Copyright
©The Author(s) 2017.
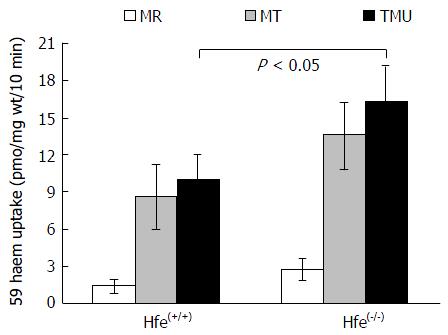
Figure 1 Tied loop mucosal uptake of 59Fe-heme (100 μmol/L) in wild type and hemochromatosis gene(-/-) mice.
Iron absorption was determined using tied-off duodenal segments. Data are means ± SD for 5 mice in each group (P < 0.05). MR: Mucosal retention; MT: Mucosal transfer; TMU: Total mucosal uptake of 59Fe from in vivo tied-off duodenal segments; Hfe: Hemochromatosis gene.
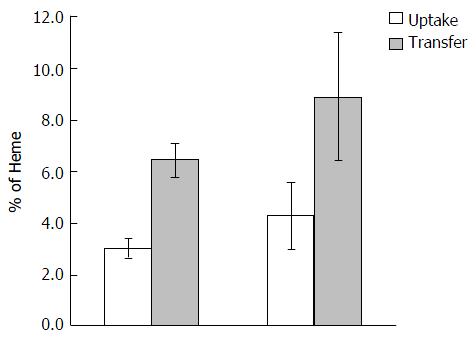
Figure 2 Heme absorption in wild type and hemochromatosis gene(-/-) mice by gavage method.
Mice were orally gavaged with 59Fe-heme (100 μmol/L) after an overnight fast. Mice were sacrificed 30 min after the oral dose was administered and tissues were collected as detailed in Methods. Values are mean ± SD, n = 5 per group.
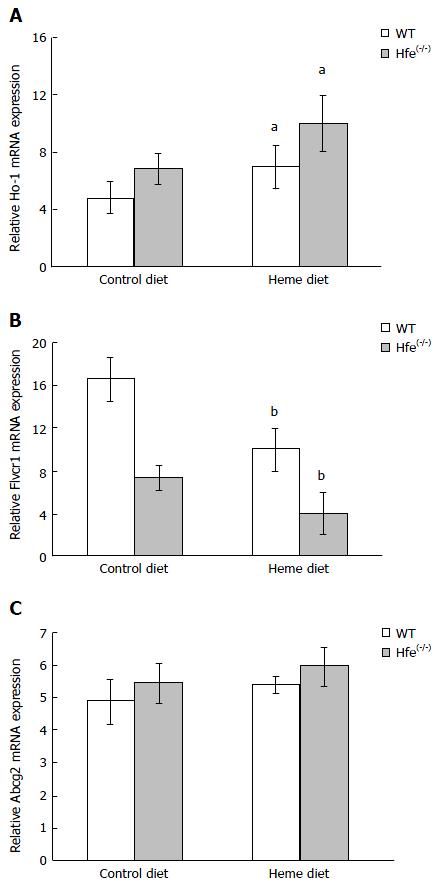
Figure 3 HO-1, Flvcr and Abcg2 mRNA expression in wild type and hemochromatosis gene(-/-) mice fed control diet or heme for 24 h.
Real-time polymerase chain reaction of mRNA of the genes from the duodenum of WT and Hfe(-/-) were determined and normalised β-actin (Actb) mRNA. Statistical analysis was performed by 2-way ANOVA with Bonferroni post-hoc test (aP < 0.05 and bP < 0.001). Hfe: Hemochromatosis gene; WT: Wild type.
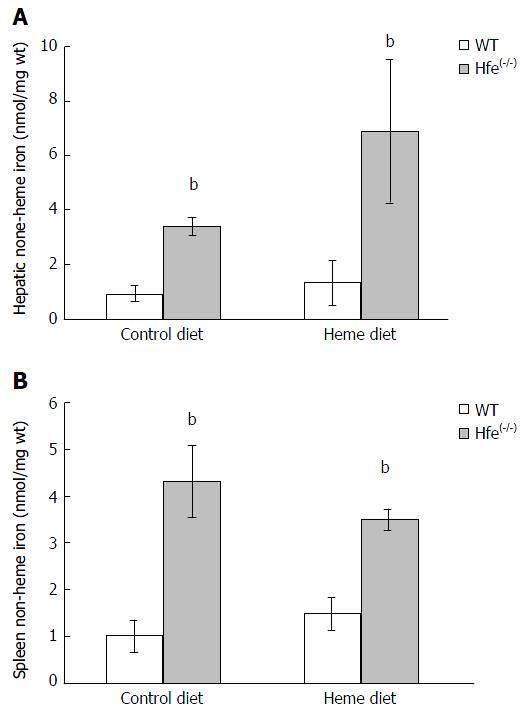
Figure 4 Tissue iron levels of mice.
Effect of 24 h heme feeding on liver (A) or spleen (B) non-heme iron levels (nmol/mg) of WT and Hfe(-/-) mice. Results are means ± SD for 6-8 mice in each group (bP < 0.005). Hfe: Hemochromatosis gene; WT: Wild type.
- Citation: Laftah AH, Simpson RJ, Latunde-Dada GO. Intestinal heme absorption in hemochromatosis gene knock-out mice. World J Hematol 2017; 6(1): 17-23
- URL: https://www.wjgnet.com/2218-6204/full/v6/i1/17.htm
- DOI: https://dx.doi.org/10.5315/wjh.v6.i1.17