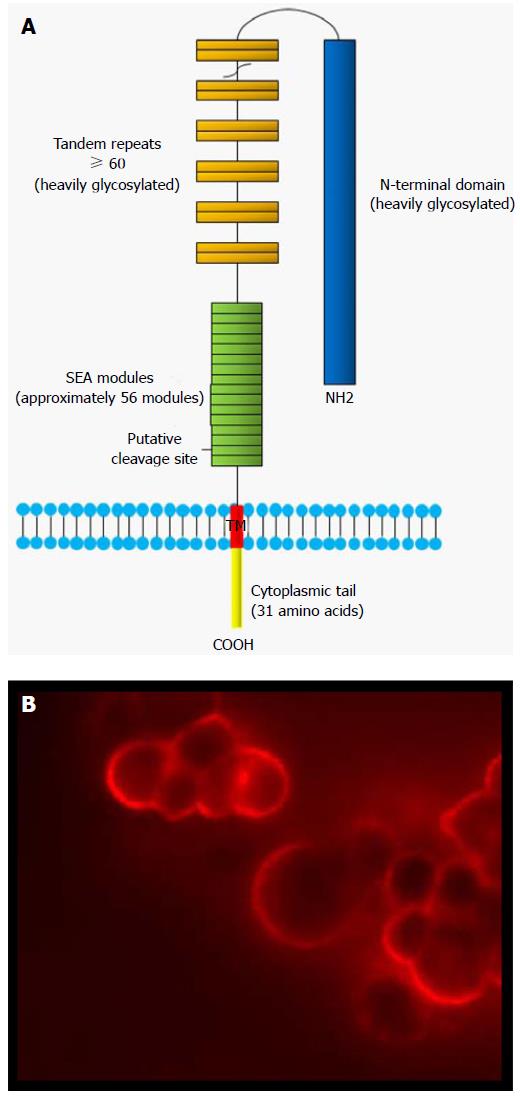
Figure 1 Schematic structure of MUC16 mucin.
A: The major domains of MUC16 include the N-terminal domain, the tandem repeat domain and the C-terminal domain. The SEA modules contain a putative proteolytic cleavage site which divides MUC16 in two subunits. The extracellular larger subunit consists of the N-terminal (> 12000 a.a.) and tandem repeat domains (156 a.a. each), and are heavily glycosylated. The smaller subunit contains SEA domains, a transmembrane domain (TM) and the cytoplasmic tail (31 a.a.); B: MUC16 is usually expressed at the apical surface of normal epithelial cells. In EOC cells, this pattern of expression is lost and MUC16 is expressed through the entire surface of the tumor cells. The micrograph represents OVCAR3 cells probed with M11 antibody. SEA: Sea-urchin sperm protein, enterokinase and agrin; a.a: Amino acids.
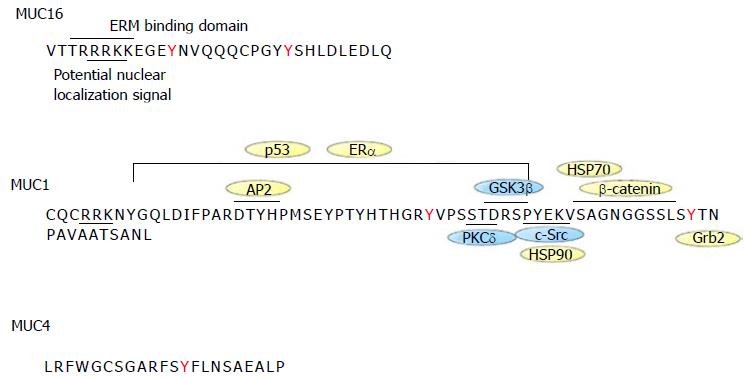
Figure 2 Sequence of MUC1, MUC4 and MUC16 cytoplasmic tails.
The intracellular sequence of the different mucins is shown along with protein interaction sites. MUC1 is the best characterized mucin. MUC1 cytoplasmic tail interacts with c-Src, GSK3β, PCKδ, β-catenin, p53, ERα, HSP70/90, Grb2, AP-2. Proteins with kinase activity are in blue whereas those without kinase activity are in yellow. HSP70 binds to MUC1 cytoplasmic tail in the same region as β-catenin. HSP90 binding to MUC1 depends on c-Src-induced Y-46 phosphorylation. MUC16 cytoplasmic tail has an ERM motif for potential interaction with the cytoskeleton. Both MUC1 and MUC16 contain a potential nuclear localization signal motif. MUC4 has no known interaction binding partners. EMR: Ezrin/radixin/moesin.
- Citation: Piché A. Pathobiological role of MUC16 mucin (CA125) in ovarian cancer: Much more than a tumor biomarker. World J Obstet Gynecol 2016; 5(1): 39-49
- URL: https://www.wjgnet.com/2218-6220/full/v5/i1/39.htm
- DOI: https://dx.doi.org/10.5317/wjog.v5.i1.39