Published online Oct 10, 2017. doi: 10.5306/wjco.v8.i5.378
Peer-review started: March 22, 2017
First decision: July 10, 2017
Revised: August 3, 2017
Accepted: September 5, 2017
Article in press: September 5, 2017
Published online: October 10, 2017
Processing time: 191 Days and 9.8 Hours
Metastasis is the major cause of mortality in cancer disease and still constitutes one of the most controversial mechanism, not yet fully understood. What is almost beyond doubt is that circulatory system is crucial for cancer propagation. Regarding this system, much attention has been recently paid to liquid biopsy. This technique is aimed to detect circulating tumor cells (CTCs) and circulating nucleic acids so it can be used as a tool for diagnostic, prognostic and follow-up of patients. Whereas CTCs tend to be scarce in serum and plasma from cancer patient, abundant circulating nucleic acids can be detected in the same location. This fact, together with the genetic origin of cancer, stands out the relevance of circulating nucleic acids and shed light into the role of nucleic acids as drivers of metastasis, a recently discovered phenomenon called Genometastasis. This innovative theory supports the transfer of oncogenes from cancer cells to normal and susceptible cells located in distant target organs through circulatory system. What is more, many biological processes haven been described to deliver and secrete circulating nucleic acids into the circulation which can allow such horizontal transfer of oncogenes. In this review, we focus not only on these mechanisms but also we demonstrate its putative role in cancer propagation and give insights about possible therapeutic strategies based on this theory. Our objective is to demonstrate how findings about cell-to-cell communications and previous results can agree with this unprecedented theory.
Core tip: Liquid biopsy not only constitutes a promising tool for cancer diagnostic and patient follow-up but also it may help in the comprehension of metastasis. This technique has revealed how circulating tumor cells are limited in blood, while circulating nucleic acids are much more abundant. This property, together with the demonstrated capability of circulating nucleic acids to transform susceptible cells, strongly support the theory of genometastasis. This theory sustains that cancer propagation relies on gene transfer from malignant cells to normal cells. We pretend to gather all these concepts, also including cell-to-cell communication mechanisms to demonstrate this phenomenon.
- Citation: García-Casas A, García-Olmo DC, García-Olmo D. Further the liquid biopsy: Gathering pieces of the puzzle of genometastasis theory. World J Clin Oncol 2017; 8(5): 378-388
- URL: https://www.wjgnet.com/2218-4333/full/v8/i5/378.htm
- DOI: https://dx.doi.org/10.5306/wjco.v8.i5.378
Traditionally, tissue biopsy has been used to diagnose and manage diseases. In cancer, biopsies are used to determine histological properties of the tumor as well as its genetic profile for diagnostic, prognostic purposes and prediction of response to therapies. However, the characteristic heterogeneity of tumors makes it necessary to analyze different parts of the same tissue which results in repeated sampling. Obtaining several tissue biopsies involves a high risk for the patient as well as economic cost for the system. As an alternative to tissue biopsy, liquid biopsy constitutes a promising and less invasive technique.
Liquid biopsy consists on the detection of cancer-derived molecular biomarkers, such as tumor cells or cell-free nucleic acids (cfNA) in biological fluids, mainly in blood. Given the non-invasiveness properties of the technique, it is possible to take repeated samples and so, to follow the progression and evolution of the disease in contrast to the static image from tissue biopsy.
The effectiveness of this approach has been demonstrated in different malignancies including breast, pancreatic and colorectal cancer (CRC)[1]. In the case of pancreatic cancer, liquid biopsy provides an advantageous technology regarding the anatomical and clinical difficulties for pancreatic tissue[2]. It would also help in the early detection of this disease, which is usually diagnosed at an advanced stage because it develops with no symptoms. For its part, CRC is mainly characterized by its heterogeneous genetic profile, in which new mutations constantly appear during tumor development[3]. These new mutations may confer proliferative capacities to tumor cells and, thus, molecular and genetic analysis of the whole tumor might be crucial during CRC follow-up. Similarly, tumor genotyping is also required in the case of anti-EGFR therapies, to which only the patients with KRAS wild-type gene respond. Thus, liquid biopsy can be conceived not only for recording tumor progression but also for selecting the most suitable treatment.
As mentioned before, liquid biopsy can be intended to detect circulating tumor cells (CTCs) and/or circulating cfNA.
CTCs can be secreted into circulation by primary and metastatic tumor deposits. In 1869, during autopsy of a breast cancer patient, CTCs were first identified as cells similar to those of the primary tumor, presented in the bloodstream[4]. These cells are mainly found in patients with malignant diseases like carcinomas, being extremely rare in healthy subjects and patients with nonmalignant diseases[5].
CTCs can be difficult to obtain given its heterogeneous morphology and its limited amount in the circulation: They constitute one cell per 1 × 109 normal bloodstream cells in patients with metastatic cancer[6]. In other terms, in 7.5 mL of blood from metastatic carcinoma patients, only 5 to 50 CTCs are presented on average[7]. This small cell number makes it difficult to detect CTCs, especially small subpopulations of tumor cells, which can harbor crucial mutation for tumorigenesis. However, many attempts and approaches have been designed to isolate CTCs. Most of them are based on antibody identification of cell surface markers, such as EpCAM, or size differences between CTCs and the rest of blood cells[8]. Once CTCs are obtained, they have to be further analyzed through genome sequencing. Nevertheless, these isolating techniques might not provide the whole spectrum of CTCs, uncovering the tumor heterogeneity. As an example, basal-like breast cancer CTCs with low levels of EpCAM may not be captured using this cell surface marker determinant[7]. On the other hand, false-positive CTC results can also be found in the case of patients with benign inflammatory disease such as Crohn disease. It has been shown that 11% to 19% of these patients present small numbers of circulating epithelial cells detectable that can be confused with CTCs[9]. In addition, although correlation between cell number and disease severity have been established in metastatic patients from breast, colon and prostate cancer[10-12], less is known about early-stage tumors and CTC number. Altogether, more studies are required to elucidate the relationship between tumor burden and the number of CTCs in order to verify the clinical utility of CTCs as prognostic markers[7].
It is also worth noting that CTCs are difficult to grow in culture, which questions the functionality of these cells. Thus, it can be hypothesized that these cells are more likely to constitute death cells, poured by tumor mass, than active cells responsible for metastasis emergence.
Regarding to circulating nucleic acids we can make a distinction between circulating cell-free DNA (cfDNA) and cell-free RNA (cfRNA).
Circulating cfDNA: The first association between cancer and the presence of circulating cfDNA was established in 1977 by Leon et al[13] who detected a higher concentration of DNA in serum from cancer patients. Ten years later, Stroun et al[14] confirmed this relation by isolating and characterizing DNA obtained from the plasma of cancer patients. Moreover, it was further shown that patients with malignant tumors have higher circulating cfDNA levels than patients suffering benign disease[15]. The tumor origin of such cfDNA was also confirmed by the identification of tumor-specific abnormalities such as loss of heterozygosity (LOH) of microsatellites and methylation of CpG islands[16,17]. In addition from tumor cells, plasma cfDNA may come from blood cells and other tissue-specific cells. However, the proportion of DNA derived from different origins widely varies. In fact, circulating tumor DNA proportion range between 0.01% and 93% in cancer patients[7,18].
cfDNA is usually found in plasma as with a double-stranded structure, although single-stranded circulating DNA has also been identified[19-21]. It should be noted that DNA molecule need to be protected by different complexes or other molecules, described in detail below, in order to avoid its degradation by serum nucleases.
Circulating cfRNA: Circulating cfRNA was first isolated in 1987 from serum of patients with malignant disorders and culture media of different malignant cell lines. It was initially found in the form of RNA-proteolipid complex[22]. As it happens with DNA, it is no surprising to detect cfRNA associated with other molecules since it alone can be very labile due to the increased amounts of RNases present in the circulation.
Circulating RNA consist of messenger RNA (mRNA) and microRNA (miRNA). Regarding to mRNA, different transcripts have been identified to be overexpressed in plasma of tumor patients, especially human transcriptase reverse telomerase (hTERT) mRNA levels in malignancies such as breast cancer or colon cancer[23-25]. miRNA molecules are fragments of 19-25 bp non-coding RNA molecules which derive from 70-100 bp hairpin precursor molecules. By posttranscriptional regulation, they modulate the expression of target genes involved in many physiological and pathological process such as development, cell proliferation, differentiation or apoptosis[26,27].
Circulating nucleic acids as biomarkers: Although the term nucleic acids refers to both types of molecules, special attention has been paid to cfDNA in the field of liquid biopsy because it carries the tumor-associated mutations and thus, it represents an attractive biomarker.
As commented before, circulating DNA gives more detailed information about the heterogeneity of the tumour because it may come from different cells with presumably different genomic alterations, which can be detected by sequencing.
Likewise, circulating cfDNA is much easier to isolate than CTCs because it is abundantly present in blood, especially in patients with advanced disease[28]. Indeed, circulating DNA extraction can be performed following a simple protocol that does not exceed 5 h[18]. Once it is isolated, PCR, followed by DNA sequencing can be used to detect tumor-specific genetic aberrations which may also help in the comprehension of tumor dynamics. In this issue, droplet digital PCR, together with genome-wide high throughput sequencing, provide a high sensitivity and specificity for detecting mutations[29]. These new tools for DNA analysis are also contributing to give a more profound insight into the presence and role of circulating DNA, among its value as a biomarker.
It would be reasonable to suspect that tumor cfDNA found in the circulation can be released by CTCs. However, the discrepancy between the number of CTCs and the quantity of circulating DNA discards this theory. Considering the average amount of circulating DNA in a ml of plasma from advanced-stage cancer patients (17 ng) and the amount of DNA contained by a single human cells (6 pg), more than 2000 CTCs would be required if CTCs were the primary source of circulating DNA. Conversely, less than 10 CTCs per 7.5 mL of blood are found on average[18]. Therefore, tumor cfDNA might come from different regions within the tumor and thus, it may better represent tumor genetic heterogeneity. This fact, together with its high concentration in blood, suggests that circulating DNA might be a better liquid biopsy-derived biomarker. In the following section, we will focus on the reasons why DNA can be released into circulation.
Depending on how they are released, circulating nucleic acids can be found in different forms including molecular or macromolecular complex, linked to serum proteins or internalized in vesicles such as exosomes or microvesicles (Figure 1). In general terms, circulating nucleic acids can be either passively released, by apoptotic and necrotic cells, or actively released by living cells.
During cell-death mechanisms, such as necrosis or apoptosis, both circulating DNA and circulating RNA can be liberated into bloodstream by dying or dead cells.
In necrosis, cellular DNA is incompletely and nonspecifically digested. In this condition, a smearing pattern would be observed when DNA is run electrophoretically in agarose gel. However, when circulating DNA is analyzed by agarose-gel electrophoresis, a ladder pattern is observed. This feature indicates that necrosis is not the major source of circulating DNA although it may be a possible contributor given the presence of DNA fragments ranging from 21 kb to 80 kb in length in blood plasma samples[14,30].
The mentioned ladder pattern is formed by fragments ranging from 180 bp to 1000 bp which matches with the fragments released from chromatin cleavage into nucleosomes, a process that occurs during apoptosis[30].
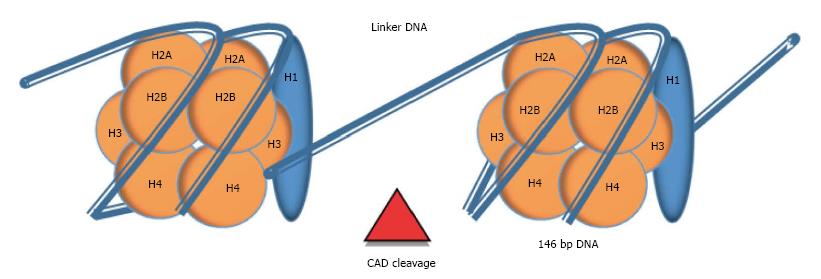
Nucleosome: Nucleosomes are molecular complexes that allow DNA stabilization and packing into the nucleus. In each nucleosome, 146 bp of double-stranded DNA are wound on an octamer of positively-charged proteins called histones (H2A, H2B, H3 and H4), through electrostatic interaction. Nucleosomes are linked by 10 to 100 bp of naked DNA, termed as linker DNA. An extra histone (H1), which is localized outside the octamer, stabilizes the tertiary structure of the chromatin chain[31] (Figure 2). Cell death by apoptosis implies the activation of a set of caspases that catalyze the hydrolysis of cellular components. Some of these caspases (e.g., Caspase-3) trigger the activation of endonucleases, especially the caspase-activated deoxyribonuclease (CAD). Endonucleases cleavage chromatin through linker DNA, the most accessible region, generating oligo- and mono-nucleosomes that are packed into vesicles called apoptotic bodies. Apoptotic bodies are subsequently released from the cells and phagocytosed by macrophages and dendritic cells. Nevertheless, in conditions when higher cellular turnover is required, such as inflammation or tumor cell proliferation, this process collapses and nucleosomes are liberated into circulation[30,31]. Then, cell-free nucleosomes can be internalized into cells by crossing plasma membrane and penetrating into the nucleus from where it can alter gene expression[32].
It is worth noting that the octamer of histones of the nucleosome protects DNA molecule from its degradation by circulating endonucleases. It also should be noted that tumor-derived circulating DNA may be more fragmented than DNA derived from healthy cells as recent publications have shown[33].
The apoptotic origin is confirmed by the existence of circulating mitochondrial DNA (mitDNA). In contrast to nuclear DNA, mitDNA is a circular and smaller (16.5 kb) molecule of DNA, not protected with histones[34]. It can be secreted to the circulation during cell death (e.g., apoptosis) and mitophagy, which consist on the elimination of damaged mitochondria through autophagy[35]. Due to its elevated copy number, circulating mitDNA may account for a high proportion of the total circulating DNA found in blood. It can be present in circulation in both protein-associated and free form[34]. Circulating mitDNA measurement and mutation analysis has been proposed to diagnose different malignancies such as breast tumors or epithelial ovarian cancer and hepatocellular or colorectal cancer, respectively[1,36].
In addition to passive secretion, circulating nucleic acids can be actively released through cell-derived vesicles, such as exosomes and microvesicles, from living cells. The phenomenon of spontaneously released DNA was first described in lymphocytes, frog auricles and cultured cell lines[37-43]. Like nucleosomes, vesicles protect cell-free nucleic acids from the circulating nucleases and hinder the recognition by the immune system[32].
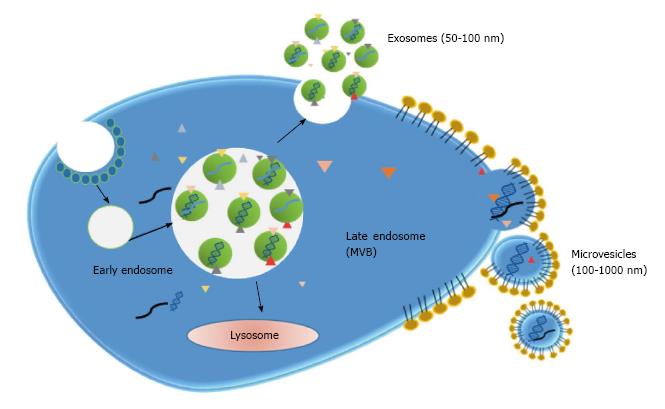
Exosomes: Exosomes are small lipid membrane vesicles (50-100 nm) secreted from various cell types including dendritic cells, B cells, T cells, tumor cells and epithelial cells[44]. Exosomes result from the recycling endosomal pathway. During endocytosis, vesicles are generated at the plasma membrane and enter into the cell forming early endosomes. These early endosomes are transformed into late endosomes which then develop multivesicular bodies (MVB). MVBs can fused with lysosomes for degradation of its content or with the plasma membrane. In this last case, internal vesicles are liberated into the extracellular space and termed exosomes[45] (Figure 3). Therefore, exosomes contain membrane and cytoplasmic components such as lipids, proteins and RNA (mainly mRNA and miRNA). Additionally, the presence of single-stranded and double-stranded DNA was further demonstrated[46].
Furthermore, exosomes are capable to enter in recipient cells by either binding to cell surface receptors through adhesion molecules or being internalized through mechanism similar to endocytosis and so can act as cellular communicators. What is more, these vesicles can travel to distant sites of the organism and release the packed biomolecules into local and remote cells. Exosomes can bear different proteins including transmembrane proteins, such as major histocompatibility complex (MHC), and other intraluminal proteins and oncoproteins such as mutant KRAS[47]. Proteins delivered by exosomes can activate or inhibit different signalling pathways, altering cell function. For its part, exosomes-derived miRNA can modulate gene expression by posttranscriptional regulation.
Particularly in cancer cells, exosomes secretion is usually increased. Tumor-derived (TD) exosomes may favour tumor growth by inhibiting apoptosis and increasing cellular proliferation. As an example, it was demonstrated that exosomes increased cellular proliferation in gastric cancer cell lines by activating Akt phosphorylation[48]. Moreover, it has been described that TD exosomes can also facilitate cancer invasion and metastasis by regulating stromal cells, remodelling the extracellular matrix and stimulating angiogenesis[47,49].
Regarding to nucleic acids, the presence of mRNAs, miRNAs and DNA highlights the role of exosomes as carriers of genetic information too. Indeed, the role of exosome-derived miRNA has been widely demonstrated. Depending on its target gene, miRNA can act either as a tumor suppressor or as a tumor enhancer. For instance, miR-198 has been demonstrated to be released by T-lymphoblast exosomes performing a tumor suppressor role in lung, liver and colorectal cancer[50-55]. Conversely, other miRNAs favour tumor progression such as miR-21, which can also be secreted through exosomes[56,57]. It should be considered that exosomes generally carry more than one kind of miRNA, so its effects depend on the combination of miRNAs presented[58].
Microvesicles: Microvesicles emerge from plasma membrane budding and the following fission of the vesicles from the plasma membrane. They have a larger size (100-1000 nm) than exosomes and membrane composition is more similar to that of plasma membrane than exosome membrane composition (Figure 3). Thus, tumor-derived microvesicles constitute a representation of the tumor proteomic signature. Microvesicles can be secreted by different cell types including hematopoietic cells, endothelial cells, mesenchymal stem cells and cancer cells[59]. It has to be taken into consideration that, despite their differences, the terms exosomes and microvesicles are usually interchanged. Moreover, in most studies, vesicles are obtained by approaches that cannot discriminate both types of vesicles and so it may be difficult to classify published information according to each type.
As well as exosomes, microvesicles are key elements in cell-to cell communication, modulating the recipient cell phenotype. For instance, it has been shown that cultured hematopoietic progenitor cells can be reprogrammed by microvesicles derived from embryonic stem cells. In fact, these microvesicles contained mRNA for several pluripotent transcription factors demonstrating an additional mechanism of horizontal transfer of genetic material[60].
In cancer scenario, microvesicles from tumor and non-tumor cells can also be secreted to transfer miRNA and other oncogenic proteins to facilitate invasion and tumor growth. Likewise, it was reported that tumor-derived microvesicles carrying surface determinants of tumor cells, like chemokine receptors, and mRNA for growth factors, such as vascular endothelial growth factor (VEGF) or hepatocyte growth factor (HGF), were able to internalize in monocytes and so, change its phenotype and biology activity[61]. Furthermore, it was also published that tumor-associated macrophages can secrete microvesicles containing miRNA that can promote breast tumor cell invasiveness[62].
Virtosomes: The existence of virtosomes was first described by Stroun and Gahan[63]. The virtosome is a macromolecular complex formed by newly synthesized DNA and RNA associated with lipoproteins, which is spontaneously released from living cells. To form this structure, newly DNA is synthesised in the nucleus and then transferred to the cytosol. In cytosol, DNA associates with a lipoprotein, which serves as a protector from nuclease digestion, and before leaving the cell, an RNA molecule is attached to the complex. The complex can exit the cell in an energy-dependent way and entering other cells by mechanism not well understood[63].
Viral nucleic acids: Viral DNA as well as viral RNA can be found in plasma and serum from patients[30]. Given the relation between some viral infections and particular malignancies, detection of viral DNA might be used as a biomarker for certain neoplastic disease. As an example, cell-free DNA from Epstein-Barr virus (EBV) serves as diagnostic and prognostic marker for nasopharyngeal carcinoma[64].
cfDNA and RNA can also be found attached to the exterior part of the plasma membrane from where they can be detached and released into circulation. DNA is usually found in the cell surface of leucocytes and erythrocytes and can be internalized by receptor recognition or remain associated with the surface[30].
Liquid biopsy has been commonly proposed as a tool for cancer diagnostic, characterisation and prognostic in patients as both CTCs and cfNA provide relevant information from the tumor. Nonetheless, very much attention has been paid for this practical application without taking full account of the possible biological roles of cfNA in blood. Although it is known how circulating nucleic acids can be presented in blood (as it has been described), its function in this location is still controversial. Considering the above commented discrepancy between CTCs number and cfDNA quantity as well as the active mechanisms of cfNA release, cfNA presence in blood does not appear to be a mere coincidence. What is more, many evidences point to cfNA as a key driver of metastasis, which is the essence of the theory of genometastasis.
Metastasis is an enormously complex process that remains to be a major problem in the management of cancer. The metastatic properties of tumor cells were extensively investigated from 1970s, although so much earlier (as soon as 1889) it was proposed the “seed an soil” theory that today is still alive and even under constant reformulation (e.g.,[65])
During the seventies, some theories were proposed, such as that most primary tumor cells have a low metastatic potential, and that during later stages of tumorigenesis rare cells acquire metastatic capacity through additional somatic mutations (reviewed in[66]). This suggested mechanism had contrary evidence in other studies that concluded that metastases are a random representation of disseminated tumor cells, all of which have the ability to form a metastasis[66]. On the whole, it might be said that the discussion of “dynamic heterogeneity” models vs “clonal dominance” theories prevailed during two decades, always under the premise of a circulatory view of cancer progression. In fact, nowadays, many authors appears to not conceive any other way, as showed in the recent literature, e.g., “Metastasis is the consequence of a cancer cell that disperses from the primary tumor, travels throughout the body, and invades and colonizes a distant site”[65].
This view does not explain some questions such as the lack of correlation between the sites of development of metastasis and the anatomic vascular filters[67]. Several million cells per gram of tumor can be shed daily into the lymphatic system or bloodstream. However, insufficient data exists to quantify the fraction of shed tumor cells that successfully seed secondary tissues. Moreover, the fate of blood borne tumor cells is controversial and many experimental evidence are contradictory: Whereas in some models most circulating cells die, in others most survive and extravasate. Nevertheless, all studies show that most cells entering the vasculature fail to form macroscopic foci at distant sites (reviewed in[68]). On the other hand, an unquestionable fact is that the identification and characterization of CTC require extremely sensitive and specific analytical methods, much more than detection of cell-free tumor DNA.
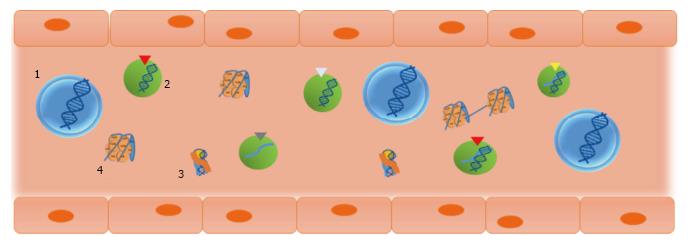
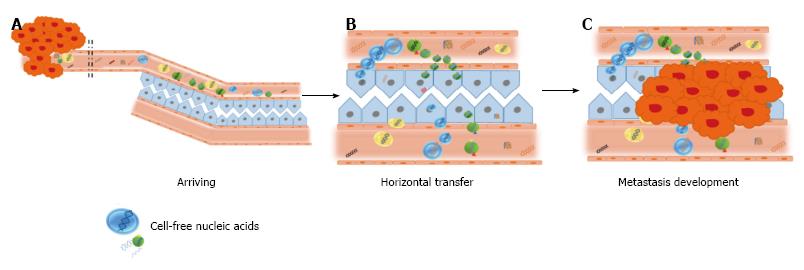
In connection with the circulatory theory, surgical maneuvers for tumor resection (particularly, gastrointestinal tumors) have classically been designed to avoid blood dissemination of cancer cells, which hypothetically results in a lower risk of recurrence and metastasis (reviewed in[69]). However, benefits of such procedures have not been fully demonstrated yet. At late 1990s, our group challenged that technical axiom-not sufficiently supported-, and performed a study in colorectal cancer patients that showed that the use of no-touch isolation techniques in colorectal cancer was not justified, based on lack of evidence indicating the detachment of cells from the tumor at surgery[70]. Apart from the clinical discussion (which has not been finished yet), the fact was that circulation of tumor cells appeared to have lesser value than attributed. In parallel, the evidence of high levels of cell-free nucleic acids in plasma of cancer patients and tumor-bearing animals led us to examine the biological role of such molecules[1,29,71]. Firstly in cancer models using immunocompetent animals and later in clinical studies with colon cancer patients, we demonstrated the biological feasibility of gene transfer and of the transformation of cells by cell-free tumor-derived nucleic acids in the plasma[41,72,73]. In the light of such results, we proposed that cell-free nucleic acids in the plasma participate in tumorigenesis and the development of metastases via transfection-like uptake of such nucleic acids by susceptible cells. This putative phenomenon was named as “genometastasis” (Figure 4).
Albeit, at first, some authors exhibited more criticism than enthusiasm for this hypothesis[74], later experimental evidence supported the existence of the genometastasis. Moreover, the assays that substantiated this theory were repeated and enlarged by other authors, who confirmed our results[75-78].
Consistently with our theory, recently Mittra et al[79] have asserted that circulating nucleic acids, far from being biologically inert particles, have significant deleterious functions in the host. According with their results, they concluded that circulating nucleic acids are ubiquitous and continuously arising, and freely can enter healthy cells integrate into their genomes, inflicting repeated damage to the somatic DNA. Moreover, the authors have suggested that the somatic genome may not be stable, but rather remains in a state of turmoil characterized by dsDNA breaks, genomic instability and apoptosis affected by integration of circulating DNA. These events may lead to deletions, duplications and rearrangements causing DNA mosaicism[79]. Once demonstrated the existence of this phenomena and connecting all previous results, it would be even naive to think that progression of cancer is not related to triggering genetic events and consequent genomic rearrangements.
Nonetheless, despite the soundness of results, some authors were still showing their reticence to accept the genometastasis as a feasible mechanism for metastasis, arguing mainly that such theory is not able to explain the tropism of metastasis[80]. In our opinion, this is an erroneous assessment perhaps motivated by a partial view of the phenomenon that we described, because, precisely, in both own and other authors’ studies, it was shown that not all kind of cells were transformed by plasma[73,75,76]. Our model is, not only incompatible with the idea of specific tropism for metastasis, but it is really proper to search tropism mechanism.
Mittra et al[79] clearly demonstrated that cellular/nuclear uptake of DNA is energy dependent and requires an active metabolic machinery of the recipient cells, which might be a first selection. However, it is possible that the key is not only in the characteristics of susceptible cells (“soil”), but also in the particles circulating in the transforming plasma (“seed”). In fact, there is an increasing stream of studies about the potential of extracellular vesicles on induction of cellular transformation and most of those observations are fully consistent with the theory of genometastasis[81-83].
Traditionally, treatments directed to prevent metastasis have been based on the use of cytotoxic substances that avoid circulation, homing and reproduction of malignant cells. If we assume that circulating nucleic acids in cancer patients have a role in the production of metastasis, a new scenario can be opened up. We can imagine a variety of strategies for interfering with these circulating nucleic acids either during their travel or during the horizontal transfer at the target organ.
Perhaps, the most immediate approach appears to be the use of enzymes to degrade circulating nucleic acids. The idea of enzymes-based therapies for cancer hovered since four decades ago[84], and in the last years, some convincing approaches have been reported. For example, Trejo-Becerril et al[85] have reported that systemic treatment with DNAse I and a protease mix in rats decreased DNA and proteins from serum and had antitumor effects. Interestingly, Patutina et al[86] have reported that tumor-bearing animals treated with RNase A and DNase I had a general systemic and immunomodulatory effect that led to a drastic suppression of metastasis development. Undoubtedly, those results support the role of the genometastasis phenomenon in the development of metastasis and encourage deepening.
Other potential therapeutic approach might be based on the use of potentially transfecting particles charged by “good sequences” of nucleic acids. It has not been enough tested but, theoretically, it is possible that such particles promote a “competitive” effect with cell-free tumor nucleic acids and, then, avoiding metastasis. In this line, virtosomes (i.e., the mentioned DNA-RNA-lipoprotein complex) might constitute a useful tool. These particles are spontaneously released from healthy human, other mammalian, avian, amphibian and plant cells in a regulated and energy-dependent manner[63]. Likewise, these released virtosomes have been demonstrated to enter other cells[87-89]. More importantly, the biology of the recipient cells may be also modified if virtosomes come from a different cell type. Experiments with virtosomes in an immunocompetent animal model of colorectal cancer, showed a virtosomal effect in blocking cell multiplication in both in vitro and in vivo studies, resulting in a scape from inhibition at times after inhibition initiation. These results could indicate the existence of a response derived from the initiation of an immune reaction[90].
In other way, some previous studies have suggested the possibility of silencing these circulating oncogenic signals through RNA interference. As an example, the use of some micro-RNA can determine a novel regulatory pathway in KRAS-driven cancers, which offers a potential therapeutic target for their eradication[91], if this microRNAs are harboured by particles such as virtosomes or exosomes.
Indeed, a lot of strategies can be suggested in order to interfere with the horizontal transfer mechanism, responsible for the transformation of healthy and normal cell into malignant cell. Nonetheless, as happens with all new paradigms, lots of further lines of research are required in this field.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Fu DL, Koutsilieris M, Sugimura H S- Editor: Qi Y L- Editor: A E- Editor: Lu YJ
| 1. | González-Masiá JA, García-Olmo D, García-Olmo DC. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 2013;6:819-832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Imamura T, Komatsu S, Ichikawa D, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H, Shiozaki A. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2016;22:5627-5641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 3. | Bettoni F, Masotti C, Habr-Gama A, Correa BR, Gama-Rodrigues J, Vianna MR, Vailati BB, São Julião GP, Fernandez LM, Galante PA. Intratumoral Genetic Heterogeneity in Rectal Cancer: Are Single Biopsies representative of the entirety of the tumor? Ann Surg. 2017;265:e4-e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 4. | Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:4. |
| 5. | Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-6904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1953] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 6. | Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid Biopsies for Cancer: Coming to a Patient near You. J Clin Med. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 7. | Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17:209-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 8. | Webb S. The cancer bloodhounds. Nat Biotechnol. 2016;34:1090-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Ignatiadis M, Lee M, Jeffrey SS. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin Cancer Res. 2015;21:4786-4800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 276] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 10. | Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3360] [Cited by in RCA: 3382] [Article Influence: 161.0] [Reference Citation Analysis (0)] |
| 11. | Cohen SJ, Alpaugh RK, Gross S, O’Hara SM, Smirnov DA, Terstappen LW, Allard WJ, Bilbee M, Cheng JD, Hoffman JP. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 151] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 12. | Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053-7058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 512] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 13. | Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. [PubMed] |
| 14. | Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23:707-712. [PubMed] |
| 15. | Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116-2120. [PubMed] |
| 16. | Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035-1037. [PubMed] |
| 17. | Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67-70. [PubMed] |
| 18. | Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1313] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 19. | Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest. 1966;45:1732-1740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 641] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 20. | Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-1665. [PubMed] |
| 21. | Koffler D, Agnello V, Winchester R, Kunkel HG. The occurrence of single-stranded DNA in the serum of patients with systemic lupus erythematosus and other diseases. J Clin Invest. 1973;52:198-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 186] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Wieczorek AJ, Sitaramam V, Machleidt W, Rhyner K, Perruchoud AP, Block LH. Diagnostic and prognostic value of RNA-proteolipid in sera of patients with malignant disorders following therapy: first clinical evaluation of a novel tumor marker. Cancer Res. 1987;47:6407-6412. [PubMed] |
| 23. | Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823-3826. [PubMed] |
| 24. | Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, Esposito G, DelBianco P, Nitti D, De Rossi A. Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res. 2008;14:7444-7451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | García-Olmo DC, Contreras JD, Picazo MG, López-Torres J, García-Olmo D. Potential clinical significance of perioperative levels of mRNA in plasma from patients with cancer of the larynx or hypopharynx. Head Neck. 2017;39:647-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087-2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 1029] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 27. | Connolly ID, Li Y, Gephart MH, Nagpal S. The “Liquid Biopsy”: the Role of Circulating DNA and RNA in Central Nervous System Tumors. Curr Neurol Neurosci Rep. 2016;16:25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159-6175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 234] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 29. | Olmedillas López S, García-Olmo DC, García-Arranz M, Guadalajara H, Pastor C, García-Olmo D. KRAS G12V Mutation Detection by Droplet Digital PCR in Circulating Cell-Free DNA of Colorectal Cancer Patients. Int J Mol Sci. 2016;17:484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Gahan PB, Stroun M. The Biology of Circulating Nucleic Acids in Plasma and Serum (CNAPS). Extracellular Nucleic Acids. 1st ed. Berlin Heidelberg: Springer-Verlag Berlin Heidelberg 2010; 167-189. |
| 31. | Holdenrieder S, Stieber P, Bodenmüller H, Busch M, Von Pawel J, Schalhorn A, Nagel D, Seidel D. Circulating nucleosomes in serum. Ann N Y Acad Sci. 2001;945:93-102. [PubMed] |
| 32. | Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 590] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 33. | Mouliere F, Thierry AR. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther. 2012;12 Suppl 1:S209-S215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 34. | Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;49:719-726. [PubMed] |
| 35. | Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 770] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 36. | Yu M. Circulating cell-free mitochondrial DNA as a novel cancer biomarker: opportunities and challenges. Mitochondrial DNA. 2012;23:329-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975;35:2375-2382. [PubMed] |
| 38. | Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci USA. 1972;69:1685-1689. [PubMed] |
| 39. | Stroun M, Anker P. Nucleic acids spontaneously released by living frog auricles. Biochem J. 1972;128:100P-101P. [PubMed] |
| 40. | Stroun M, Anker P, Gahan P, Henri J. Spontaneous release of newly synthesized DNA from frog auricles. Arch Sci Geneva. 1977;30:230-241. |
| 41. | van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. 2007;53:2215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 127] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 42. | Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139-142. [PubMed] |
| 43. | Abolhassani M, Tillotson J, Chiao J. Characterization of the release of DNA by a human leukemia-cell line hl-60. Int J Oncol. 1994;4:417-421. [PubMed] |
| 44. | Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 594] [Cited by in RCA: 706] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 45. | Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 592] [Cited by in RCA: 671] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 46. | Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 1254] [Article Influence: 114.0] [Reference Citation Analysis (0)] |
| 47. | Weidle UH, Birzele F, Kollmorgen G, Rüger R. The Multiple Roles of Exosomes in Metastasis. Cancer Genomics Proteomics. 2017;14:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 48. | Gu H, Ji R, Zhang X, Wang M, Zhu W, Qian H, Chen Y, Jiang P, Xu W. Exosomes derived from human mesenchymal stem cells promote gastric cancer cell growth and migration via the activation of the Akt pathway. Mol Med Rep. 2016;14:3452-3458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 49. | Wang Z, Chen JQ, Liu JL, Tian L. Exosomes in tumor microenvironment: novel transporters and biomarkers. J Transl Med. 2016;14:297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 50. | Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1165] [Cited by in RCA: 1509] [Article Influence: 137.2] [Reference Citation Analysis (0)] |
| 51. | Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495:103-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 52. | Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, Jiang C, Chen X, Wang Y, Du Y. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol. 2016;37:5193-5202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Elfimova N, Sievers E, Eischeid H, Kwiecinski M, Noetel A, Hunt H, Becker D, Frommolt P, Quasdorff M, Steffen HM. Control of mitogenic and motogenic pathways by miR-198, diminishing hepatoma cell growth and migration. Biochim Biophys Acta. 2013;1833:1190-1198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Wang M, Wang J, Kong X, Chen H, Wang Y, Qin M, Lin Y, Chen H, Xu J, Hong J. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 55. | Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 56. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 645] [Article Influence: 58.6] [Reference Citation Analysis (1)] |
| 57. | Fabbri M, Paone A, Calore F, Galli R, Croce CM. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013;10:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 58. | Yu X, Odenthal M, Fries JW. Exosomes as miRNA Carriers: Formation-Function-Future. Int J Mol Sci. 2016;17:E2028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 299] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 59. | Zandberga E, Kozirovskis V, Ābols A, Andrējeva D, Purkalne G, Linē A. Cell-free microRNAs as diagnostic, prognostic, and predictive biomarkers for lung cancer. Genes Chromosomes Cancer. 2013;52:356-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 60. | Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1312] [Cited by in RCA: 1246] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 61. | Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Brański P, Ratajczak MZ, Zembala M. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 304] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 62. | Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 491] [Cited by in RCA: 556] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 63. | Gahan PB, Stroun M. The virtosome-a novel cytosolic informative entity and intercellular messenger. Cell Biochem Funct. 2010;28:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 64. | Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1954] [Cited by in RCA: 2142] [Article Influence: 153.0] [Reference Citation Analysis (0)] |
| 65. | de Groot AE, Roy S, Brown JS, Pienta KJ, Amend SR. Revisiting Seed and Soil: Examining the Primary Tumor and Cancer Cell Foraging in Metastasis. Mol Cancer Res. 2017;15:361-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 66. | Weigelt B, Peterse JL, van’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1569] [Cited by in RCA: 1628] [Article Influence: 81.4] [Reference Citation Analysis (0)] |
| 67. | Garcia-Olmo D, Garcia-Rivas M, Garcia-Olmo D, Ontanon J. The site of injection of tumor cells in rats does not influence the subsequent distribution of metastases. Oncol Rep. 2003;10:903-907. [PubMed] |
| 68. | Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742-1757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 592] [Cited by in RCA: 563] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 69. | Takii Y, Maruyama S, Nogami H. Can the prognosis of colorectal cancer be improved by surgery? World J Gastrointest Surg. 2016;8:574-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 70. | García-Olmo D, Ontañón J, García-Olmo DC, Vallejo M, Cifuentes J. Experimental evidence does not support use of the “no-touch” isolation technique in colorectal cancer. Dis Colon Rectum. 1999;42:1449-1456; discussion 1454-1456. [PubMed] |
| 71. | Garcia-Olmo DC, Gutierrez-Gonzalez L, Samos J, Picazo MG, Atienzar M, Garcia-Olmo D. Surgery and hematogenous dissemination: Comparison between the detection of circulating tumor cells and of tumor DNA in plasma before and after tumor resection in rats. Ann Surg Oncol. 2006;13:1136-1144. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 72. | Garcia-Olmo D, Garcia-Olmo D, Ontanon J, Martinez E, Vallejo M. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol. 1999;14:1159-1164. [PubMed] |
| 73. | Garcia-Olmo DC, Dominguez C, Garcia-Arranz M, Anker P, Stroun M, Garcia-Verdugo JM, Garcia-Olmo D. Cell-Free Nucleic Acids Circulating in the Plasma of Colorectal Cancer Patients Induce the Oncogenic Transformation of Susceptible Cultured Cells. Cancer Res. 2010;70:560-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 198] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 74. | Hunter KW, Crawford NP, Alsarraj J. Mechanisms of metastasis. Breast Cancer Res. 2008;10 Suppl 1:S2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 75. | Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, Anker P, Herrera-Goepfert R, Medina-Velázquez LA, Hidalgo-Miranda A, Pérez-Montiel D, Chávez-Blanco A, Cruz-Velázquez J. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One. 2012;7:e52754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 76. | Abdouh M, Zhou S, Arena V, Arena M, Lazaris A, Onerheim R, Metrakos P, Arena GO. Transfer of malignant trait to immortalized human cells following exposure to human cancer serum. J Exp Clin Cancer Res. 2014;33:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 77. | Hamam D, Abdouh M, Gao ZH, Arena V, Arena M, Arena GO. Transfer of malignant trait to BRCA1 deficient human fibroblasts following exposure to serum of cancer patients. J Exp Clin Cancer Res. 2016;35:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 78. | Arena GO, Arena V, Arena M, Abdouh M. Transfer of malignant traits as opposed to migration of cells: A novel concept to explain metastatic disease. Med Hypotheses. 2017;100:82-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 79. | Mittra I, Khare NK, Raghuram GV, Chaubal R, Khambatti F, Gupta D, Gaikwad A, Prasannan P, Singh A, Iyer A. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J Biosci. 2015;40:91-111. [PubMed] |
| 80. | Ghasemi R, Grassadonia A, Tinari N, Piccolo E, Natoli C, Tomao F, Iacobelli S. Tumor-derived microvesicles: the metastasomes. Med Hypotheses. 2013;80:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 81. | Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun. 2014;451:295-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 156] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 82. | Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SI. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 560] [Cited by in RCA: 694] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 83. | Kreger BT, Dougherty AL, Greene KS, Cerione RA, Antonyak MA. Microvesicle Cargo and Function Changes upon Induction of Cellular Transformation. J Biol Chem. 2016;291:19774-19785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 84. | Hall IH, Ishaq KS, Piantadosi C. Role of deoxyribonuclease in cancer chemotherapy. J Pharm Sci. 1974;63:625-626. [PubMed] |
| 85. | Trejo-Becerril C, Pérez-Cardenas E, Gutiérrez-Díaz B, De La Cruz-Sigüenza D, Taja-Chayeb L, González-Ballesteros M, García-López P, Chanona J, Dueñas-González A. Antitumor Effects of Systemic DNAse I and Proteases in an In Vivo Model. Integr Cancer Ther. 2016;15:NP35-NP43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 86. | Patutina O, Mironova N, Ryabchikova E, Popova N, Nikolin V, Kaledin V, Vlassov V, Zenkova M. Inhibition of metastasis development by daily administration of ultralow doses of RNase A and DNase I. Biochimie. 2011;93:689-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 87. | Anker P, Jachertz D, Stroun M, Brögger R, Lederrey C, Henri J, Maurice PA. The role of extracellular DNA in the transfer of information from T to B human lymphocytes in the course of an immune response. J Immunogenet. 1980;7:475-481. [PubMed] |
| 88. | Anker P, Lyautey J, Lefort F, Lederrey C, Stroun M. [Transformation of NIH/3T3 cells and SW 480 cells displaying K-ras mutation]. C R Acad Sci III. 1994;317:869-874. [PubMed] |
| 89. | Adams DH, Diaz N, Gahan PB. In vitro stimulation by tumour cell media of [3H]-thymidine incorporation by mouse spleen lymphocytes. Cell Biochem Funct. 1997;15:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 90. | Garcia-Arranz M, Garcia-Olmo D, Vega-Clemente L, Stroun M, Gahan PB. Non-dividing Cell Virtosomes Affect In Vitro and In Vivo Tumour Cell Replication. Adv Exp Med Biol. 2016;924:43-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 91. | Zhou Y, Dang J, Chang KY, Yau E, Aza-Blanc P, Moscat J, Rana TM. miR-1298 Inhibits Mutant KRAS-Driven Tumor Growth by Repressing FAK and LAMB3. Cancer Res. 2016;76:5777-5787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |