Published online May 24, 2025. doi: 10.5306/wjco.v16.i5.104138
Revised: February 21, 2025
Accepted: April 15, 2025
Published online: May 24, 2025
Processing time: 149 Days and 2.7 Hours
High-grade serous ovarian carcinoma (HGSOC) is among the most lethal gynecological malignancies, characterized by late-stage diagnosis, extensive peritoneal dissemination, and limited treatment options, resulting in poor survival out
To characterize the functional heterogeneity and tissue-specific distributions of CD4+ T cell subtypes in HGSOC and identify biomarkers for therapy.
We analyzed single-cell RNA sequencing (scRNA-seq) data from 42 HGSOC patients, examining samples collected from adnexal tissues and ascites. CD4+ T cells were identified and classified into subtypes using unsupervised clustering and marker gene analysis. Functional profiling was performed using pathway enrichment, differential expression analysis, and functional signature scoring. Kaplan-Meier survival and Cox proportional hazards modeling were conducted to evaluate the prognostic value of CD4+ T cell subtypes.
Distinct distributions of CD4+ T cell subtypes were identified between adnexal tissues and ascites. Naive CD4+ T cells were predominant in ascites, while Tregs and CXCL13-expressing CD4+ T cells were enriched in adnexal tissues. Tregs were further categorized into four subtypes (Treg1, Treg2, Treg3, and TISG), each exhibiting unique molecular signatures and tissue-specific adaptations. Treg3 cells, enriched in adnexal tissues, were characterized by high levels of activation and exhaustion markers, correlating with poor clinical outcomes in HGSOC patients.
Treg3 cells drive immune suppression and tumor progression in HGSOC, making them a key immunotherapy target. Their adnexal enrichment highlights the need for tissue-specific immune profiling in precision treatment.
Core Tip: This study uncovers the functional heterogeneity of CD4+ T cell subtypes in high-grade serous ovarian carcinoma (HGSOC), revealing their tissue-specific distributions and roles within the tumor microenvironment. Regulatory T cells (Tregs), particularly Treg3 cells, were identified as key immunosuppressive players enriched in adnexal tissues, correlating with poor prognosis and reduced immunotherapy response. By integrating single-cell RNA sequencing data, we provide novel insights into immune evasion mechanisms and identify Treg3 cells as potential therapeutic targets. These findings advance our understanding of HGSOC immunity and offer pathways for developing personalized immunotherapies.
- Citation: Zhang BL, Gao W, He L, Liu XT, Wang ZM, Tan L. Functional heterogeneity and clinical implications of CD4+ T cell subtypes in high-grade serous ovarian carcinoma. World J Clin Oncol 2025; 16(5): 104138
- URL: https://www.wjgnet.com/2218-4333/full/v16/i5/104138.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i5.104138
High-grade serous ovarian carcinoma (HGSOC) remains one of the most lethal gynecological malignancies, characterized by late diagnosis, extensive peritoneal dissemination, and frequent chemoresistance[1]. Despite standard treatments including surgery and platinum-based chemotherapy, the 5-year survival rate remains poor, highlighting an urgent need for novel therapeutic strategies[2,3].
The immune landscape of HGSOC is complex and dynamic, involving intricate interactions between immune cells, cancer cells, and stromal components[4,5]. Among these, CD4+ T cells play pivotal roles in both anti-tumor immunity and immune suppression[6-8]. These cells exhibit remarkable plasticity, differentiating into various subtypes including naive T cells, regulatory T cells (Tregs), and specialized effector cells, each with distinct functions in tumor immunity[9]. Current immunotherapeutic approaches, particularly immune checkpoint inhibitors, have shown limited efficacy in HGSOC patients[10,11]. This limited success may be attributed to our incomplete understanding of the immune microenvironment, especially regarding the distribution and function of different CD4+ T cell populations within various anatomical sites[12].
Recent studies have highlighted the complex roles of CD4+ T cells in ovarian cancer progression and immunotherapy response. CD4+ T cells exhibit remarkable plasticity and can differentiate into various subtypes with distinct functions in tumor immunity[13,14]. A critical gap in our knowledge is the lack of comprehensive understanding of tissue-specific immune responses, particularly the functional heterogeneity of CD4+ T cells between different anatomical sites in HGSOC. While previous research has shown that CD4+ T cells, including Tregs, play essential roles in modulating immune responses[14], their precise distribution patterns and functional states across different tumor locations remain poorly characterized. Understanding these functional differences is crucial for developing more effective immunotherapeutic strategies that can overcome the current limitations of treatment.
In this study, we conducted a comprehensive analysis of CD4+ T cell populations in HGSOC using single-cell RNA sequencing (scRNA-seq) data from 42 patients. Through extensive transcriptional profiling of samples from both adnexal tissues and ascites, we revealed distinct CD4+ T cell phenotypes and their tissue-specific distributions, providing novel insights into the functional heterogeneity of immune responses within different anatomical sites of ovarian cancer.
Publicly available scRNA-seq datasets were acquired to examine CD4+ T cells across diverse tissue contexts. Quality control, annotation, and integration of CD4+ T cell datasets were performed based on previously established protocols. Raw data, including unique molecular identifier (UMI) counts and annotations, were obtained directly from original publications.
Data normalization was conducted in Seurat by standardizing UMI counts for each cell and scaling with a factor of 10000. When UMI data were unavailable, counts were converted to count-per-million or transcripts-per-million, followed by log2 transformation to facilitate comparability. We used reciprocal principal component analysis to reduce batch effects and integrate datasets across experiments. Specifically, we applied the Normalize Data and Find Variable Features functions to identify highly variable genes, followed by Select Integration Features to select 1000 informative genes for integration. Genes linked to cell cycle phases were excluded to minimize cell cycle-driven batch effects. Integration was performed using the Find Integration Anchors and Integrate Data functions, aligning datasets based on shared principal components. Final adjustments included scaling and dimensional reduction to ensure harmonization.
To address batch variability, we utilized single-cell Variational Inference (scVI), a probabilistic deep learning model designed for scRNA-seq analysis. The scVI model was trained with batch identifiers as covariates, systematically correcting for batch-specific variation while preserving underlying biological signals. Corrected data were integrated, followed by dimensionality reduction and visualization using UMAP projections, ensuring consistent distribution across different batches, cell types, and experimental conditions.
We applied Seurat's Find Neighbors and Find Clusters functions to build a shared nearest-neighbor graph and perform unsupervised clustering, using varying resolution parameters to optimize fidelity. Multiple rounds of clustering helped identify major CD4+ T cell subtypes and their specific transcriptional states. The optimal number of principal components was determined via Elbow Plot, with cluster purity validated using ROGUE, an entropy-based metric. UMAP projections were generated for visual representation, and cluster assignments were validated using canonical marker gene expression.
For the classification of Treg subpopulations, we employed an unsupervised clustering approach based on scVI. The scVI model was initialized with the following parameters: N_layers = 2 (ensuring model depth without overfitting), n_latent = 30 (to capture sufficient biological variability), and gene_likelihood = "nb" (negative binomial, optimal for RNA-seq data). Training was performed until convergence. The latent space representation of the cells was extracted using vae.get_latent_representation(), followed by graph-based clustering using the Leiden algorithm (sc.tl.leiden) with nearest-neighbor graph construction based on scVI latent embeddings [sc.pp.neighbors(use_rep = "X_scVI")]. Subclustering was performed to refine the classification of Treg subpopulations based on marker gene expression patterns.
To refine T cell classification, we identified differentially expressed genes (DEGs) using Seurat's Find All Markers function, selecting genes with a minimum fold-change of 1.2, expressed in at least 20% of cells within a cluster, and with an adjusted P-value < 0.05. Feature and bubble plots were generated for the top 50 DEGs to confirm the identity of each CD4+ T cell subtype.
For Treg subpopulations, differential gene expression analysis was employed to identify key marker genes that define each subtype. Based on these markers, Treg cells were categorized into four distinct subpopulations, each characterized by unique gene expression signatures. The use of scVI ensured an unbiased classification while preserving the biological heterogeneity of the dataset.
Differential gene expression between ascites and adnexa CD4+ T cell populations was conducted using the sc.tl. rank_genes_groups function in SCANpy, with Wilcoxon rank-sum tests applied for statistical comparisons. Gene set enrichment analysis (GSEA) was conducted using GSEApy with gene ontology sets from KEGG, GO, Hallmark, and MSigDB. A minimum of 2,000 permutations was used to ensure statistical robustness, with enrichment significance defined as |normalized enrichment score| > 1 and adjusted P-value < 0.05. Functional gene signatures were calculated using the scanpy.tl.score_genes function, enabling comparison of functional profiles within and across clusters.
Kaplan-Meier survival analysis and Cox proportional hazards modeling were used to assess the association between T cell transcriptional signatures and patient survival. Cutoff points for stratifying high- and low-risk groups were determined using the surv_cutpoint function from the survminer package, based on maximally selected rank statistics.
All analyses were conducted using R (v4.3.1) and Python (v3.10.9). Statistical significance was set at P < 0.05. Where multiple comparisons were made, false discovery rate correction was applied. Data processing and analysis utilized Python packages including Scanpy, Pandas, NumPy, and SciPy. Detailed methods are described in the relevant figure legends.
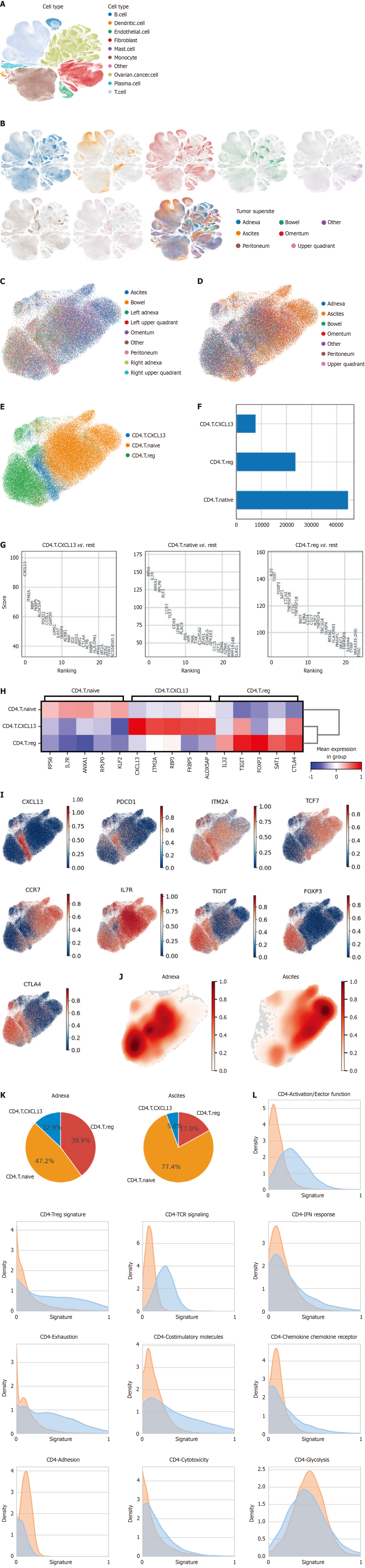
To systematically characterize CD4+ T cell populations in HGSOC, we analyzed public scRNA-seq data from 42 patients undergoing upfront diagnostic laparoscopic or debulking surgery[15]. The data were collected from fresh tumor tissues from 42 patients at the time of upfront diagnostic laparoscopic or debulking surgery. Cell types were annotated based on the method described in the previous study[15] (Figure 1A) (Supplementary material). The samples included ascites and tumor tissues from both primary and metastatic sites, systematically collected from various locations such as bilateral adnexa, omentum, pelvic peritoneum, bilateral upper quadrants, and bowel, with a median of four tissue samples per patient (Figure 1B). Initially, all T cells were isolated, followed by the selection of CD4+ T cells for subsequent analysis.
Initial analysis of CD4+ T cells revealed distinct transcriptional profiles between adnexal tissues and ascites. UMAP visualization showed markedly different distributions between adnexal tissues and ascites for isolated CD4+ T cells (Figure 1C and D). Subtype analysis revealed three main subtypes: Naive T cells, Tregs, and CXCL13-expressing cells (Figure 1E). Naive cells represented the highest cell counts, followed by Tregs, and CXCL13-expressing CD4+ cells constituted the lowest proportions (Figure 1F). We performed differential gene expression analysis to identify upre
The relative proportions of CD4+ T cell subtypes varied significantly between adnexal tissues and ascites. Naive cells were the majority in ascites, while adnexal tissues contained higher proportions of Tregs and CXCL13-expressing cells (Figure 1J and K). Notably, adnexal tissues exhibited a striking enrichment of CXCL13-expressing CD4+ T cells, which are often associated with tertiary lymphoid structure formation and anti-tumor immune responses[7]. Conversely, the pre
To better understand the functional differences between CD4+ T cells in adnexal tissues and ascites, we analyzed various functional signatures, including TCR signaling, Treg signature, co-stimulatory molecule expression, activation markers, effector function, and metabolic states (Figure 1L). CD4+ T cells from adnexa displayed significantly enhanced TCR signaling, activation signatures, and expression of co-stimulatory molecules, indicating a more activated immune profile. In contrast, CD4+ T cells from ascites showed elevated glycolytic signatures and reduced expression of adhesion markers, consistent with a less activated and more metabolically stressed phenotype (Figure 1L).
These results underscore the importance of tissue-specific immune profiling to understand localized immune responses within the peritoneal cavity and their implications for immunotherapeutic strategies in HGSOC.
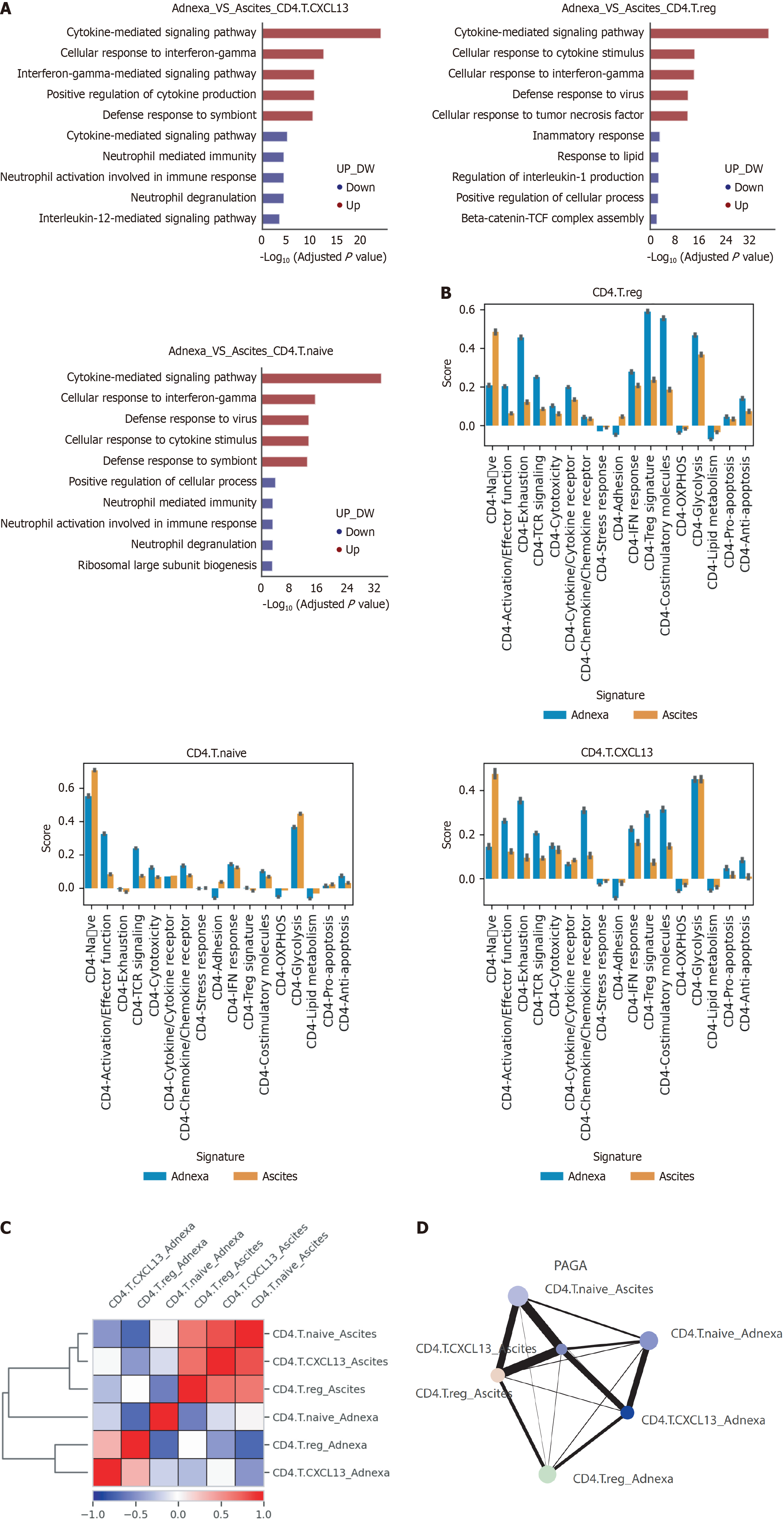
To explore potential drivers behind these observed differences, we performed differential pathway analysis for each CD4+ T cell subtype between adnexa and ascites (Figure 2A). In CXCL13-expressing CD4+ T cells from adnexa, several pathways were upregulated compared to cells from ascites, including cytokine-mediated signaling, cellular response to interferon-gamma, and defense response to symbiont, while pathways related to neutrophil activation and degranulation were notably downregulated. For Tregs, upregulated pathways in adnexa included cytokine-mediated signaling and defense responses, whereas pathways like regulation of interleukin-1 production were consistently downregulated. Naive CD4+ T cells from adnexa showed increased cytokine-mediated signaling and responses to interferon-gamma, while pathways related to neutrophil mediated immunity were downregulated. These findings reflect the distinct roles and functional states of these T cell subsets in cancer immunity.
We also conducted comparative analyses of gene signatures in CD4+ T cell subtypes between adnexa and ascites (Figure 2B). In CD4+ Tregs, signatures such as naive state, lipid metabolism, and adhesion were higher in ascites, whereas activation, TCR signaling, and cytotoxicity were more prominent in adnexa, suggesting enhanced immune activation in adnexa. In naive cells, glycolysis and adhesion were more pronounced in ascites, while activation and exhaustion markers were higher in adnexa. CXCL13-expressing CD4+ T cells exhibited higher activation and co-stimulatory sig
Correlation analysis of these CD4+ subsets between adnexa and ascites revealed distinct expression profiles (Figure 2C). Heatmap and hierarchical clustering indicated that the similarity between CD4+ subsets was higher in ascites compared to adnexa. CXCL13 CD4+T and Tregs from adnexa showed no significant correlation with their counterparts in ascites, highlighting unique tissue-specific characteristics.
Partition-based Graph Abstraction analysis (Figure 2D) illustrated the connectivity and lineage relationships among CD4+ T cell subsets in both environments. Strong connectivity was observed among subsets within ascites, whereas connections within adnexa were weaker. Cross-environment connectivity was also weak, indicating distinct immune networks influenced by the tumor microenvironments in adnexa and ascites.
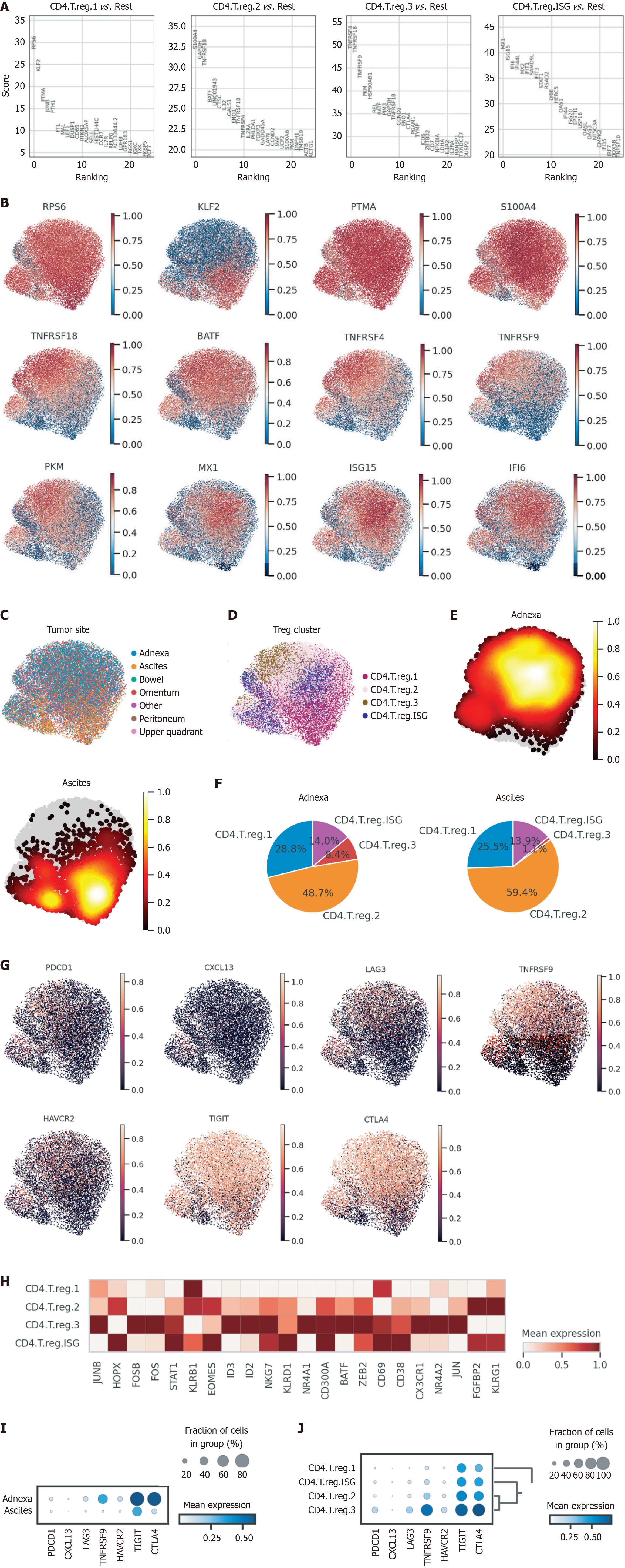
Following the isolation of Tregs from the tumor microenvironment, we performed cluster analysis to categorize these cells into four distinct subtypes—Treg1, Treg2, Treg3, and Tisg (interferon-stimulated gene-expressing Tregs)—each characterized by unique marker genes. Treg1 cells were defined by high expression of RPS6 and KLF2, while S100A4 emerged as a defining marker for Treg2. Treg3 cells were enriched in TNFRSF4, TNFRSF18, and PKM, indicating a potentially activated regulatory phenotype[17,18], and Tisg cells (interferon-stimulated gene-expressing Tregs) were distinguished by high expression of MX1, ISG15, and IFI6 (Figure 3A and B). These marker genes provide molecular signatures that reflect the distinct functional adaptations and roles of each Treg subtype in shaping the immune landscape. To delineate the functional heterogeneity of Treg subtypes across microenvironments, we conducted differential gene expression analysis to characterize subtype-specific transcriptional profiles of Treg subpopulations (Treg1, Treg2, Treg3) and interferon-stimulated gene-expressing Tregs, with comprehensive results detailed in Supplementary material.
We observed a striking differential distribution of Treg subtypes between the ascitic and adnexal tumor microenvironments. Treg2 cells were significantly more abundant in ascites, whereas Treg3 cells predominated in adnexal tissue (Figure 3C-F). This selective enrichment suggests that Treg subtypes are not only distinct in their molecular profiles but also adapt uniquely to different tumor microenvironments, likely in response to specific immune or stromal cues within these niches. The higher prevalence of Treg2 cells in ascites may indicate a specialization in immunosuppressive functions within the peritoneal fluid, whereas the accumulation of Treg3 cells in adnexa could suggest a role in local immune modulation and tissue-specific immune interactions.
UMAP analysis revealed that these Treg subtypes exhibit distinct exhaustion-associated gene expression profiles (Figure 3G). Further analysis of gene expression in Treg3 cells revealed elevated levels of transcription factors and activation-associated genes, including JUNB, FOSB, FOS, ID3, ID2, NR4A1, CX3CR1, NR4A2, and JUN (Figure 3H). Additionally, adnexal Treg cells exhibited heightened expression of exhaustion-associated genes such as TNFRSF9, TIGIT, and CTLA4, reflecting a differentiated phenotype (Figure 3I). Consistent with this profile, Treg3 cells also showed high levels of other exhaustion markers, including HAVCR2, LAYN, LAG3, and PDCD1, suggesting a potentially exhausted state that likely enhances their immunosuppressive function within the tumor microenvironment (Figure 3J). This nuanced profiling of Treg subtypes enhances our understanding of their functional heterogeneity and implications for immune regulation in these specific anatomical contexts.
To further assess the clinical relevance of our findings, we examined immunohistochemistry data from the human protein atlas database (Supplementary material). This analysis confirmed that key marker genes of Treg3, such as TNFRSF4 (OX40) and TNFRSF18 (GITR), are strongly expressed in ovarian cancer tissues, particularly in the tumor stroma and immune cell compartments. These findings reinforce the hypothesis that Treg3 plays an essential role in the immune microenvironment of ovarian cancer.
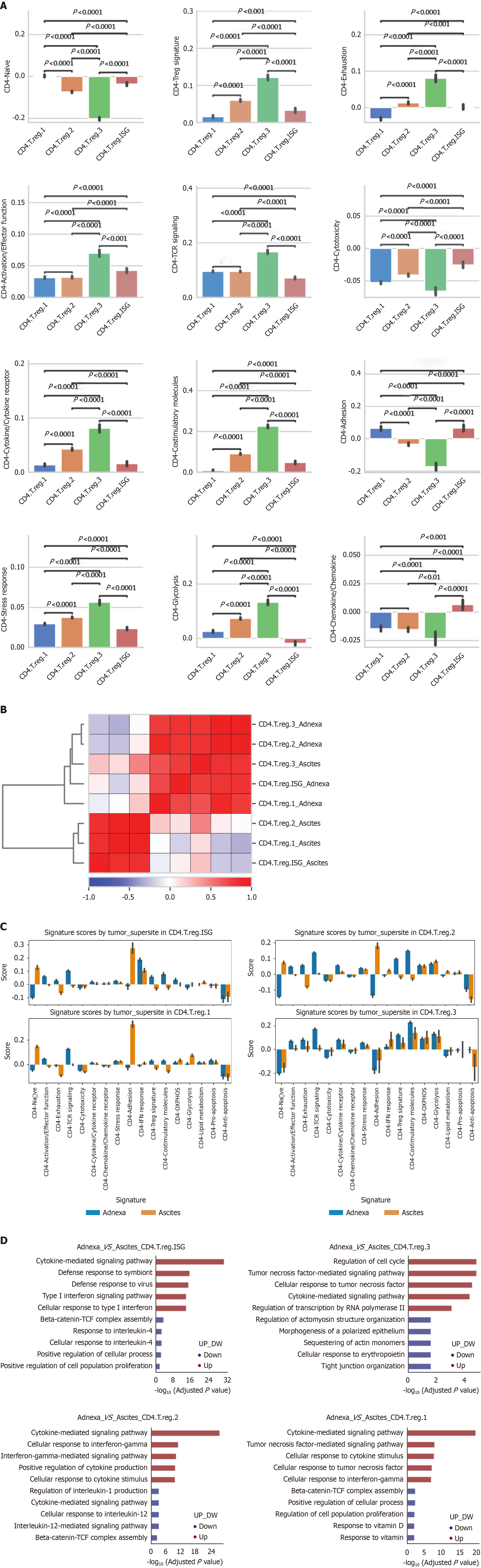
Our analysis of Treg subtypes within the tumor microenvironment revealed substantial functional diversity, under
Hierarchical clustering analysis further demonstrated that Treg subtypes from the same microenvironment, such as Treg3 and Treg. ISG in adnexa, cluster closely based on gene expression patterns (Figure 4B). This clustering indicates that environmental context strongly influences the transcriptional landscape of Treg cells, with adnexal Tregs sharing higher expression correlations within their environment compared to ascitic Tregs, and vice versa. This suggests that each microenvironment uniquely shapes the functional and transcriptional characteristics of Treg cells, potentially enhancing their adaptability to specific tumor niches.
When comparing Treg subtype functionality across tumor microenvironments, significant adaptations were observed between adnexa and ascites (Figure 4C). CD4.T.reg.ISG cells in adnexa exhibited enhanced adhesion and anti-apoptosis scores, supporting their retention and survival within this tissue. Conversely, CD4.T.reg.1 cells displayed higher exhaustion and activation scores in ascites, indicating a more differentiated, suppressive phenotype. CD4.T.reg.2 cells showed increased TCR signaling and activation in ascites, indicative of heightened regulatory activity, while exhibiting elevated adhesion and pro-apoptosis markers in adnexa, suggesting a transient regulatory role. Notably, CD4.T.reg.3 cells in ascites demonstrated the highest exhaustion and metabolic activity, positioning them as a highly suppressive, metabolically resilient phenotype suited to the demanding ascitic microenvironment.
Differential pathway analysis further underscored these environment-specific adaptations, aligning with distinct functional roles for Treg subtypes in each microenvironment (Figure 4D). In adnexa, CD4.T.reg.ISG cells upregulated antiviral and cytokine signaling pathways, enhancing their inflammatory response capacity. CD4.T.reg.2 cells de
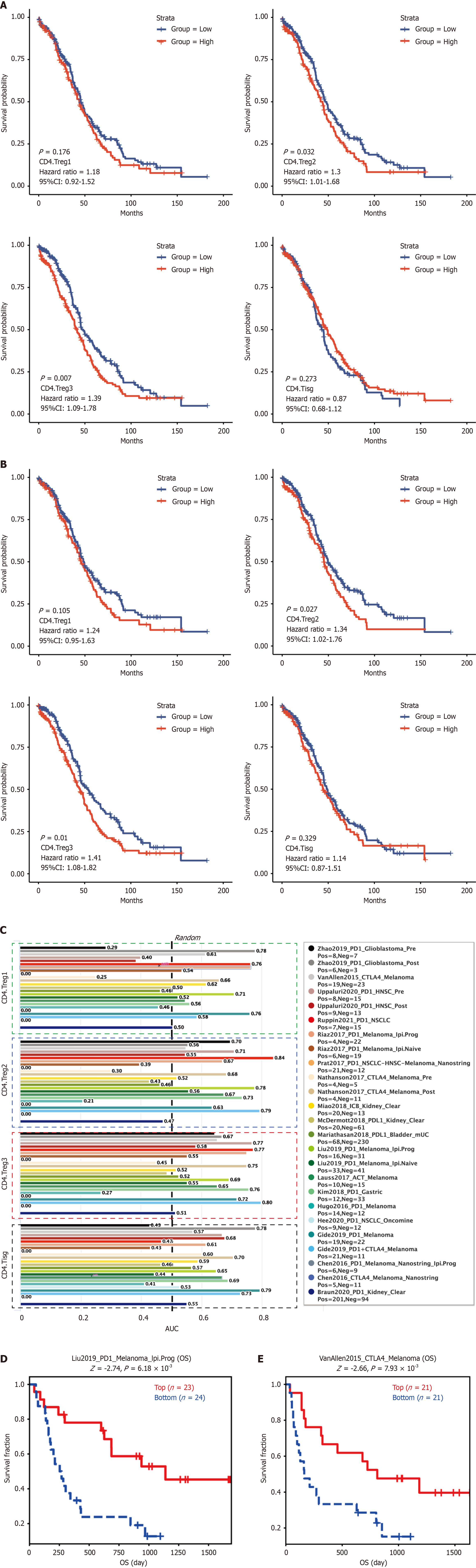
Our study analyzed the prognostic and predictive significance of CD4+ Treg subsets, particularly CD4.Treg3, in relation to overall survival (OS) and disease-specific survival (DSS) among cancer patients. Kaplan-Meier survival curves were generated to compare high and low expression levels of each Treg subset, highlighting their impact on patient outcomes (Figure 5A and B).
For OS, elevated expression of CD4.Treg3 [hazard ratio (HR): 1.39, 95%CI: 1.09–1.78, P = 0.007, Figure 5A] and CD4. Treg2 (HR: 1.30, 95%CI: 1.01–1.68, P = 0.032, Figure 5A) was significantly associated with worse survival outcomes, suggesting these subsets as potential prognostic markers. In contrast, CD4.Treg and CD4.TISg showed no significant associations with OS (Figure 5A). Similarly, for DSS, higher levels of CD4.Treg3 (HR: 1.41, 95%CI: 1.08–1.82, P = 0.010, Figure 5B) and CD4.Treg2 (HR: 1.34, 95%CI: 1.02–1.76, P = 0.027, Figure 4B) were linked to poorer outcomes, further underscoring their prognostic relevance. CD4.TISg, however, did not demonstrate a significant impact on DSS (Fi
We also evaluated the predictive potential of CD4.Treg3 in relation to immunotherapy outcomes. Across multiple datasets, including treatments targeting CTLA-4, PD-1, and PD-L1, CD4.Treg3 demonstrated strong predictive value for immunotherapy response (AUC > 0.7, Figure 5C). For example, in the Liu2019_PD1_Melanoma dataset, high CD4.Treg3 expression was associated with improved survival in patients treated with PD-1 inhibitors (P = 6.18 × 10-3, Figure 5D). Similarly, the VonAllen2015 cohort showed that high CD4.Treg3 expression correlated with significantly better survival outcomes compared to the low-expression group (P = 7.93 × 10-3, Figure 5E).
These findings suggest that elevated CD4.Treg3 expression may serve as both a prognostic and predictive biomarker. In particular, it can help identify patients less likely to benefit from current immunotherapy regimens, thereby guiding personalized treatment strategies and emphasizing the predictive relevance of CD4.Treg3 in immune checkpoint inhibitor response. Overall, our findings emphasize the dual role of CD4.Treg3 as both a prognostic and predictive marker in cancer. Elevated levels of CD4.Treg3 are associated with poorer survival and reduced responses to immunotherapy, highlighting the clinical importance of this subtype in guiding therapeutic decisions and potentially improving patient outcomes through tailored treatment approaches.
Our study presents a comprehensive analysis of the immune microenvironments in adnexal tissues and ascites in HGSOC, revealing distinct differences in CD4+ T cell phenotypes and their functional roles. The findings demonstrate that naive CD4+ T cells dominate in ascites, reflecting an immunosuppressive environment that facilitates immune evasion[16], whereas adnexal tissues are enriched with Tregs and CXCL13-expressing CD4+ T cells, suggesting a more activated and locally adaptive immune response. The presence of CXCL13-expressing CD4+ T cells in adnexa points to their involvement in forming tertiary lymphoid structures (TLS), which have been associated with favorable anti-tumor responses in other cancers[7,17-20]. This suggests that adnexa may initially mount an anti-tumor response, which is ultimately evaded by the tumor as it progresses.
Our characterization of four Treg subtypes (Treg1, Treg2, Treg3, and TISG) revealed considerable functional heterogeneity that aligns with their anatomical localization. Treg3 cells, enriched in adnexal tissues, displayed both activation and exhaustion markers such as CTLA4, LAG3, and PDCD1. This suggests that Treg3 cells may initially help control inflammation but are later co-opted to contribute to tumor immune evasion, similar to other solid tumors[21,22]. The coexistence of activation and exhaustion markers in Treg3 cells reflects a dynamic regulation of immune activity that can shift from protective to suppressive as the tumor evolves. In contrast, Treg2 cells were more prevalent in ascites and showed enhanced lipid metabolism and adhesion signatures, indicating a phenotype adapted to survive in a metabolically challenging environment. Such metabolic adaptations are reminiscent of observations in other cancers where Tregs adapt their metabolic profiles to maintain suppressive functions under nutrient-poor conditions[23,24]. These findings emphasize the need to consider the functional plasticity of different Treg subtypes in designing targeted therapies.
Our results have significant therapeutic implications. The association of Treg3 and Treg2 cells with poorer survival outcomes positions these subtypes as potential prognostic markers for HGSOC. High expression of Treg3 correlated with reduced OS and DSS, underscoring their role in promoting an immunosuppressive tumor microenvironment. This aligns with studies suggesting that Treg-mediated suppression is a major barrier to successful immunotherapy[25,26]. There
While our study provides valuable insights into the immune heterogeneity of HGSOC, it also has limitations. The reliance on scRNA-seq data provides a static snapshot of the immune landscape, whereas immune-tumor interactions are inherently dynamic. Future longitudinal studies that incorporate time-course analyses could offer a more nuanced understanding of how immune cell phenotypes evolve during disease progression and treatment. Additionally, fun
Overall, our study emphasizes the profound impact of anatomical sites on shaping the immune landscape in HGSOC. The enrichment and functional specialization of CD4+ T cell subtypes in adnexal tissues vs ascites highlight the importance of spatial context in immune regulation. Treg3 cells, in particular, emerge as a key immunosuppressive population that limits anti-tumor immunity and correlates with poor clinical outcomes, making them an important target for future therapies. Addressing the unique immune pressures present in different tumor niches and drawing from insights into other cancers will be critical for developing more effective, personalized treatments aimed at overcoming immune suppression and improving survival outcomes for patients with HGSOC.
In this study, we comprehensively analyzed the functional heterogeneity and tissue-specific adaptations of CD4+ T cell subtypes in HGSOC using scRNA-seq. Our findings reveal distinct immune landscapes between adnexal tissues and ascites, highlighting the critical role of Tregs, particularly the Treg3 subset, in shaping the tumor microenvironment. Treg3 cells were significantly enriched in adnexal tissues and displayed a highly activated yet exhausted phenotype, correlating with poorer clinical outcomes. Their elevated expression of immunosuppressive markers suggests a crucial role in tumor progression and immune evasion.
Moreover, our analysis demonstrated that CD4+ T cell subtypes exhibit unique metabolic and transcriptional adaptations depending on their microenvironment, emphasizing the need for site-specific immune profiling. The prognostic significance of Treg3 cells, coupled with their predictive value for immunotherapy response, underscores their potential as a novel therapeutic target in HGSOC. Targeting Treg3 cells while preserving overall immune homeostasis may enhance the efficacy of immune checkpoint inhibitors and other immunotherapies.
Our study provides critical insights into the immune landscape of HGSOC and paves the way for precision immunotherapy approaches. Future research should focus on validating these findings through in vivo models and clinical trials to assess the feasibility of targeting specific Treg subsets in ovarian cancer treatment. Understanding the dynamic interplay between CD4+ T cell subtypes and the tumor microenvironment will be essential for developing more effective, personalized therapeutic strategies that improve patient outcomes.
We sincerely thank the researchers and institutions that generated and shared the publicly available scRNA-seq datasets utilized in this study. Their contributions have been invaluable in advancing our understanding of the tumor immune microenvironment in HGSOC. We are also grateful to our colleagues for their insightful discussions and constructive feedback, which have enriched this study. Additionally, we acknowledge the support of the bioinformatics team for their assistance with data processing and analysis. Finally, we appreciate the open-access resources and computational tools that facilitated the analysis and visualization of the data in this study.
| 1. | Webb PM, Jordan SJ. Global epidemiology of epithelial ovarian cancer. Nat Rev Clin Oncol. 2024;21:389-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 2. | Kandalaft LE, Dangaj Laniti D, Coukos G. Immunobiology of high-grade serous ovarian cancer: lessons for clinical translation. Nat Rev Cancer. 2022;22:640-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 90] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 3. | Govindarajan M, Wohlmuth C, Waas M, Bernardini MQ, Kislinger T. High-throughput approaches for precision medicine in high-grade serous ovarian cancer. J Hematol Oncol. 2020;13:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 4. | Rajtak A, Ostrowska-Leśko M, Żak K, Tarkowski R, Kotarski J, Okła K. Integration of local and systemic immunity in ovarian cancer: Implications for immunotherapy. Front Immunol. 2022;13:1018256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Preston CC, Goode EL, Hartmann LC, Kalli KR, Knutson KL. Immunity and immune suppression in human ovarian cancer. Immunotherapy. 2011;3:539-556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Liu S, Tao Z, Lou J, Li R, Fu X, Xu J, Wang T, Zhang L, Shang W, Mao Y, Wang F. CD4(+)CCR8(+) Tregs in ovarian cancer: a potential effector Tregs for immune regulation. J Transl Med. 2023;21:803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 7. | Ukita M, Hamanishi J, Yoshitomi H, Yamanoi K, Takamatsu S, Ueda A, Suzuki H, Hosoe Y, Furutake Y, Taki M, Abiko K, Yamaguchi K, Nakai H, Baba T, Matsumura N, Yoshizawa A, Ueno H, Mandai M. CXCL13-producing CD4+ T cells accumulate in the early phase of tertiary lymphoid structures in ovarian cancer. JCI Insight. 2022;7:e157215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 109] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 8. | Cassar E, Kartikasari AER, Plebanski M. Regulatory T Cells in Ovarian Carcinogenesis and Future Therapeutic Opportunities. Cancers (Basel). 2022;14:5488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 9. | Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1163] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 10. | Demircan NC, Boussios S, Tasci T, Öztürk MA. Current and future immunotherapy approaches in ovarian cancer. Ann Transl Med. 2020;8:1714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 11. | Pawłowska A, Rekowska A, Kuryło W, Pańczyszyn A, Kotarski J, Wertel I. Current Understanding on Why Ovarian Cancer Is Resistant to Immune Checkpoint Inhibitors. Int J Mol Sci. 2023;24:10859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 12. | Yoon WH, DeFazio A, Kasherman L. Immune checkpoint inhibitors in ovarian cancer: where do we go from here? Cancer Drug Resist. 2023;6:358-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 13. | Hua T, Liu DX, Zhang XC, Li ST, Yan P, Zhao Q, Chen SB. CD4+ conventional T cells-related genes signature is a prognostic indicator for ovarian cancer. Front Immunol. 2023;14:1151109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 14. | Johnson RL, Cummings M, Thangavelu A, Theophilou G, de Jong D, Orsi NM. Barriers to Immunotherapy in Ovarian Cancer: Metabolic, Genomic, and Immune Perturbations in the Tumour Microenvironment. Cancers (Basel). 2021;13:6231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Vázquez-García I, Uhlitz F, Ceglia N, Lim JLP, Wu M, Mohibullah N, Niyazov J, Ruiz AEB, Boehm KM, Bojilova V, Fong CJ, Funnell T, Grewal D, Havasov E, Leung S, Pasha A, Patel DM, Pourmaleki M, Rusk N, Shi H, Vanguri R, Williams MJ, Zhang AW, Broach V, Chi DS, Da Cruz Paula A, Gardner GJ, Kim SH, Lennon M, Long Roche K, Sonoda Y, Zivanovic O, Kundra R, Viale A, Derakhshan FN, Geneslaw L, Issa Bhaloo S, Maroldi A, Nunez R, Pareja F, Stylianou A, Vahdatinia M, Bykov Y, Grisham RN, Liu YL, Lakhman Y, Nikolovski I, Kelly D, Gao J, Schietinger A, Hollmann TJ, Bakhoum SF, Soslow RA, Ellenson LH, Abu-Rustum NR, Aghajanian C, Friedman CF, McPherson A, Weigelt B, Zamarin D, Shah SP. Ovarian cancer mutational processes drive site-specific immune evasion. Nature. 2022;612:778-786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 145] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 16. | Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188:4135-4140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 234] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 17. | Ronchetti S, Ricci E, Petrillo MG, Cari L, Migliorati G, Nocentini G, Riccardi C. Glucocorticoid-induced tumour necrosis factor receptor-related protein: a key marker of functional regulatory T cells. J Immunol Res. 2015;2015:171520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 18. | Davar D, Zappasodi R. Targeting GITR in cancer immunotherapy - there is no perfect knowledge. Oncotarget. 2023;14:614-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Sautès-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19:307-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 1138] [Article Influence: 189.7] [Reference Citation Analysis (0)] |
| 20. | Gao SH, Liu SZ, Wang GZ, Zhou GB. CXCL13 in Cancer and Other Diseases: Biological Functions, Clinical Significance, and Therapeutic Opportunities. Life (Basel). 2021;11:1282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 21. | Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 2012;72:2162-2171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 643] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 22. | Sarkar T, Dhar S, Sa G. Tumor-infiltrating T-regulatory cells adapt to altered metabolism to promote tumor-immune escape. Curr Res Immunol. 2021;2:132-141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 23. | Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, Wang Z, Quinn WJ 3rd, Kopinski PK, Wang L, Akimova T, Liu Y, Bhatti TR, Han R, Laskin BL, Baur JA, Blair IA, Wallace DC, Hancock WW, Beier UH. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017;25:1282-1293.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 876] [Article Influence: 109.5] [Reference Citation Analysis (0)] |
| 24. | Yan Y, Huang L, Liu Y, Yi M, Chu Q, Jiao D, Wu K. Metabolic profiles of regulatory T cells and their adaptations to the tumor microenvironment: implications for antitumor immunity. J Hematol Oncol. 2022;15:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 122] [Reference Citation Analysis (0)] |
| 25. | Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2349] [Cited by in RCA: 3830] [Article Influence: 478.8] [Reference Citation Analysis (0)] |
| 26. | Kim JH, Kim BS, Lee SK. Regulatory T Cells in Tumor Microenvironment and Approach for Anticancer Immunotherapy. Immune Netw. 2020;20:e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 27. | Fattori S, Roux H, Connen E, Robert L, Gorvel L, Le Roy A, Houacine J, Foussat A, Chretien AS, Olive D. Therapeutic Targeting of Tumor-Infiltrating Regulatory T Cells in Breast Cancer. Cancer Res. 2022;82:3868-3879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 28. | van Wilpe S, Tolmeijer SH, Koornstra RHT, de Vries IJM, Gerritsen WR, Ligtenberg M, Mehra N. Homologous Recombination Repair Deficiency and Implications for Tumor Immunogenicity. Cancers (Basel). 2021;13:2249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | Silva SB, Wanderley CWS, Colli LM. Immune Checkpoint Inhibitors in Tumors Harboring Homologous Recombination Deficiency: Challenges in Attaining Efficacy. Front Immunol. 2022;13:826577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |