Published online May 24, 2025. doi: 10.5306/wjco.v16.i5.103830
Revised: February 10, 2025
Accepted: March 28, 2025
Published online: May 24, 2025
Processing time: 164 Days and 13.4 Hours
Bladder cancer (BLCA) is a common urological tumor. Homeobox C6 (HOXC6) is an HOX family gene that has an oncogenic effect in various malignancies.
To investigate the expression and function of HOXC6 in BLCA.
This study employed immunohistochemistry, along with global chip and sequen
The immunohistochemistry results, sequencing data, and microarray data revealed that both the mRNA and protein expressions of HOXC6 in BLCA were notably greater than the expressions in non-cancerous tissues. Knocking down the expression of HOXC6 considerably limited the function of cell proliferation, migration, and invasion abilities of BLCA cells, elevated cell apoptosis, and triggered the G0/G1 phase blockade. The potential target genes of HOXC6 were enriched in pathways such as chemical carcinogenesis and reactive oxygen species. A notable positive correlation between HOXC6 mRNA expression and its target gene timeless circadian regulator
HOXC6 is upregulated in BLCA and may influence the cellular functions of BLCA by regulating the expression of the target gene TIMELESS.
Core Tip: Homeobox C6 (HOXC6) is an HOX family gene that has an oncogenic effect in various malignancies. Our study showed the increased expression levels of HOXC6 at the protein and mRNA levels. Down-regulated HOXC6 considerably limited the function of cell proliferation, migration, and invasion abilities of bladder cancer (BLCA) cells, elevated cell apoptosis, and triggered the G0/G1 phase blockade. HOXC6 may influence the cellular functions of BLCA by regulating the expression of the target gene timeless circadian regulator (TIMELESS), with the evidence of positive correlation between HOXC6 and TIMELESS and the binding peak signals for HOXC6 in the promoter region of TIMELESS.
- Citation: Lu DJ, Wang HR, Xu YS, Huang HB, Zhong QG, Luo YN, Qi JF, Wu HC, Pei JY, Zhang K, Xu CX, Wang TX, Zhang W, Zhou YH, Huang ZG, Wang FB. Homeobox C6 plays an oncogenic role in bladder cancer. World J Clin Oncol 2025; 16(5): 103830
- URL: https://www.wjgnet.com/2218-4333/full/v16/i5/103830.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i5.103830
Bladder cancer (BLCA) is one of the most common urological tumors, ranking eighth in incidence and mortality of men in China[1]. According to the latest tumor statistics for 2024, BLCA ranks as the ninth major cancer burden globally, with a higher incidence in males than in females[2]. The occurrence and development of BLCA are influenced by both genetic factors, such as mutations in oncogenes and deletions of tumor suppressor genes, and environmental factors, including long-term smoking, exposure to harmful chemicals and prolonged irritation from urinary system inflammation. In treatment, in addition to surgical resection, chemotherapy, and radiotherapy, immunotherapy and targeted therapy have shown promising efficacy, reflecting a trend toward precision medicine[3]. Therefore, exploring the mutation profile and molecular mechanisms of BLCA is crucial for its early detection, diagnosis, and treatment.
Homeobox (HOX) genes comprise a family of genes that includes four subtypes: A, B, C, and D, which are primarily expressed during embryonic development and in tumors. Existing research has revealed that expression dysregulation of HOX genes is associated with the development of numerous malignancies through mechanisms that interfere with cell differentiation, proliferation, angiogenesis, autophagy, apoptosis, invasion, and metastasis[4]. HOXC6, one of the HOX family genes, has oncogenic effects in several malignancies, including prostate cancer, glioma, cervical cancer, gastric cancer, hepatocellular carcinoma, and esophageal cancer[5-10]. However, as no studies have confirmed the involvement of HOXC6 in the progression of BLCA, a more comprehensive exploration of the HOXC6 gene in BLCA is warranted.
Based on the existing research gap, in this study, we integrated immunohistochemical staining experiments and high-throughput sequencing datasets of BLCA from around the world to thoroughly investigate the expression characteristics, potential molecular mechanisms, and clinical value of the HOXC6 gene in BLCA.
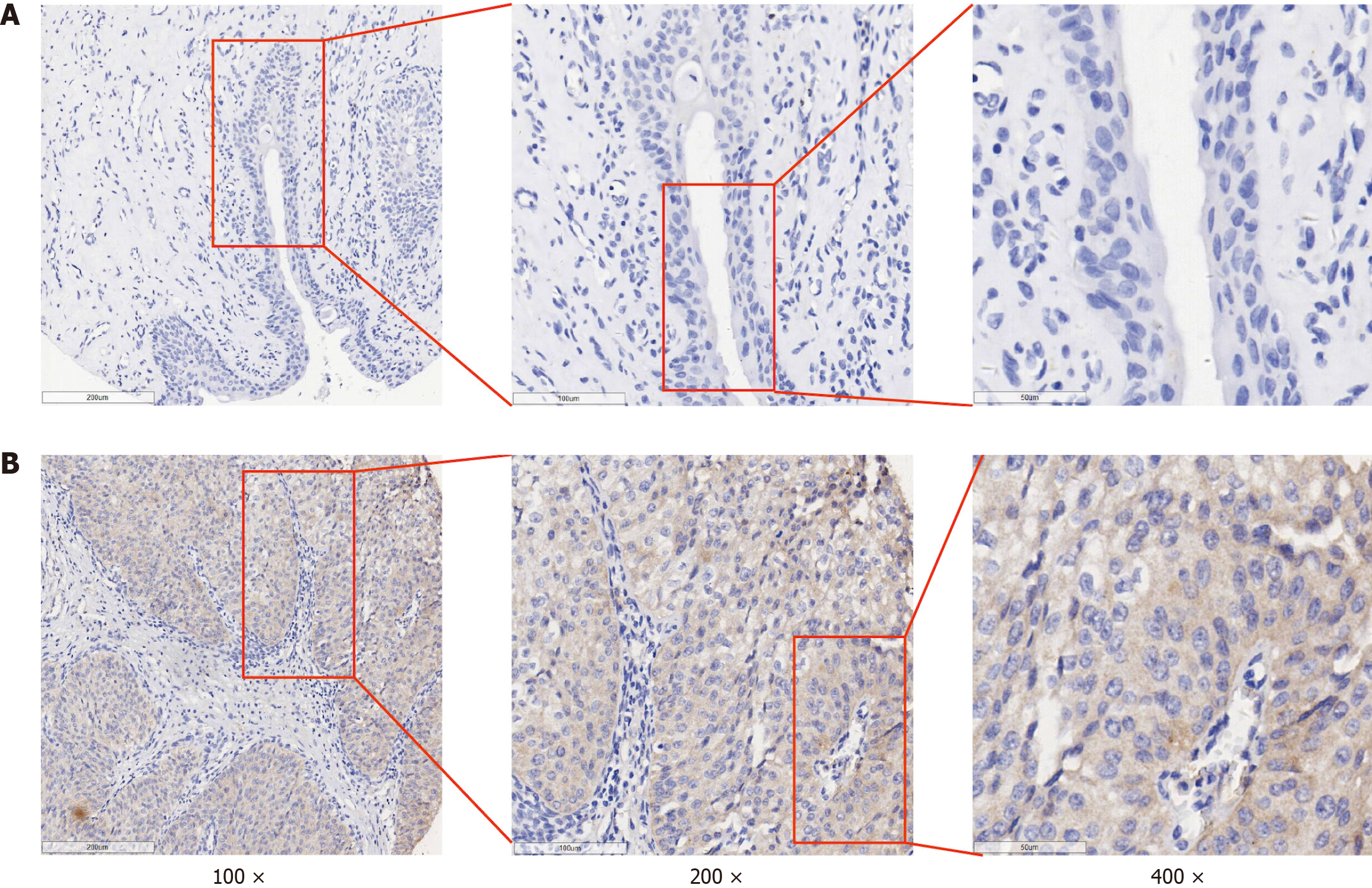
Materials: The BLCA tissue microarrays used in this experiment (BLC481, BLC1021, BLC1501, BLC242) were purchased from the Guilin Pansum Bio-Technology Company (Guangxi Zhuang Autonomous Region, China). The primary antibody for HOXC6 was obtained from Thermo Fisher Scientific (PA5-41479, United States, dilution ratio 1:100). The secondary antibody was purchased from Changdao Biotechnology Co., Ltd. (D-3004-15, China), and the immunohistochemical experiment was strictly conducted based on the kit instructions.
Immunohistochemistry results interpretation: The IHC results of HOXC6 in BLCA were measured by two pathologists in a double-blind manner according to the following scoring criteria. First, the staining intensity was categorized into no staining, weak, moderate, and strong, corresponding to scores of 0, 1, 2, and 3, respectively. Next, five random regions were selected to evaluate the percentage of positively stained cells, with percentages categorized as follows: < 5%, 5%-25%, 26%-50%, 51%-75% and > 75% corresponding to scores of 0, 1, 2, 3 and 4, respectively. Finally, the IHC scores of the samples were the product of the staining intensity score and the percentage of positively stained cells score[11].
RNA sequencing and gene expression array datasets related to BLCA available globally as of January 1, 2024, were retrieved from GEO, ArrayExpress, SRA, BioProject, and TCGA databases. The inclusion criteria for the datasets were as follows: (1) Species must be Homo sapiens; (2) Experimental method must be RNA sequencing or gene expression array; (3) Sample type must be tissue; and (4) The number of cancer samples and control samples must each be ≥ 3. The exclusion criteria were: (1) The presence of metastatic cancer; and (2) Samples that had undergone drug treatment. Finally, the datasets included in the study were optimized by merging datasets from the same sequencing platform and removing batch effects by the remove Batch Effect function of the limma package in R.
The standardized mean difference (SMD) of HOXC6 gene transcript expression levels across all datasets was calculated using Stata v15.1 (TX, United States). This integrated SMD was used to comprehensively assess the expression characteristics of HOXC6 in BLCA. During the integrated analysis, I² values were employed to evaluate the degree of data heterogeneity. If I² was higher than 50%, a random effects model was utilized for integration due to the high data heterogeneity. Conversely, a fixed-effects model was selected when I² was 50% or lower. A summary receiver operating characteristic curve (SROC) was used to analyze the diagnostic efficacy of HOXC6 expression for BLCA. Egger’s test was employed to examine publication bias. Statistically significant publication bias was considered when P < 0.05.
The BLCA cell lines used in this experiment (EJ1, 5637, J82, and HT-1376) were all purchased from the Chinese Typical Culture Preservation Center. The EJ1 and J82 cell lines were cultured with MEM medium, while the 5637 cell line was cultured with RPMI 1640 medium, and the J82 cell line was cultured with DMEM medium. The complete medium was composed of 89% basal medium, 10% fetal bovine serum (purchased from Life Technologies, Inc.), and 1% penicillin-streptomycin (purchased from Gibco). The mRNA expression level of HOXC6 was detected via reverse transcription-quantitative PCR (RT-qPCR) after reverse transcription with total RNA. The cell lines that exhibited relatively high expressions of HOXC6 were selected to construct the HOXC6 Low-expression cell models. Information about the primers is shown in Table 1.
| Primer | Sequence (5’-3’) |
| Hoxc6 (human)-F | CCAGAAAGCCAGTATCCA |
| Hoxc6 (human)-R | TTCTCCAGTTCCAGGGTC |
| Gapdh (human)-F | GGAGCGAGATCCCTCCAAAAT |
| Gapdh (human)-R | GGCTGTTGTCATACTTCTCATGG |
The siRNA used in this experiment was synthesized by Jiangsu Genecefe Biotechnology Co., Ltd. (Jiangsu, China), with sequence information provided in Table 2. The transfection mixture was composed of 125 μL of basic culture medium, 100 pmol of siRNA, and 4 μL of Lipo8000 and was incubated at room temperature for 20 minutes. To each well of a six-well plate cultured with cells, 5 μL of the transfection mixture was added and incubated for 24 hours. Three replicates were set up for each group, including the control group (CK group), negative control group (NC group), HOXC6-siRNA-1 group, HOXC6-siRNA-2 group and HOXC6-siRNA-3 group. The mRNA expression of HOXC6 was detected by RT-qPCR after RNA extraction and reverse transcription. The siRNA with the most noticeable knockdown effect was chosen for subsequent cellular function experiments.
| siRNA | Sense sequence (5’ to 3’) | Anti-sense sequence (5’ to 3’) |
| HOXC6-siRNA-1 | CCUCAAUCGCUCAGGAUUUTT | AAAUCCUGAGCGAUUGAGGTT |
| HOXC6-siRNA-2 | CAGAAGAGGAGAAGCAGAATT | UUCUGCUUCUCCUCUUCUGTT |
| HOXC6-siRNA-3 | CUCUCAAACUGCAGACAAATT | UUUGUCUGCAGUUUGAGAGTT |
Cells were adjusted to a density of 5 × 104 cells/mL with complete medium, and 100 μL of the cell solution was transferred to each well in a 96-well plate. Three replicates were set up for the 5637 group, 5637 + NC group and 5637 + HOXC6-siRNA group. Then, 4 pmol of si-NC or si-HOXC6 and 0.16 μL of Lipo8000™ transfection reagent were added together, gently pipetted to ensure thorough mixing, and incubated for 24, 48, or 72 hours. Afterward, 10 μL of CCK-8 reagent from Vazyme (China) was added to each well. A BioTek microplate reader was used to detect the absorbance value at 450 nm after 2 hours.
The 3 × 105 cells were transferred to a 3.5 cm dish and cultured until cells grew all over the dish. Three replicates were set up for the 5637 group, 5637 + NC group and 5637 + HOXC6-siRNA group. A 200 μL pipette tip was utilized to create a vertical scratch in the culture dish. Phosphate-buffered saline (PBS) was employed to wash the dish three times. We took photographs to record the 0-hour results. The dish was then returned to the incubator, and after 24 hours, the cells were photographed again to observe cell migration. The distance of cell migration was assessed using ImageJ software. Finally, we calculated the cell migration rate as previously reported[12].
Cells were adjusted to a concentration of 3 × 105 cells/mL with complete medium. Three replicates were set up for the 5637 group, 5637 + NC group and 5637 + HOXC6-siRNA group. To each well of a 24-well plate, 0.8 mL of complete medium was pre-added, and the transwell chambers were placed in. Next, 200 μL cell suspension was added to the upper transwell chamber. After 1 hour, the suspension was moved to a cell culture incubator for 72 hours. The transwell chambers were removed and washed with PBS. An ice-cold 70% ethanol solution was employed to fix with chambers for 1 hour. Then, 0.5% crystal violet solution was used to stain the cells in the chambers for 20 minutes at 25 °C. The cells were photographed under a microscope after being washed with PBS.
Three replicates were set up for the 5637 group, 5637 + NC group and 5637 + HOXC6-siRNA group. We digested cells in the dish using trypsin without EDTA, centrifuged the cell suspension for 5 minutes, and removed the supernatant. The cells were then resuspended in precooled 1 × PBS (4 °C), centrifuged, and had the supernatant removed again. We added 300 μL 1 × binding buffer for resuspension and 5 μL Annexin V-FITC for staining. We mixed the cell suspension gently, and put it in the dark for 15 minutes. Then, we added 10 μL propidium iodide (PI) to the cell suspension and incubated the cell suspension at 25 °C in the dark for another 10 minutes. Apoptosis was determined with flow cytometry, and FlowJo 7.6 software was employed for the results analysis.
Three replicates were set up for the 5637 group, 5637 + NC group and 5637 + HOXC6-siRNA group. We used 0.25% trypsin without EDTA for cell digestion, centrifuged the cell suspension, and removed the supernatant. The cells were resuspended with 500 μL of PBS. After centrifuging and discarding the supernatant, we added 100 μL of PBS to resuspend the cells. Cells were then fixed at 4 °C for 4 hours with 700 μL of precooled 80% ethanol. After that, we centrifuged and washed the cells with precooled PBS twice. Then, 100 μL of RNase (50 μg/mL) was added to the cell suspension, which was then incubated at 37 °C for 30 minutes. Following this, 400 μL of PI (50 μg/mL) was added for staining, and the suspension was kept at 4 °C for 30 minutes. The analysis was performed using flow cytometry.
The Cistrome Data Browser is a publicly available database that compiles Chromatin Immunoprecipitation Sequencing (ChIP-seq) information for humans and mice[13]. We utilized data from the Cistrome Data Browser to predict the po
Statistical analysis was conducted using the software of Statistical Package for the Social Sciences (SPSS, version 25, IBM Corporation, Armonk, NY, United States). A t-test was employed for statistical analysis of the two groups, if the data met the following conditions: (1) Approximately following a normal distribution; (2) A continuous variable; (3) Showing equal variances between the two groups; and (4) Independent samples. If the data does not meet these conditions, non-parametric methods such as the Wilcoxon rank-sum test were used for statistical analysis. The results were presented as mean ± SD. The Pearson correlation coefficient method was used to analyze the correlation between the expression levels of the two genes. Statistical significance was indicated by P < 0.05.
Up to 154 BLCA tissues and 46 adjacent tissues were included in the IHC experiment. Representative images of HOXC6 immunohistochemical detection are shown in Figure 1. The IHC score of HOXC6 in the BLCA samples was significantly higher than the IHC score in adjacent samples (9.517 ± 0.171 vs 3.170 ± 0.263, P < 0.001; Table 3). In addition, the expression level of HOXC6 protein in BLCA samples from patients under 60 years old (n = 59) was significantly higher than that in samples from patients aged 60 and above (n = 95) (9.946 ± 2.184 vs 9.251 ± 2.165, P = 0.048; Table 3). There was no statistically significant difference in the expression of HOXC6 protein in BLCA tissues from male and female patients. Additionally, the expression of the HOXC6 protein showed no statistically significant difference among patients with different T stages and pathological grades in BLCA tissues (Table 3).
| Clinicopathological parameters | n | HOXC6 expression | P value | |
| mean | SD | |||
| Group | ||||
| Cancer | 154 | 9.517 | 2.121 | d |
| Non-cancer | 46 | 3.170 | 1.784 | |
| Gender | ||||
| Male | 133 | 9.504 | 2.128 | 0.848 |
| Female | 21 | 9.600 | 2.132 | |
| Age (years) | ||||
| < 60 | 59 | 9.946 | 1.992 | a |
| ≥ 60 | 95 | 9.251 | 2.165 | |
| Pathological T stage | ||||
| T1 + T2 | 124 | 9.565 | 2.184 | 0.573 |
| T3 + T4 | 30 | 9.320 | 1.859 | |
| Pathological grade | ||||
| I-II | 92 | 9.544 | 2.098 | 0.850 |
| III-IV | 62 | 9.477 | 2.172 | |
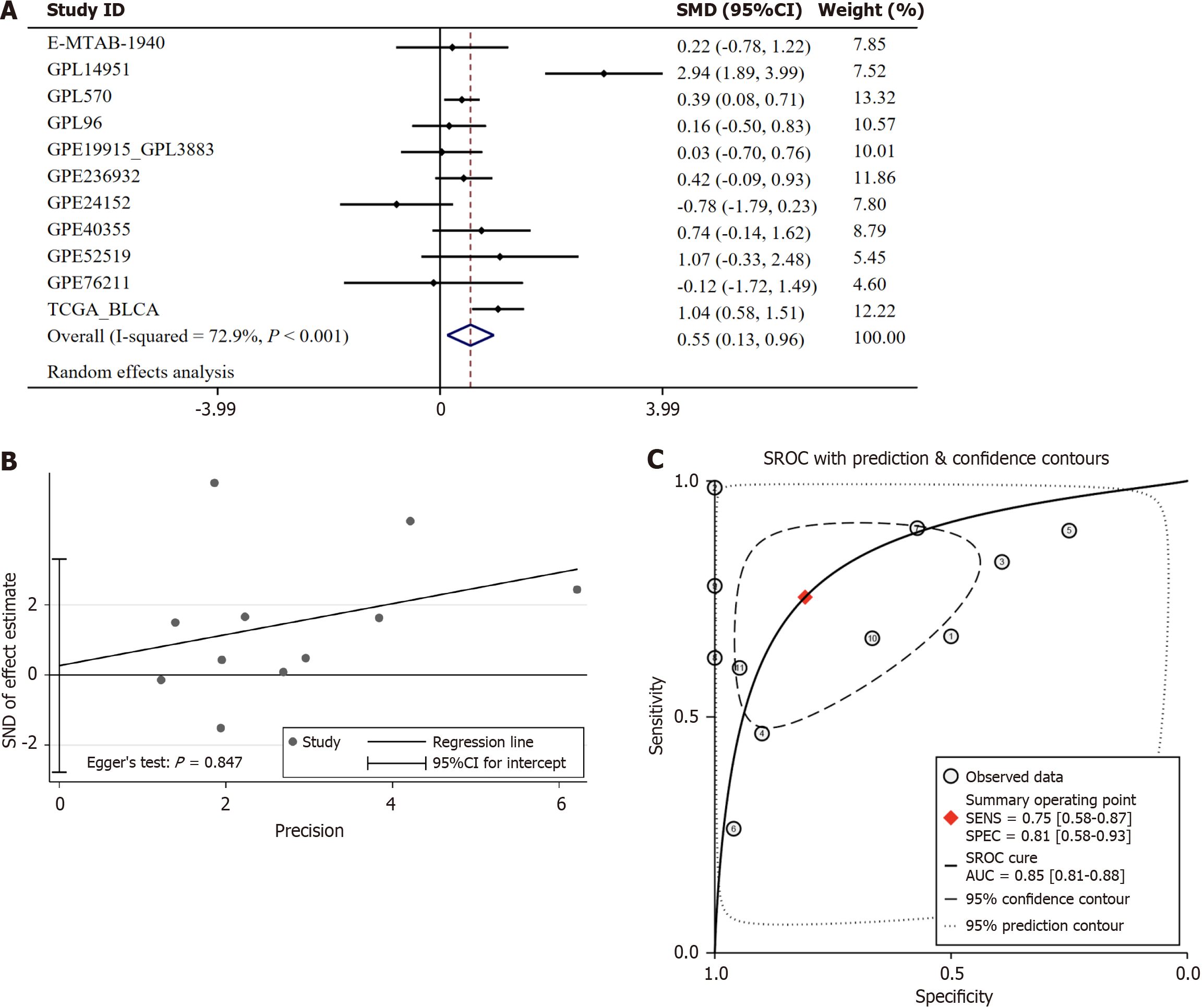
HOXC6 mRNA is also highly expressed in BLCA. We included a total of 11 datasets containing expression values of HOXC6, which comprised 1028 cancer tissues and 142 adjacent or noncancerous tissues. Because I² = 72.9%, indicating high heterogeneity in the data, we employed a random-effects model to integrate the SMD. The SMD was 0.55 (95%CI: 0.13-0.96, P = 0.01; Figure 2A). Egger’s test displayed no extraordinary publication bias (P = 0.847; Figure 2B). The SROC curve was 0.85 (95%CI: 0.81-0.88), with a sensitivity of 0.75 (95%CI: 0.58-0.87) and a specificity of 0.81 (95%CI: 0.58-0.93) (Figure 2C), suggesting that HOXC6 has high efficacy in distinguishing BLCA from non-cancerous tissues.
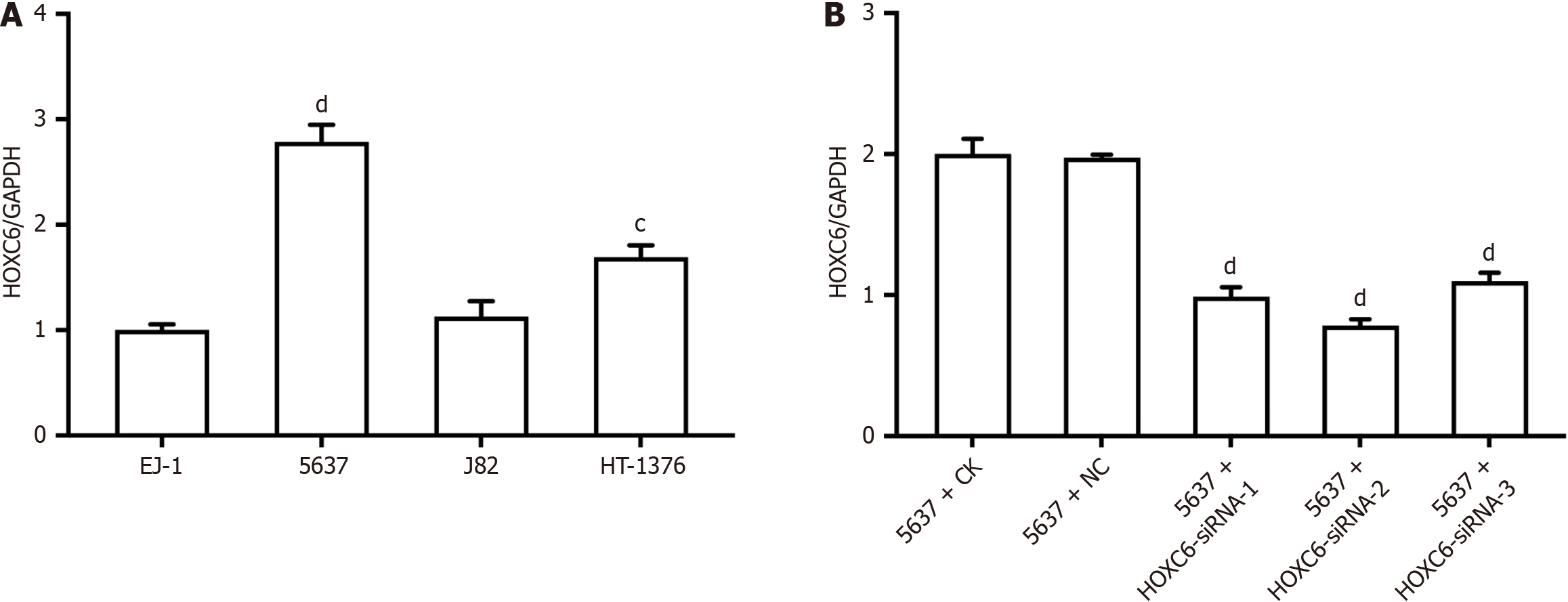
Detection revealed that HOXC6 expression was highest in the 5637 cell line (Figure 3A), and subsequent knockdown of HOXC6 expression was conducted on this cell line. Following siRNA transfection, no considerable changes were noticed in the NC group compared to the CK group; however, the gene expression of HOXC6 was significantly downregulated after siRNA transfection, with siRNA-2 showing the best knockdown efficiency (Figure 3B). Therefore, siRNA-2 was chosen for further transfection experiments.
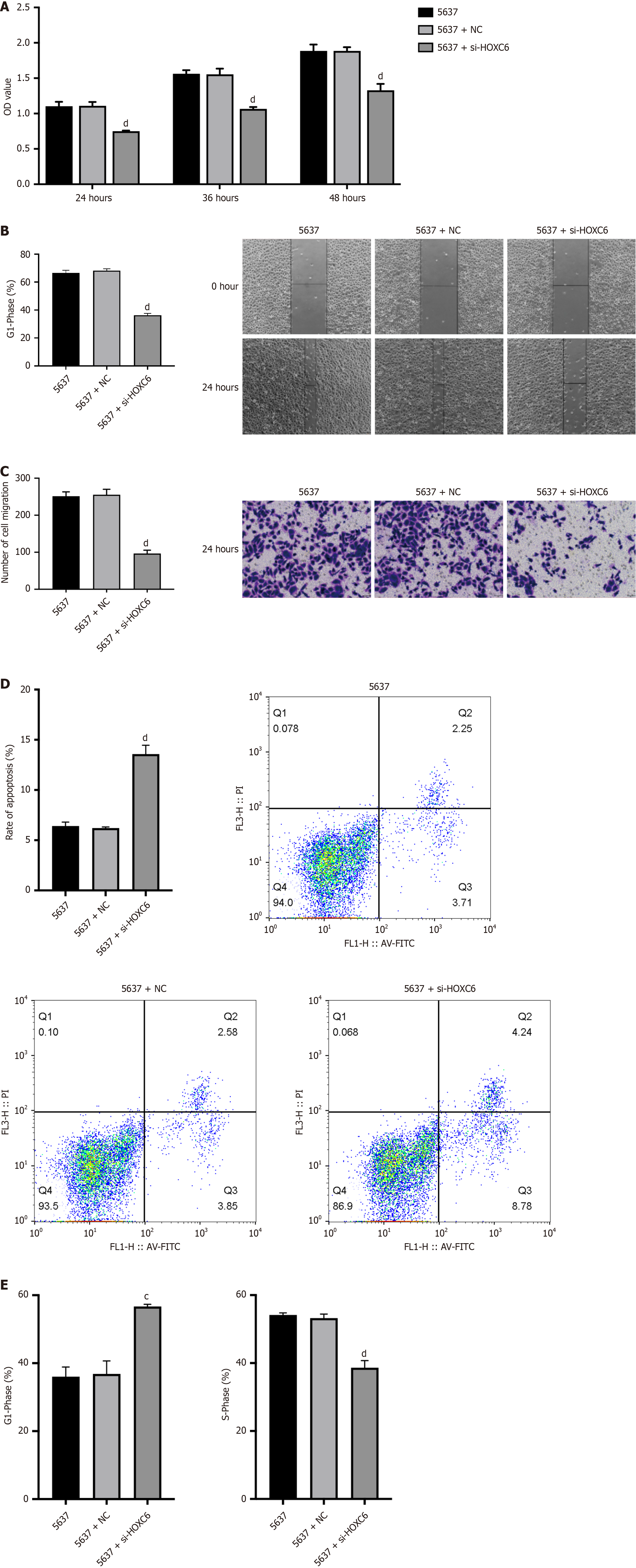
The results of the CCK-8 experiment indicate that, at the same time point, cell viability was significantly downregulated following the knockdown of HOXC6 expression compared to the 5637 group, with the most pronounced effect observed at the 24-hour mark (Figure 4A). The cell scratch assay results showed no considerable differences between the 5637 and NC groups. However, after HOXC6 knockdown, the cell migration rate was notably slower in the 5637 + siHOXC6 group compared to the NC group (Figure 4B). The transwell assay results indicated that transfection with NC had no considerable effect on cell migration compared to the 5637 cell group, whereas HOXC6 knockdown remarkably inhibited cell migration (Figure 4C). The apoptosis assay results revealed that the NC group showed no considerable changes in apoptosis rate compared to the 5637 group, while HOXC6 knockdown significantly promoted apoptosis (Figure 4D). Cell cycle analysis indicated that transfection with siRNA NC had no critical effect on the cell cycle compared to the 5637 group. However, after HOXC6 knockdown, the percentage of cells in the S phase was markedly reduced, with tumor cells being arrested in the G1/G0 phase, resulting in hampered cell proliferation (Figure 4E).
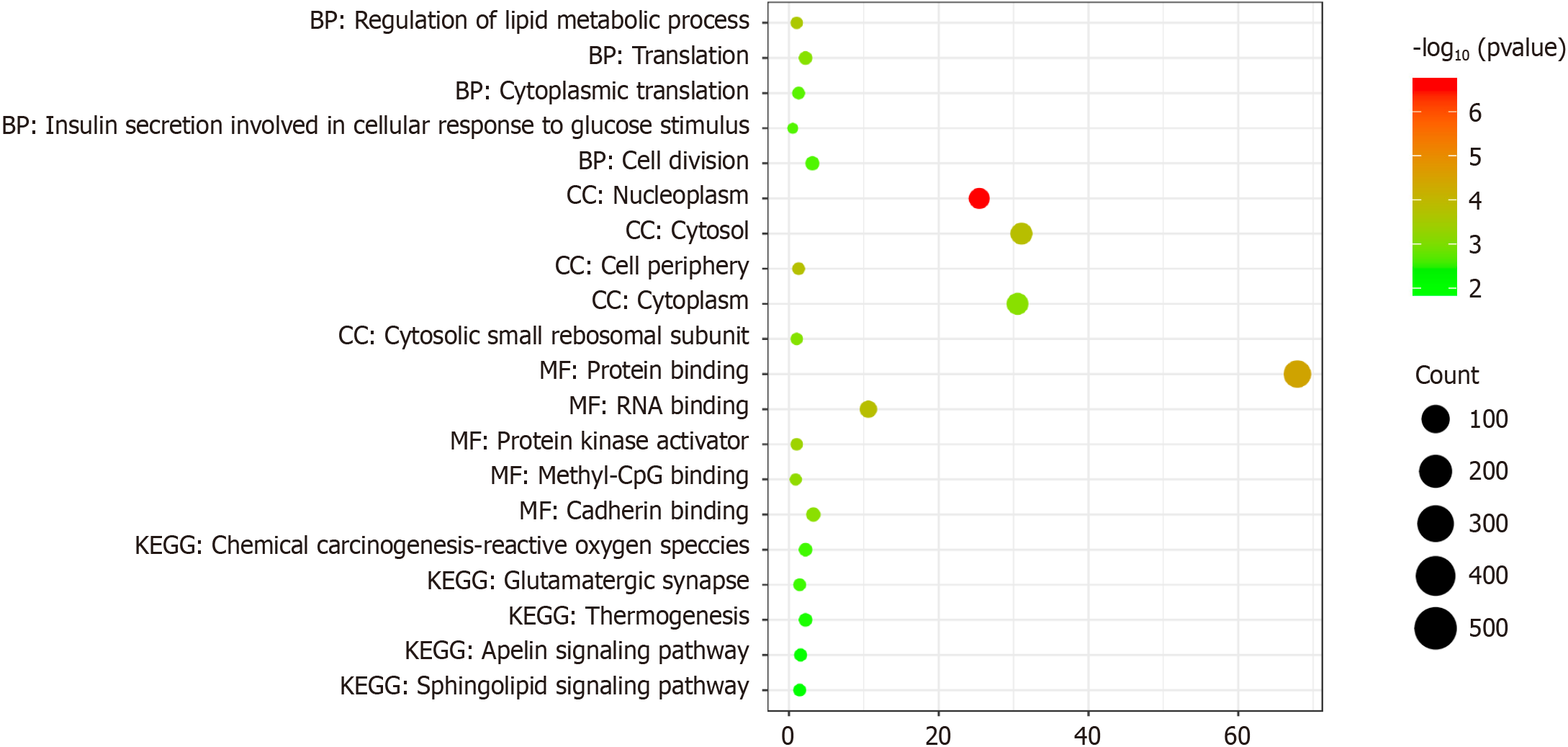
The potential function of HOXC6 in BLCA remains unknown. We tried to explore this issue through pathway enrichment analysis. The data demonstrated that 773 potential target genes of HOXC6 were primarily enriched in pathways related to the “regulation of lipid metabolic processes” at the biological process level. At the cellular component level, these genes were chiefly enriched in “nucleoplasm”. At the molecular function level, they were primarily enriched in “protein binding.” In terms of the KEGG analysis, the target genes were primarily enriched in the pathway of “chemical carcinogenesis-reactive oxygen species” (Figure 5).
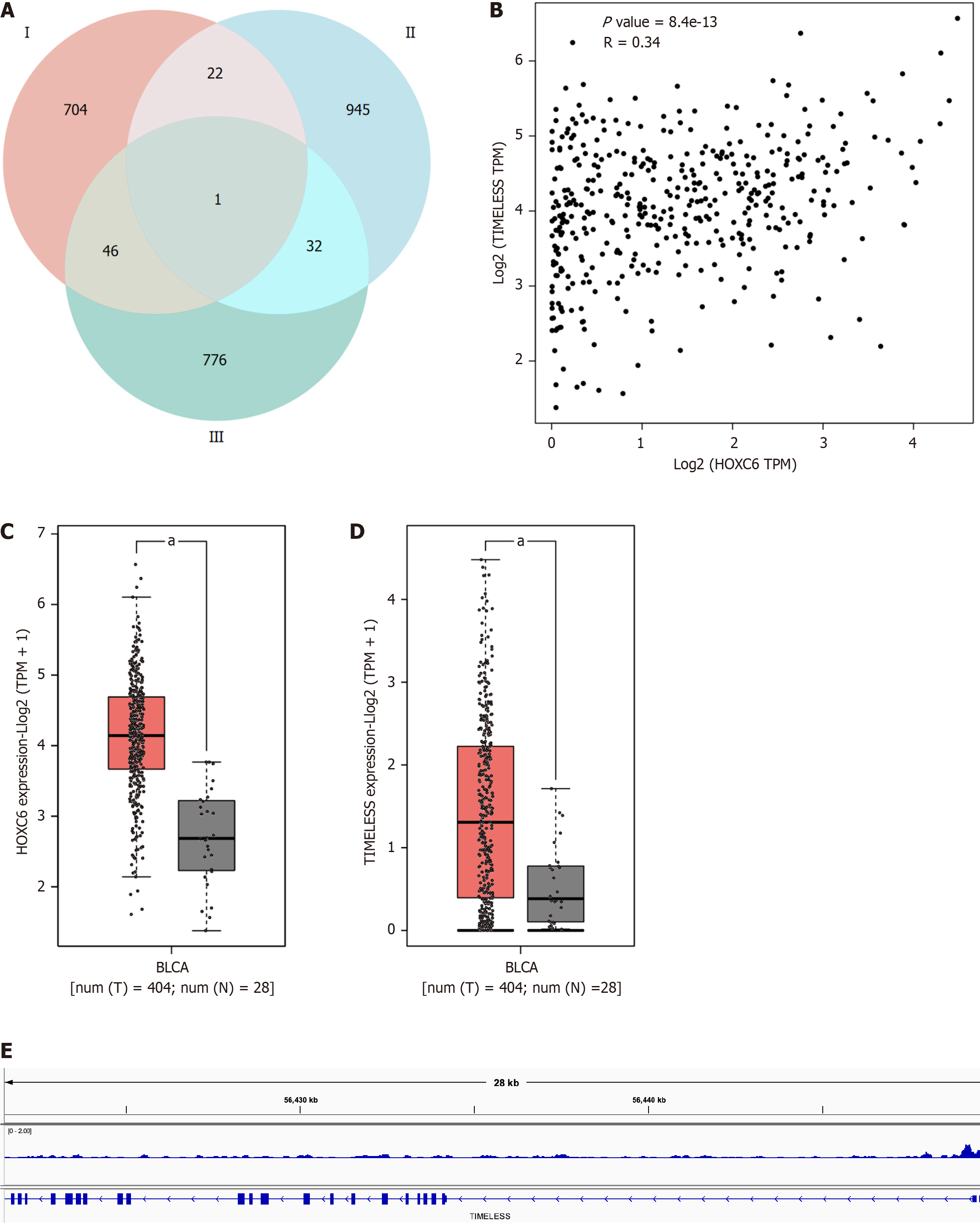
We explored the potential mechanism of the target genes of HOXC6 in depth. By integrating the potential target genes of HOXC6, genes positively correlated with HOXC6 expression in BLCA, and genes significantly upregulated in BLCA, we ultimately identified a unique gene, TIMELESS (Figure 6A). The expressions of HOXC6 and TIMELESS were found to be significantly correlated (r = 0.34, P = 8.4e-13; Figure 6B). Both HOXC6 and TIMELESS mRNA expressions were significantly higher in BCLA groups than in noncancerous groups (Figure 6C and D). The ChIP-seq results for HOXC6 demonstrated significant binding to the promoter region of TIMELESS (Figure 6E). The above data reveal that TIMELESS is likely a target gene of HOXC6 and that HOXC6 promotes BLCA by regulating the expression of TIMELESS.
Until now, no study has reported on the expression and mechanistic exploration of HOXC6 in BLCA. This study utilized IHC to establish that the HOXC6 protein was highly expressed in BLCA. Additionally, data collected from 11 BLCA microarray and sequencing studies globally demonstrated high mRNA expression of HOXC6 in BLCA. Knockdown of HOXC6 expression significantly inhibited the various biological functions of cells, including proliferation, migration, apoptosis, and cell cycle progression. Furthermore, HOXC6 may influence the tumorigenesis of BLCA by targeting and regulating TIMELESS expression via the “chemical carcinogenesis-reactive oxygen species” pathway.
Research has shown that HOXC6 is highly expressed in multiple malignancies, including pancreatic ductal adenocarcinoma[16], prostate cancer[17,18], oral squamous cell carcinoma[19], glioma[6], gastric cancer[20] and cervical cancer[7]. In these tumor types, upregulated HOXC6 is significantly related to poor prognosis, particularly in prostate cancer[18], gastric cancer[20], and cervical cancer[7]. These studies suggest that HOXC6 potentially contributes to tumorigenesis and progression. In the present study, we observed that both the mRNA and protein expressions of HOXC6 in BLCA samples were remarkably greater than those in adjacent noncancerous samples, providing new perspectives on its potential role in BLCA. Furthermore, the high expression of HOXC6 exhibits good discriminatory ability, effectively distinguishing BLCA from noncancerous tissues. This finding indicates that HOXC6 may function as a biomarker for BLCA, contributing to early diagnosis and thereby improving patient prognosis.
HOXC6 has clearly demonstrated biological functions across various tumors. Previous investigations have indicated that the knockdown of HOXC6 expression can effectively block the G0/G1 phase in glioma cells and elevated the cell apoptosis rate, thereby hindering cell proliferation and colony formation ability[6,21]. In prostate cancer, low expression of HOXC6 increased the occurrence of apoptosis[17]. In laryngeal cancer studies, silencing HOXC6 significantly inhibited epithelial-mesenchymal transition and survival, migration, and invasion capabilities[22]. These results are consistent with our observations in BLCA cells, where we found that decreased expression of HOXC6 not only hindered the ability of cell proliferation and migration but also increased the percentage of apoptotic cells and arrested cells in the G1/G0 phase. These findings suggest that HOXC6 plays a critical regulatory role in the biological functions of BLCA cells; however, the exact mechanisms involved still require further exploration.
In various tumor types, HOXC6 affects cellular biological functions by regulating a series of target genes. In pancreatic ductal adenocarcinoma, HOXC6 promotes tumor growth by upregulating MSK1 expression and inhibiting the tumor suppressor protein PPP2R2B[16]. Overexpression of HOXC6 enhances BCL2-dependent anti-apoptotic effects, promoting cell cycle progression and proliferation in cervical cancer[7]. In gastric cancer, HOXC6 can upregulate MMP9 expression, thereby raising tumor cell migration and invasion[8]. In prostate cancer, HOXC6 mediates apoptosis by regulating the expression of NEP and IGFBP-3[17]. Our study reveals that the potential target genes of HOXC6 are primarily enriched in pathways, including “chemical carcinogenesis-reactive oxygen species”. This suggests that HOXC6 may alter the biological functions of BLCA cells via these pathways. We observed remarkable binding peaks of HOXC6 in the promoter region of the target gene TIMELESS and a notable positive correlation between the mRNA expressions of HOXC6 and TIMELESS, indicating that TIMELESS may engage in HOXC6-mediated tumorigenesis. However, the specific regulatory mechanisms by which HOXC6 operates in BLCA still need to be validated through more in-depth experiments.
In summary, this study verified the high expression of HOXC6 in BLCA at both the protein and mRNA levels. We demonstrated that HOXC6 participates in the cellular functions of BLCA, including cell proliferation, migration, cell apoptosis, and cell cycle. We also found that HOXC6 may influence the cellular functions of BLCA cells by managing the expression of the target gene TIMELESS.
| 1. | Zheng RS, Chen R, Han BF, Wang SM, Li L, Sun KX, Zeng HM, Wei WW, He J. [Cancer incidence and mortality in China, 2022]. Zhonghua Zhong Liu Za Zhi. 2024;46:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 153] [Reference Citation Analysis (0)] |
| 2. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8084] [Article Influence: 8084.0] [Reference Citation Analysis (2)] |
| 3. | Lopez-Beltran A, Cookson MS, Guercio BJ, Cheng L. Advances in diagnosis and treatment of bladder cancer. BMJ. 2024;384:e076743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 122] [Reference Citation Analysis (0)] |
| 4. | Paço A, Aparecida de Bessa Garcia S, Leitão Castro J, Costa-Pinto AR, Freitas R. Roles of the HOX Proteins in Cancer Invasion and Metastasis. Cancers (Basel). 2020;13:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 5. | Zhou J, Yang X, Song P, Wang H, Wang X. HOXC6 in the prognosis of prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:2715-2720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Huang H, Huo Z, Jiao J, Ji W, Huang J, Bian Z, Xu B, Shao J, Sun J. HOXC6 impacts epithelial-mesenchymal transition and the immune microenvironment through gene transcription in gliomas. Cancer Cell Int. 2022;22:170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 7. | Wang Y, Wang C, Liu N, Hou J, Xiao W, Wang H. HOXC6 promotes cervical cancer progression via regulation of Bcl-2. FASEB J. 2019;33:3901-3911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 8. | Chen SW, Zhang Q, Xu ZF, Wang HP, Shi Y, Xu F, Zhang WJ, Wang P, Li Y. HOXC6 promotes gastric cancer cell invasion by upregulating the expression of MMP9. Mol Med Rep. 2016;14:3261-3268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Li PD, Chen P, Peng X, Ma C, Zhang WJ, Dai XF. HOXC6 predicts invasion and poor survival in hepatocellular carcinoma by driving epithelial-mesenchymal transition. Aging (Albany NY). 2018;10:115-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 10. | Tang L, Cao Y, Song X, Wang X, Li Y, Yu M, Li M, Liu X, Huang F, Chen F, Wan H. HOXC6 promotes migration, invasion and proliferation of esophageal squamous cell carcinoma cells via modulating expression of genes involved in malignant phenotypes. PeerJ. 2019;7:e6607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Zhang W, Chen XS, Wei Y, Wang XM, Chen XJ, Chi BT, Huang LQ, He RQ, Huang ZG, Li Q, Chen G, He J, Wu M. Overexpressed KCNK1 regulates potassium channels affecting molecular mechanisms and biological pathways in bladder cancer. Eur J Med Res. 2024;29:257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Fan Q, Wu GB, Chen M, Zheng L, Li HJ, Xiang LZ, Luo M. Analysis of disulfidptosis- and cuproptosis-related LncRNAs in modulating the immune microenvironment and chemosensitivity in colon adenocarcinoma. IET Syst Biol. 2024;18:55-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 13. | Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen CH, Brown M, Zhang X, Meyer CA, Liu XS. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47:D729-D735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 513] [Cited by in RCA: 574] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 14. | Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 291] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 15. | Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7078] [Article Influence: 884.8] [Reference Citation Analysis (0)] |
| 16. | Malvi P, Chava S, Cai G, Hu K, Zhu LJ, Edwards YJK, Green MR, Gupta R, Wajapeyee N. HOXC6 drives a therapeutically targetable pancreatic cancer growth and metastasis pathway by regulating MSK1 and PPP2R2B. Cell Rep Med. 2023;4:101285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 17. | Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA, Moreno CS. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 18. | Hamid AR, Hoogland AM, Smit F, Jannink S, van Rijt-van de Westerlo C, Jansen CF, van Leenders GJ, Verhaegh GW, Schalken JA. The role of HOXC6 in prostate cancer development. Prostate. 2015;75:1868-1876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | You X, Zhou Z, Chen W, Wei X, Zhou H, Luo W. MicroRNA-495 confers inhibitory effects on cancer stem cells in oral squamous cell carcinoma through the HOXC6-mediated TGF-β signaling pathway. Stem Cell Res Ther. 2020;11:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Jung J, Jeong S, Jeong H, Oh HE, Choi JW, Lee ES, Kim YS, Kwak Y, Kim WH, Lee JH. Increased HOXC6 mRNA expression is a novel biomarker of gastric cancer. PLoS One. 2020;15:e0236811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Yan TF, Wu MJ, Xiao B, Hu Q, Fan YH, Zhu XG. Knockdown of HOXC6 inhibits glioma cell proliferation and induces cell cycle arrest by targeting WIF-1 in vitro and vivo. Pathol Res Pract. 2018;214:1818-1824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Chen L, Sun DZ, Fu YG, Yang PZ, Lv HQ, Gao Y, Zhang XY. Upregulation of microRNA-141 suppresses epithelial-mesenchymal transition and lymph node metastasis in laryngeal cancer through HOXC6-dependent TGF-β signaling pathway. Cell Signal. 2020;66:109444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |