Published online Feb 10, 2017. doi: 10.5306/wjco.v8.i1.67
Peer-review started: June 18, 2016
First decision: August 16, 2016
Revised: November 11, 2016
Accepted: December 27, 2016
Article in press: December 28, 2016
Published online: February 10, 2017
Processing time: 237 Days and 17.8 Hours
To investigate if the down-regulation of N-myc Downstream Regulated Gene 2 (NDRG2) expression in colorectal carcinoma (CRC) is due to loss of the NDRG2 allele(s).
The following were investigated in the human colorectal cancer cell lines DLD-1, LoVo and SW-480: NDRG2 mRNA expression levels using quantitative reverse transcription-polymerase chain reaction (qRT-PCR); interaction of the MYC gene-regulatory protein with the NDRG2 promoter using chromatin immunoprecipitation; and NDRG2 promoter methylation using bisulfite sequencing. Furthermore, we performed qPCR to analyse the copy numbers of NDRG2 and MYC genes in the above three cell lines, 8 normal colorectal tissue samples and 40 CRC tissue samples.
As expected, NDRG2 mRNA levels were low in the three colorectal cancer cell lines, compared to normal colon. Endogenous MYC protein interacted with the NDRG2 core promoter in all three cell lines. In addition, the NDRG2 promoter was heavily methylated in these cell lines, suggesting an epigenetic regulatory mechanism. Unaltered gene copy numbers of NDRG2 were observed in the three cell lines. In the colorectal tissues, one normal and three CRC samples showed partial or complete loss of one NDRG2 allele. In contrast, the MYC gene was amplified in one cell line and in more than 40% of the CRC cases.
Our study suggests that the reduction in NDRG2 expression observed in CRC is due to transcriptional repression by MYC and promoter methylation, and is not due to allelic loss.
Core tip: NDRG2 is a putative tumor suppressor gene whose expression is reduced in many cancer forms, including colorectal carcinoma (CRC). We set out therefore to investigate if down-regulation of NDRG2 expression was due to loss of one or both alleles and/or to other mechanisms. In our paper, we show that allelic loss of NDRG2 is a rare event in CRC. To our knowledge, this is the first study that has specifically investigated gene copy number of NDRG2 in CRC. Furthermore, our results suggest that MYC is amplified in more than 40% of CRC cases. MYC is known to repress transcription of NDRG2. Our results lead us to suggest that it is the transcriptional control of NDRG2 expression, including repression by MYC and epigenetic regulation, that results in decreased NDRG2 mRNA levels in CRC, rather than allelic loss of NDRG2.
- Citation: Lorentzen A, Mitchelmore C. NDRG2 gene copy number is not altered in colorectal carcinoma. World J Clin Oncol 2017; 8(1): 67-74
- URL: https://www.wjgnet.com/2218-4333/full/v8/i1/67.htm
- DOI: https://dx.doi.org/10.5306/wjco.v8.i1.67
N-myc downstream regulated gene 2 (NDRG2) is one of four genes belonging to the NDRG gene family. Common for these genes is an NDR domain, a protein motif covering almost the entire protein, but the cellular functions of these genes are currently unclear[1,2]. NDRG2 expression has been found to be down-regulated in several human cancers including colorectal carcinoma (CRC), hepatocellular carcinoma, glioblastoma and thyroid cancer[3-7]. NDRG2 is a candidate tumor suppressor gene, with a better overall survival for CRC, hepatocellular carcinoma and glioma patients displaying expression of the gene compared to low or no expression[8-12]. Further evidence of the tumor suppressor function of NDRG2 comes from the observation that NDRG2-lacking mice develop various types of tumors, and from xenograft studies showing that NDRG2-expressing tumor cells implanted in nude mice form smaller tumors and fewer metastases than control cells[13-15]. NDRG2 has a number of downstream targets, including activation of phosphatase and tensin homolog, a known tumor suppressor in the PI3K-AKT pathway[13,16].
Several mechanisms have been suggested as possible regulators of NDRG2 expression, of which epigenetic silencing, due to promoter hypermethylation, is the most widely observed[4,8,9,13,14,17]. However, other regulatory mechanisms may also play a role. One example could be the transcription factor MYC, which is characterised as a proto-oncogene often altered in human cancers[18]. The biological function of MYC seems to be to either activate or repress the transcription of target genes[19,20]. Zhang et al[21] have previously shown that ectopically expressed MYC is able, via Miz-1, to interact with and to repress transcription from the NDRG2 promoter. Moreover, correlation of high MYC with reduced NDRG2 expression has been observed in different cancers and cancer cell lines[15,22-24]. However, an inverse relation between MYC levels and NDRG2 expression seems not to apply to all cancer types[25].
CRC is, like most other cancers, a malignant disease with a combination of both genetic and epigenetic changes. One of these changes is chromosome instability, which affects one or several chromosomal regions. Many groups have analysed changes in gene copy numbers in CRC by different approaches and found numerous chromosomal gains and losses[26-29]. In the study by Lagerstedt et al[29], the status of CRC samples classified as Dukes stages A-D was analysed, showing an increasing frequency of allelic losses at more severe stages (Dukes C and D). According to their data, allelic deletions in chromosome 14, containing the NDRG2 gene, is already found at earlier stages (Dukes A and B) and becomes more frequent at the later stages. Although chromosome 14 is not considered one of the deletion hot spot regions, such as chromosome 8p or 18q[27,28,30,31], we hypothesised that deletions in chromosome 14 could lead to loss of one or both of the NDRG2 alleles. On the other hand, the MYC gene is found on chromosome 8q, and gains of this large chromosome arm are frequently found in CRC[26,28,32]. Analysing the gene copy number of MYC is therefore of interest with regards to its possible regulatory effect on NDRG2.
In this study, we demonstrate a frequent increase in the gene copy number of MYC in CRC. In contrast, we find that changes in the copy number of the NDRG2 locus are rare in CRC, and we suggest that reduced expression of NDRG2 in CRC is due to epigenetic and MYC-related transcriptional repression.
The DLD-1, LoVo and SW-480 colorectal cancer cell lines were a gift from Associate Professor Ole Vang, Roskilde University. Cells lines were incubated and maintained at 37 °C in an environment of humidified air with 5% CO2 in McCoy’s 5A + GlutaMaxTM-1 media with 10% Fetal Bovine Serum and 1% Penicillin-Streptomycin (Invitrogen). RNA from cell lines was purified with the SV total RNA isolation kit (Promega) and genomic DNA was purified by ethanol precipitation after an overnight Proteinase K treatment. Reference human genomic DNA, purified from blood lymphocytes, was obtained from Roche Diagnostics, United States (Cat. No.11691112001). As a normal colonic control we used commercially available DNA (BioChain Institute Inc., D4234090). Human colon genomic DNA from tissue classified as either normal or tumorigenic was obtained from BioChain Inc, United States (Cat. no. D8235090-1; Supplementary Table S1). The commercial supplier confirms that tissue and data collection were ethically approved by their Institutional Regulatory Board and that informed consent was obtained from all human subjects.
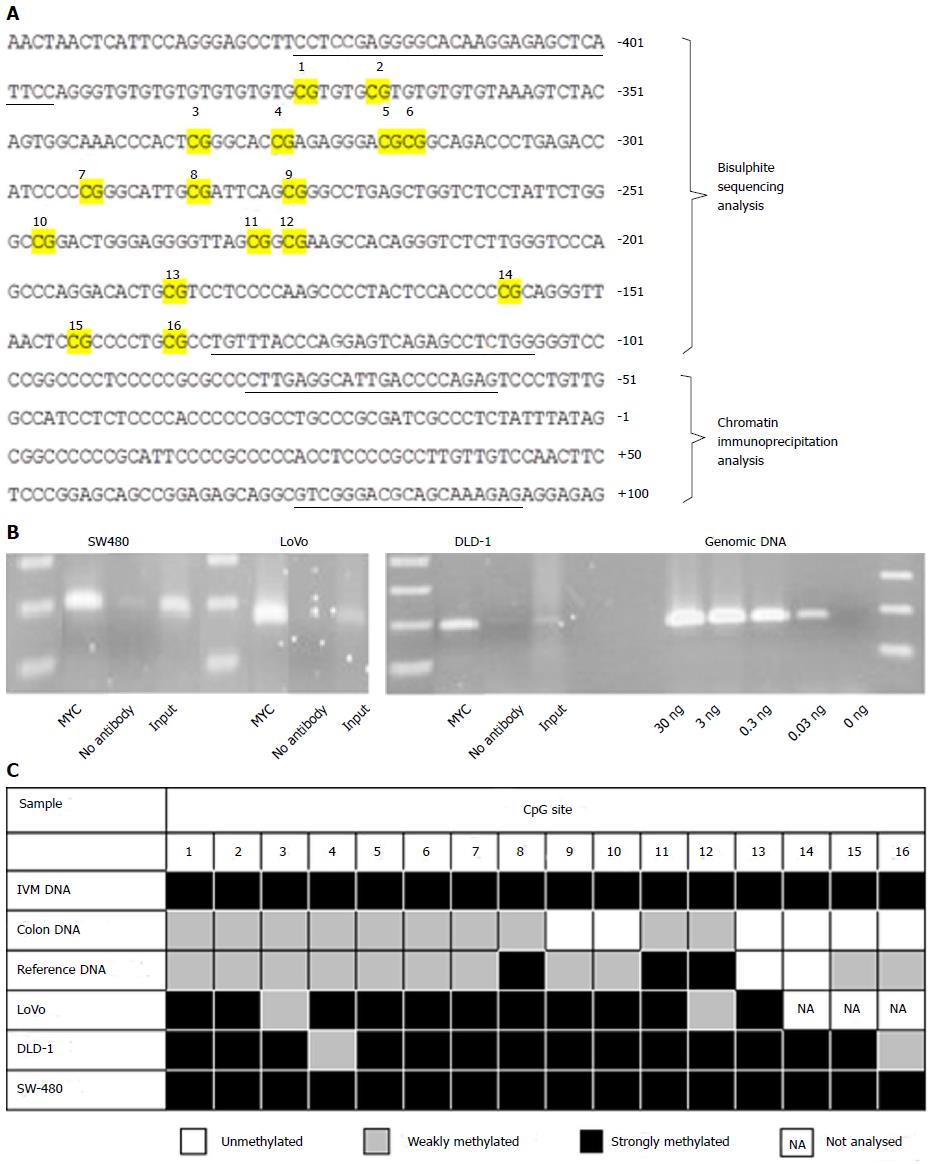
The chromatin immunoprecipitation (ChIP) kit from Abcam (Ab500) was used according to the instructions, with inclusion of a final ethanol precipitation to increase the DNA concentration. Antibody against MYC (Abcam, ab56-100) was used at a concentration of 5 µg per reaction. The primers used in the PCR step were designed to cover the core promoter region in NDRG2 (-80 to +93, Figure 1A) and their sequences were (5’-3’): CTTGAGGCATTGACCCCAGAG and CTCTTTGCTGCGTCCCGAC.
Bisulfite treatment of genomic DNA was performed as previously described[33], using glycogen as carrier, and the precipitated DNA was redissolved in TE buffer, amplified by PCR and sequenced directly. The primers were designed to cover 16 CpG sites in the promoter region in NDRG2 (Figure 1A) and their sequences were (5’-3’): TTTTCGAGGGGTATAAGGAGAGTTTATTTT and CCAAAAACTCTAACTCCTAAATAAACA[34]. A positive control with in vitro methylated (IVM) DNA was prepared by mixing 2 μL NEB2 buffer, 1 μL 20 x S-adenosylmethionine (New England Biolabs, B9003S), 200 ng reference human genomic DNA and 1 µL SssI methyltransferase (New England BioLabs, M0226S) in a total of 20 μL. Samples were incubated at 37 °C overnight with occasional addition of 2 μL 20 x S-adenosylmethionine to ensure sufficient methyl-donor substrate. The following description was used for each CpG site: Unmethylated (no methylation signal); weakly methylated (methylation signal was less than or approximately equal to unmethylated signal); and strongly methylated (methylation signal was greater than unmethylated signal).
Determination of gene copy number was based on the LightCycler technology using SYBR Green. The sequences of the primers were (5’-3’): NDRG2 (5’ end): CCCCTTGCCTTCTAACTTCCCA and ACAGCCCCTCCTCCCACCTT; NDRG2 (3’ end): GGGGTGAACGAAGAACAAAACAAAG and CGAGGGAGACGGTGAGATGAGG; MYC: CCAGAGGAGGAACGAGCTAA and TTGGACGGACAGGATGTATG; GFAP: TGACCCTCTCCACCCCATAGTGAC and CAGCAGCAGTGCCCTGAAGATTAG; and MECP2: TCAGAGGGTGTGCAGGTGAA and TTGAAAAGGCATCTTGACAAGGA. In a validation experiment using a control sample, a dilution series was produced and assayed for NDRG2, MYC, GFAP and MECP2. When Ct values were plotted against log dilution it was shown that the assays are quantitative over a range of 625-fold dilution for NDRG2 (5’ end), NDRG2 (3’ end), MYC, MECP2 and 125 for GFAP. All samples were quantified in triplicates and mean Ct values were normalised to GFAP and used to calculate delta delta Ct (ddCt) relative to the reference human genomic DNA[35]. Copy number was defined as a loss for ddCt < 0.75 and as a gain for ddCt > 1.25. Quantification of NDRG2 mRNA expression levels in colorectal cancer cell lines, using qRT-PCR and normalisation to β-actin, was carried out as previously described[25].
All statistical tests were carried out using GraphPad Prism 4 software and P values of < 0.05 were considered significant. An unpaired two-tailed t-test was used to compare the means of normal-distributed data for the two groups (normal vs tumor). The null hypothesis is that there is no difference between the two groups. When data of the two groups did not have equal variance, by F test analysis, we used a Mann-Whitney test.
In order to examine how NDRG2 expression is regulated in colorectal cancer, we chose to work with three cell lines. First of all, we quantified NDRG2 mRNA levels in the three colorectal cancer cell lines DLD-1, LoVo and SW-480 and observed no or very low expression of NDRG2, when normalised to β-actin and compared to human colon mRNA from healthy controls (Table 1).
We were interested in seeing whether endogenous MYC was bound to the NDRG2 promoter in these cell lines, since ectopically expressed MYC is a transcriptional repressor of NDRG2[21]. A ChIP experiment did indeed show binding of endogenous MYC protein to the core promoter region of NDRG2 in all three colorectal cancer cell lines (Figure 1B).
In silico analysis of the NDRG2 promoter predicted a CpG island between -380 and +1471 relative to the transcriptional start site (%GC = 66.3, observed/expected CpG = 0.673, cpgislands.usc.edu/cpg.aspx). To establish the methylation status of the NDRG2 proximal promoter in all three cell lines, we carried out bisulfite treatment and sequencing of the region from -426 to -107, which contains 16 CpG sequences. Bisulfite treatment converts all unmethylated cytosines into uracils, while cytosines with a methyl group attached remain unaltered. As controls, we compared our results with healthy colon genomic DNA, reference genomic DNA from normal blood lymphocytes, and IVM genomic DNA. As presented in Figure 1C, the normal colon genomic DNA and reference genomic DNA sample were predominantly weakly methylated, whereas the in vitro methylated control was completely methylated at all cytosines. The three colorectal cancer cell lines, LoVo, DLD-1 and SW-480, displayed strong methylation at the majority of CpG sites (Figure 1C).
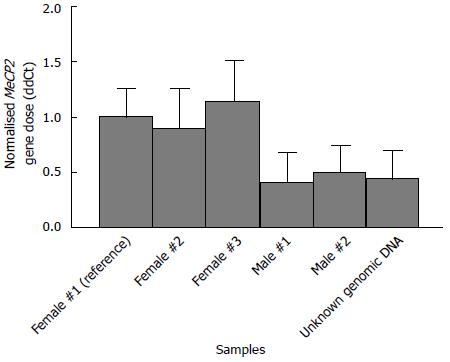
We wished to determine the allelic copy numbers of both NDRG2 and MYC in human colorectal carcinoma. By combining qPCR with the mathematical delta delta Ct equation (ddCt), we were able to quantify both losses and gains of these genes. Our experimental setup was validated by analysing the copy numbers of the X-chromosome linked MECP2 gene in males and females - with the expected one and two X-chromosomes, respectively. As visualised in Figure 2, DNA from 3 females were scored with a ddCt value close to 1.00, which means that the same gene copy ratio between MeCP2 and GFAP was present in both the analysed samples and the reference female genomic sample. A ddCt value of 1.00 therefore represents the normal two alleles. On the contrary, males displayed a ddCt value of approximately 0.50, which represents one allele. Finally, we tested our setup on an unknown sample clearly showing the pattern for male DNA. The conclusion was, therefore, that our setup clearly could differentiate between females and male, i.e., one and two alleles, and has the potential to analyse the copy numbers of NDRG2 and MYC.
We have previously published data showing a statistically significant down-regulation of NDRG2 mRNA in CRC[3], and the main aim in the present study has therefore been to analyse if allelic loss of NDRG2 could explain cases of decreased NDRG2 mRNA levels. For a thorough investigation of NDRG2, we selected two regions of the genomic sequence of NDRG2, one lying in the 5’ part of the sequence and the other lying in the 3’ end. We first analysed the three colorectal cancer cell lines for both NDRG2 and MYC and found no changes in the copy number of NDRG2, in contrast to MYC, for which we observed copy number loss in the LoVo cell line, the normal two alleles in DLD-1 cells and a clear copy number gain in SW-480 (Table 2). This latter result is in agreement with a previous study showing a 5 to 10-fold genomic amplification of MYC in SW-480 cells[36].
| Cell line | NDRG2 - 5’ end | NDRG2 - 3’ end | MYC | |||
| ddCt ± SD | Copy number | ddCt ± SD | Copy number | ddCt ± SD | Copy number | |
| LoVo | 1.23 ± 0.47 | 2 | 1.12 ± 0.51 | 2 | 0.91 ± 0.31 | 2 |
| DLD-1 | 1.04 ± 0.23 | 2 | 1.08 ± 0.50 | 2 | 0.74 ± 0.22 | Loss |
| SW-480 | 1.04 ± 0.26 | 2 | 0.94 ± 0.44 | 2 | 4.88 ± 0.30 | Gain |
We next analysed 8 normal and 40 CRC tissue samples. In one case out of the eight normal samples, our data indicated copy number loss at the 5’ end of the NDRG2 gene; otherwise, none of the samples showed any copy number alterations for NDRG2 (Table 3). As summarised in Table 3, 29 out of the 40 CRC samples (72%) had an unaltered copy number, 2 samples showed loss at either the 5’ or the 3’ end of NDRG2, and only in one case did we observe loss at both ends of the gene. In contrast, we found complete copy number gain of NDRG2 in 3 cases and partial gain in 9 cases (Supplementary Table S2).
Finally, we determined the copy numbers of MYC in the same 8 normal and 40 CRC samples, and observed one case of genomic amplification in the normal samples. Otherwise, we did not find any allelic changes in the normal samples (Table 3). For the 40 CRC samples, we observed copy number loss in 4 cases, the normal two copies in nearly half the cases (19 out of 40), and copy number gains of the MYC gene in the remaining 17 samples (42.5%) (Supplementary Table S1). However, the observed differences in copy number between normal and CRC tissue did not reach statistical significance (Mann-Whitney test, Table 3).
We and others have previously published data showing a statistically significant reduction in NDRG2 mRNA levels in CRC compared to normal colorectal tissue samples[3,12,23]. Similar findings have been observed in other cancers including gliomas, hepatocellular carcinoma, breast cancer, thyroid cancer and meningioma[5-7,25,37]. Exactly how and why NDRG2 expression is reduced is not fully understood, but repression by the MYC transcription factor is likely to be involved in some cases, just as promoter hypermethylation seems to play an important role[4,14,21,34]. Here, we show that 16 potential methylation sites in the proximal promoter of NDRG2 are heavily methylated in all three colorectal cancer cell lines tested. Methylation of the analysed region from -426 to -107 could reduce accessibility to the transcription factors WT1 and HIF1α, which have binding sites in this region[38,39] and/or result in transcriptional silencing. In support of this, previous studies have shown that reversal of methylation by 5-aza-2’-deoxycytidine treatment leads to increased NDRG2 mRNA levels in the colorectal cancer cell lines CaCo2, HCT116 and SW480[34]. Furthermore, DNA methylation at the NDRG2 promoter was shown to be significantly higher in CRC tissue compared to normal colonic tissue from the same patients[14,34].
Our ChIP experiments on three colorectal cancer cell lines showed that endogenous MYC interacts with the NDRG2 core promoter. Although MYC is considered a classical transcription factor, it is also involved in the maintenance of chromatin structure[40,41]. For example, MYC has been shown to recruit DNA methyltransferase 3a to the promoter region of a gene to exert its repressive activity[42]. Thus, we suggest that MYC could be involved in the regulation of NDRG2 by recruitment of other proteins to produce an epigenetic silencing of NDRG2.
However, the suggested regulatory mechanisms cannot explain all cases of down-regulation of NDRG2 expression, and we were therefore interested in looking at allelic loss to see if this genetic event could contribute to the decreased NDRG2 mRNA levels observed in CRC. To investigate this question, we designed an experimental setup making it possible to quantify the copy numbers of any gene. In a validation experiment, we could easily differentiate between one or two copies of the X-chromosome linked gene MECP2. Our data indicate that allelic loss at the NDRG2 locus is not very frequent in CRC. On the contrary, a subset of CRC cases showed gains of one or both ends of the NDRG2 gene, which might lead to elevated levels of NDRG2 mRNA. These findings were unexpected, since allelic losses in chromosome 14 are more frequently observed than gains[27,28]. Although we have only looked at copy number changes in CRC, our results might be applied to other cancers and could explain why we observed an increase in NDRG2 levels in approximately 8% of 154 paired normal and tumor samples analysed from 19 different tumor types[25].
The proto-oncogene MYC is located on chromosome 8 at the q24.12 region, and several groups have shown amplification of chromosome 8q[27,28,43]. Indeed, we observed an increase in MYC gene copy numbers in nearly every second CRC sample, confirming a frequent gain at this particular gene locus. However, we did not detect the same high percentage of MYC amplification as a previous study focusing on the 8q24 region, which revealed that nearly 80% of the cases analysed had some kind of gene amplification[32]. Since MYC has the potential to repress NDRG2 transcription[21], increased copy numbers of the MYC gene could lead to higher levels of MYC protein and thereby a reduced level of NDRG2 mRNA.
Finally, copy number loss of the 5’ end of NDRG2 and a gain of MYC were observed in separate normal samples and might indicate a rare, but real, genomic alteration in healthy tissue. An alternative explanation is that since all normal samples were obtained from patients diagnosed with CRC and classified as normal, the tissue might be at an early pre-malignant stage with no visual changes, but where genetic abnormalities had already occurred.
In conclusion, we observed NDRG2 promoter hypermethylation and interaction of endogenous MYC with the core promoter in three colorectal cancer cell lines, together with absent or low NDRG2 mRNA expression. Frequent allelic loss was not found at the NDRG2 locus in the colorectal cancer cell lines and tissue samples from either normal or tumor tissues. In contrast, we observed partial or complete NDRG2 copy number gains in more than 25% of the CRC cases, compared to none in the normal samples. We also found that more than 40% of CRC cases displayed MYC amplification, which indicates that the level of MYC mRNA is elevated in CRC. We conclude that epigenetic silencing and transcriptional repression by MYC are likely to be more important than copy number loss for the reduced levels of NDRG2 mRNA observed in CRC.
A frequent change observed in colorectal carcinoma (CRC) is chromosomal instability, in which gain or loss of chromosomal regions affects levels of gene expression. Thus, loss of one or both alleles could explain the reduced expression of tumor suppressor genes, such as NDRG2, that is observed in CRC. Alternatively, NDRG2 down-regulation could be due to transcriptional and epigenetic mechanisms.
In order to understand the origin of CRC, it is important to investigate changes at the epigenetic, genetic and transcriptional level. This study investigated regulation of NDRG2 gene expression using bisulfite-sequencing to study gene methylation, quantitative polymerase chain reaction to study gene copy number as well as chromatin immunoprecipitation to study DNA-binding of the endogenous gene-regulatory protein MYC.
This study shows for the first time that gene copy number for NDRG2 is unaltered in CRC cell lines and clinical samples.
The authors describe a validated approach to determine gene copy number, relative to a control gene, using the comparative (ddCt) approach. Future approaches could focus on re-activating expression of NDRG2 in CRC.
NDRG2 is a newly described tumor suppressor gene that is down-regulated in a large range of cancers, including CRC. Interest in NDRG2 as a therapeutic target is supported by studies showing a better prognosis in patients having higher NDRG2 expression in tumor tissues.
The paper is very good.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Denmark
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kopljar M, Kozovska Z, Tomuleasa C S- Editor: Kong JX L- Editor: A E- Editor: Wu HL
| 1. | Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruïne AP, Baldwin HS, van Engeland M. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. 2010;24:4153-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 2. | Lorentzen A, Mitchelmore C. NDRG2: A candidate tumor suppressor gene in search of a function. Cancer Reports. 2012;2:9-17. |
| 3. | Lorentzen A, Vogel LK, Lewinsky RH, Saebø M, Skjelbred CF, Godiksen S, Hoff G, Tveit KM, Lothe IM, Ikdahl T. Expression of NDRG2 is down-regulated in high-risk adenomas and colorectal carcinoma. BMC Cancer. 2007;7:192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 4. | Shen L, Qu X, Ma Y, Zheng J, Chu D, Liu B, Li X, Wang M, Xu C, Liu N. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-β via EMT inhibition in colorectal cancer. Oncogenesis. 2014;3:e86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Zheng J, Li Y, Yang J, Liu Q, Shi M, Zhang R, Shi H, Ren Q, Ma J, Guo H. NDRG2 inhibits hepatocellular carcinoma adhesion, migration and invasion by regulating CD24 expression. BMC Cancer. 2011;11:251: 1-251: 9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Mordalska A, Latek J, Ferenc T, Pomorski L, Gałecka E, Zygmunt A, Lewiński A. Evaluation of NDRG2 gene expression in primary papillary thyroid carcinoma and in metastases of this neoplasm to regional lymph nodes. Thyroid Res. 2010;3:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Zhou B, Tang Z, Deng Y, Hou S, Liu N, Lin W, Liu X, Yao L. Tumor suppressor candidate gene, NDRG2 is frequently inactivated in human glioblastoma multiforme. Mol Med Rep. 2014;10:891-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Lee DC, Kang YK, Kim WH, Jang YJ, Kim DJ, Park IY, Sohn BH, Sohn HA, Lee HG, Lim JS. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res. 2008;68:4210-4220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Skiriutė D, Steponaitis G, Vaitkienė P, Mikučiūnas M, Skauminas K, Tamašauskas A, Kazlauskas A. Glioma Malignancy-Dependent NDRG2 Gene Methylation and Downregulation Correlates with Poor Patient Outcome. J Cancer. 2014;5:446-456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Hu W, Yang Y, Fan C, Ma Z, Deng C, Li T, Lv J, Yao W, Gao J. Clinical and pathological significance of N-Myc downstream-regulated gene 2 (NDRG2) in diverse human cancers. Apoptosis. 2016;21:675-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 11. | Kim YJ, Kang HB, Yim HS, Kim JH, Kim JW. NDRG2 positively regulates E-cadherin expression and prolongs overall survival in colon cancer patients. Oncol Rep. 2013;30:1890-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Chu D, Zhang Z, Li Y, Wu L, Zhang J, Wang W, Zhang J. Prediction of colorectal cancer relapse and prognosis by tissue mRNA levels of NDRG2. Mol Cancer Ther. 2011;10:47-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Nakahata S, Ichikawa T, Maneesaay P, Saito Y, Nagai K, Tamura T, Manachai N, Yamakawa N, Hamasaki M, Kitabayashi I. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat Commun. 2014;5:3393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 14. | Hong SN, Kim SJ, Kim ER, Chang DK, Kim YH. Epigenetic silencing of NDRG2 promotes colorectal cancer proliferation and invasion. J Gastroenterol Hepatol. 2016;31:164-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Li R, Yu C, Jiang F, Gao L, Li J, Wang Y, Beckwith N, Yao L, Zhang J, Wu G. Overexpression of N-Myc downstream-regulated gene 2 (NDRG2) regulates the proliferation and invasion of bladder cancer cells in vitro and in vivo. PLoS One. 2013;8:e76689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Hu W, Fan C, Jiang P, Ma Z, Yan X, Di S, Jiang S, Li T, Cheng Y, Yang Y. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget. 2016;7:209-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Chang X, Li Z, Ma J, Deng P, Zhang S, Zhi Y, Chen J, Dai D. DNA methylation of NDRG2 in gastric cancer and its clinical significance. Dig Dis Sci. 2013;58:715-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 2585] [Article Influence: 198.8] [Reference Citation Analysis (0)] |
| 19. | Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer. 2010;1:580-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Lüscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494:145-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Zhang J, Li F, Liu X, Shen L, Liu J, Su J, Zhang W, Deng Y, Wang L, Liu N. The repression of human differentiation-related gene NDRG2 expression by Myc via Miz-1-dependent interaction with the NDRG2 core promoter. J Biol Chem. 2006;281:39159-39168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Zhao H, Zhang J, Lu J, He X, Chen C, Li X, Gong L, Bao G, Fu Q, Chen S. Reduced expression of N-Myc downstream-regulated gene 2 in human thyroid cancer. BMC Cancer. 2008;8:303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Shi H, Jin H, Chu D, Wang W, Zhang J, Chen C, Xu C, Fan D, Yao L. Suppression of N-myc downstream-regulated gene 2 is associated with induction of Myc in colorectal cancer and correlates closely with differentiation. Biol Pharm Bull. 2009;32:968-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Yu C, Wu G, Dang N, Zhang W, Zhang R, Yan W, Zhao Y, Gao L, Wang Y, Beckwith N. Inhibition of N-myc downstream-regulated gene 2 in prostatic carcinoma. Cancer Biol Ther. 2011;12:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Lorentzen A, Lewinsky RH, Bornholdt J, Vogel LK, Mitchelmore C. Expression profile of the N-myc Downstream Regulated Gene 2 (NDRG2) in human cancers with focus on breast cancer. BMC Cancer. 2011;11:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Postma C, Koopman M, Buffart TE, Eijk PP, Carvalho B, Peters GJ, Ylstra B, van Krieken JH, Punt CJ, Meijer GA. DNA copy number profiles of primary tumors as predictors of response to chemotherapy in advanced colorectal cancer. Ann Oncol. 2009;20:1048-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Poulogiannis G, Ichimura K, Hamoudi RA, Luo F, Leung SY, Yuen ST, Harrison DJ, Wyllie AH, Arends MJ. Prognostic relevance of DNA copy number changes in colorectal cancer. J Pathol. 2010;220:338-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Nakao M, Kawauchi S, Furuya T, Uchiyama T, Adachi J, Okada T, Ikemoto K, Oga A, Sasaki K. Identification of DNA copy number aberrations associated with metastases of colorectal cancer using array CGH profiles. Cancer Genet Cytogenet. 2009;188:70-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Lagerstedt KK, Kristiansson E, Lönnroth C, Andersson M, Iresjö BM, Gustafsson A, Hansson E, Kressner U, Nordgren S, Enlund F. Genes with relevance for early to late progression of colon carcinoma based on combined genomic and transcriptomic information from the same patients. Cancer Inform. 2010;9:79-91. [PubMed] |
| 30. | Ogino S, Nosho K, Irahara N, Shima K, Baba Y, Kirkner GJ, Meyerhardt JA, Fuchs CS. Prognostic significance and molecular associations of 18q loss of heterozygosity: a cohort study of microsatellite stable colorectal cancers. J Clin Oncol. 2009;27:4591-4598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 31. | Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB, Hamilton SR. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 639] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 32. | Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, Fink SR, Foster NR, Kabat BF, Halling KC, Cunningham JM. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2492-2500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 33. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [PubMed] |
| 34. | Piepoli A, Cotugno R, Merla G, Gentile A, Augello B, Quitadamo M, Merla A, Panza A, Carella M, Maglietta R. Promoter methylation correlates with reduced NDRG2 expression in advanced colon tumour. BMC Med Genomics. 2009;2:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Johnson MR, Wang K, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 290] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 36. | Suàrez HG, Nardeux PC, Andéol Y, Sarasin A. Multiple activated oncogenes in human tumors. Oncogene Res. 1987;1:201-207. [PubMed] |
| 37. | Skiriute D, Tamasauskas S, Asmoniene V, Saferis V, Skauminas K, Deltuva V, Tamasauskas A. Tumor grade-related NDRG2 gene expression in primary and recurrent intracranial meningiomas. J Neurooncol. 2011;102:89-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Svensson E, Vidovic K, Olofsson T, Vallon-Christersson J, Borg A, Gullberg U. The Wilms’ tumor gene 1 (WT1) induces expression of the N-myc downstream regulated gene 2 (NDRG2). DNA Cell Biol. 2007;26:589-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Wang L, Liu N, Yao L, Li F, Zhang J, Deng Y, Liu J, Ji S, Yang A, Han H. NDRG2 is a new HIF-1 target gene necessary for hypoxia-induced apoptosis in A549 cells. Cell Physiol Biochem. 2008;21:239-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 40. | Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723-2734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 314] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 41. | Varlakhanova NV, Knoepfler PS. Acting locally and globally: Myc’s ever-expanding roles on chromatin. Cancer Res. 2009;69:7487-7490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 317] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 43. | Kurashina K, Yamashita Y, Ueno T, Koinuma K, Ohashi J, Horie H, Miyakura Y, Hamada T, Haruta H, Hatanaka H. Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci. 2008;99:1835-1840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |