Published online Mar 24, 2025. doi: 10.5306/wjco.v16.i3.101725
Revised: November 4, 2024
Accepted: December 6, 2024
Published online: March 24, 2025
Processing time: 118 Days and 19.4 Hours
Colorectal cancer (CRC) is a prevalent malignant tumor characterized by a high mortality rate, with significant challenges persisting in the identification and management of its metastatic stage. The role of checkpoint kinase 1 (CHEK1), a cell cycle checkpoint kinase, in CRC has not been fully clarified. We hypothesize that the upregulation of CHEK1 may enhance the proliferation of CRC cells, indicating its potential as a novel therapeutic target for CRC therapy.
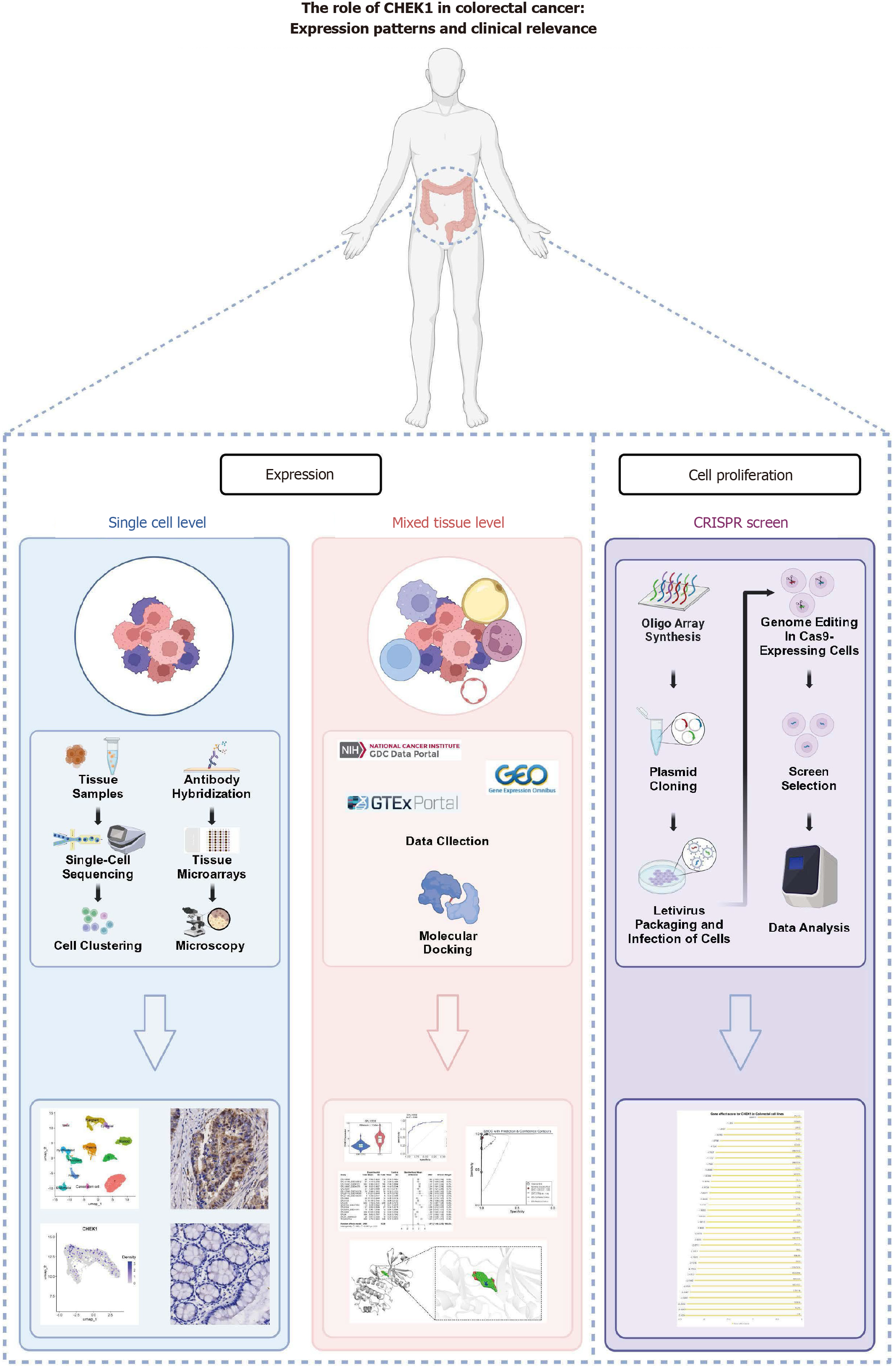
To investigate the expression and function of CHEK1 in CRC, this study utilizes single-cell RNA sequencing and tissue microarray data.
Single-cell RNA sequencing technology was employed to analyze CRC cells from the GSE144735 dataset, and immunohistochemistry was conducted to confirm the expression of CHEK1 in CRC and adjacent tissues. We also integrated mRNA expression data from multiple public databases to assess global CHEK1 expre
We found comparatively elevated CHEK1 expression in the malignant epithelial cells of CRC, with marked upregulation of its mRNA levels in CRC tissues. Immunohistochemical analysis further confirmed the high expression of CHEK1 in CRC tissues, and the receiver operating characteristic curve demonstrated high accuracy (area under the curve = 0.964) for CHEK1 as a biomarker. Analysis of global datasets indicated a statistically significant overexpression of CHEK1 in CRC (standard mean difference = 1.81, P < 0.01), with summary receiver operating characteristic analysis yielding sensitivity and specificity values of 0.83 and 0.88, respectively. Molecular docking studies indicated that NC specifically targeted CHEK1, while clustered regularly interspaced short palindromic repeats knockout experiments demonstrated that CHEK1 promoted CRC cell proliferation.
Upregulation of CHEK1 promotes CRC cell proliferation. However, the dataset's diversity is limited, requiring further investigation into its specific mechanisms.
Core Tip: This study demonstrates that checkpoint kinase 1 (CHEK1) is comparatively overexpressed in colorectal cancer (CRC) and is closely associated with tumor proliferation. Based on single-cell RNA sequencing and immunohistochemistry, the findings highlight CHEK1’s potential as a novel biomarker and therapeutic target in CRC. Furthermore, the research uncovers nitidine chloride as a specific CHEK1 inhibitor, suggesting promising avenues for targeted therapies to improve patient outcomes. These insights underscore the clinical relevance of CHEK1 in CRC management and the need for further exploration of its mechanistic roles.
- Citation: Pang YY, Chen ZY, Zeng DT, Li DM, Li Q, Huang WY, Li B, Luo JY, Chi BT, Huang Q, Feng ZB, He RQ. Checkpoint kinase 1 in colorectal cancer: Upregulation of expression and promotion of cell proliferation. World J Clin Oncol 2025; 16(3): 101725
- URL: https://www.wjgnet.com/2218-4333/full/v16/i3/101725.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i3.101725
Colorectal cancer (CRC) is the second most significant contributor to cancer-related deaths and is the third most commonly diagnosed malignancy around the world[1]. In many countries, there is an increasing incidence of CRC among younger individuals, with the risk of developing the disease rising with age. In an ageing society, the threat posed by CRC to public health is steadily increasing[2]. Although early-stage CRC is preventable and treatable, the diagnostic efficacy for metastatic CRC is low during the metastatic phase, and current treatment options remain limited. Immunotherapy and targeted therapies show certain limitations in the treatment of CRC, as only a subset of eligible patients exhibit favorable responses[3]. Approximately 50% of patients with CRC develop liver metastases, resulting in a poor prognosis. The immune tolerance characteristics of the liver contribute to the establishment of an immunosuppressive tumor microenvironment, which underscores the necessity for in-depth research to inform treatment strategies, particularly in the context of targeted interventions that combine therapeutic targets[4].
CRC has emerged as a significant global health challenge in recent years, with its complex pathogenesis compelling researchers to seek novel therapeutic targets. Suppressor of cytokine signaling (SOCS) has been shown to be a critical negative regulatory factor that plays an essential role in the progression of CRC[5]. The absence of SOCS can lead to the triggering of pro-cancer pathways, offering new insights into the biological processes underlying CRC. Matrix metalloproteinase (MMP)-2 and MMP-9 play critical roles in the extracellular matrix of CRC. Elevated levels of MMP-2 ex
Checkpoint kinase 1 (CHEK1), a class of serine/threonine kinases involved in cell cycle checkpoints, include two subtypes: CHEK1 and CHK2. CHEK1, a serine/threonine protein kinase, plays a critical role at the S-phase checkpoint by preventing the activation of late replication origins when DNA synthesis is inhibited and maintaining the stability of replication forks[7]. As a protein-coding gene, CHEK1 is related with fertilized egg or embryo maturation stasis, and maternal heterozygous mutations in CHEK1 can result in mitotic arrest of human fertilized eggs[8]. Research on resistance to poly (ADP-ribose) polymerase inhibitors (PARPi) and platinum-based drugs in high-grade serous ovarian cancer has identified an interaction between homologous recombination deficiency and CHEK1. This finding offers a potential direction for targeted therapeutic strategies that incorporate CHEK1 inhibitors, which may substantially influence the treatment of various tumor subtypes[9]. Bioinformatics analyses have revealed a strong association between CHEK1 and breast cancer, particularly the luminal A subtype[10].
Furthermore, dysregulation of CHEK1 has been observed in multiple immune cell types, correlating considerably with tumor infiltration, suggesting its research potential in breast cancer progression[11]. Although adult mammals possess limited cardiac regenerative capabilities, CHEK1 has demonstrated significant potential in promoting cardiomyocyte survival and proliferation. Studies indicate that recombinant human CHEK1 facilitates metabolic reprogramming through the activation of pyruvate kinase M2, thereby enhancing cardiac repair. This finding presents a novel target for the treatment of cardiac diseases, especially in the context of ischemia/reperfusion injury[12]. Motor dysfunction resulting from spinal cord injury is closely linked to the pathological mechanisms of DNA damage. Research shows that CHEK1 is crucial not only for the DNA damage response DNA damage but may also act as a novel regulatory factor in the pathology of spinal cord injuries. Targeting CHEK1 could offer a new approach to treating such neurotraumatic conditions[13]. Moreover, CHEK1 is crucial in modulating the DNA damage response and is intricately linked to the pathological mechanisms underlying acute lung injury. Recent investigations have shown that Apelin-13 can alleviate lipopolysaccharide-induced acute lung injury while modulating DNA damage and repairing the damaged pulmonary epithelial barrier through the induction of autophagic degradation of CHEK1. This suggests that CHEK1 may serve as a potential therapeutic target for acute lung injury[14].
In CRC research, CHEK1 has been implicated in targeted therapies for p53-deficient, DNA replication-stressed, and hyperdiploid CRC stem cells; however, the expression of CHEK1 in CRC and its impact on cell proliferation remain inadequately explored, and predictive insights for CHEK1-targeted drugs are lacking[15]. The expression levels and alterations of CHEK1 in CRC have been confirmed through western blot and ELISA analyses; however, validation at the single-cell level has yet to be conducted[16]. Although there has been varying research progress regarding CHEK1 in different cancer types, studies specifically addressing its role in CRC remain insufficient, indicating its potential research value.
This study seeks to thoroughly investigate the expression profile of CHEK1 in CRC by synthesizing single-cell RNA sequencing data and tissue cohort gene expression analyses. This approach initially revealed high expression levels at the single-cell level, followed by validation through immunohistochemical techniques. The potential impact of mesenchymal components of the tumor microenvironment, such as fibrous tissue, blood vessels, adipose, and immune cells, on CHEK1 expression was considered. Our integrated tissue hybrid gene chip and RNA sequencing data further support the observed overexpression of CHEK1 and provide new insights and strategies for its application as a clinical biomarker.
To explore the expression and biological roles of CHEK1 at the cellular level in CRC, a single-cell RNA sequencing (scRNA-seq) analysis was conducted using data from the GSE144735 dataset, which included 19653 cells. Quality control measures filtered the original scRNA data by selecting cells with number of RNA features greater than 200 and less than 2500, as well as a percentage of mitochondrial (percent.MT) RNA less than 5%. Following global normalization using the Log Normalize method, the top 10 most variable genes were identified through variance stabilizing transformation. Dimensionality reduction of the scRNA-seq data was performed using uniform manifold approximation and projection, with manual annotation of marker gene expression conducted through the CellMarker database[17].
The expression of CHEK1 at the cellular level in CRC was validated using clinical samples by obtaining tissue specimens from patients at Yulin Red Cross Hospital. A total of 208 samples were collected, comprising CRC tissues and adjacent noncancerous tissues. Twelve representative tissue microarrays were constructed, allowing for the scoring of CHEK1 expression. Of the CHEK1 expression values, 208 belonged to CRC tissues, and 208 points belonged to the corresponding adjacent noncancerous tissues. Tissue processing and immunohistochemistry were performed according to the manufacturer’s protocols, and all processes and results were verified by pathologists. The assessment involved randomly counting 100 positive cells in each field at the highest magnification. Scoring ranges were assigned according to the number of positively stained cells: A score of 0 for 0–5 cells, 1 for 6–15 cells, 2 for 16–50 cells, 3 for 51–75 cells and 4 for more than 75 cells. The staining intensity was evaluated on a scale of 0 to 3, representing no staining, weak staining, moderate staining, and strong staining, respectively. For each tissue sample, the analysis was performed on 10 randomly selected fields to derive the immunohistochemistry score using the following formula: (Σ positive cell percentage score × staining intensity score)/10. The expression level of CHEK1 mRNA in CRC was visualized through violin plots and receiver operating characteristic (ROC) curves.
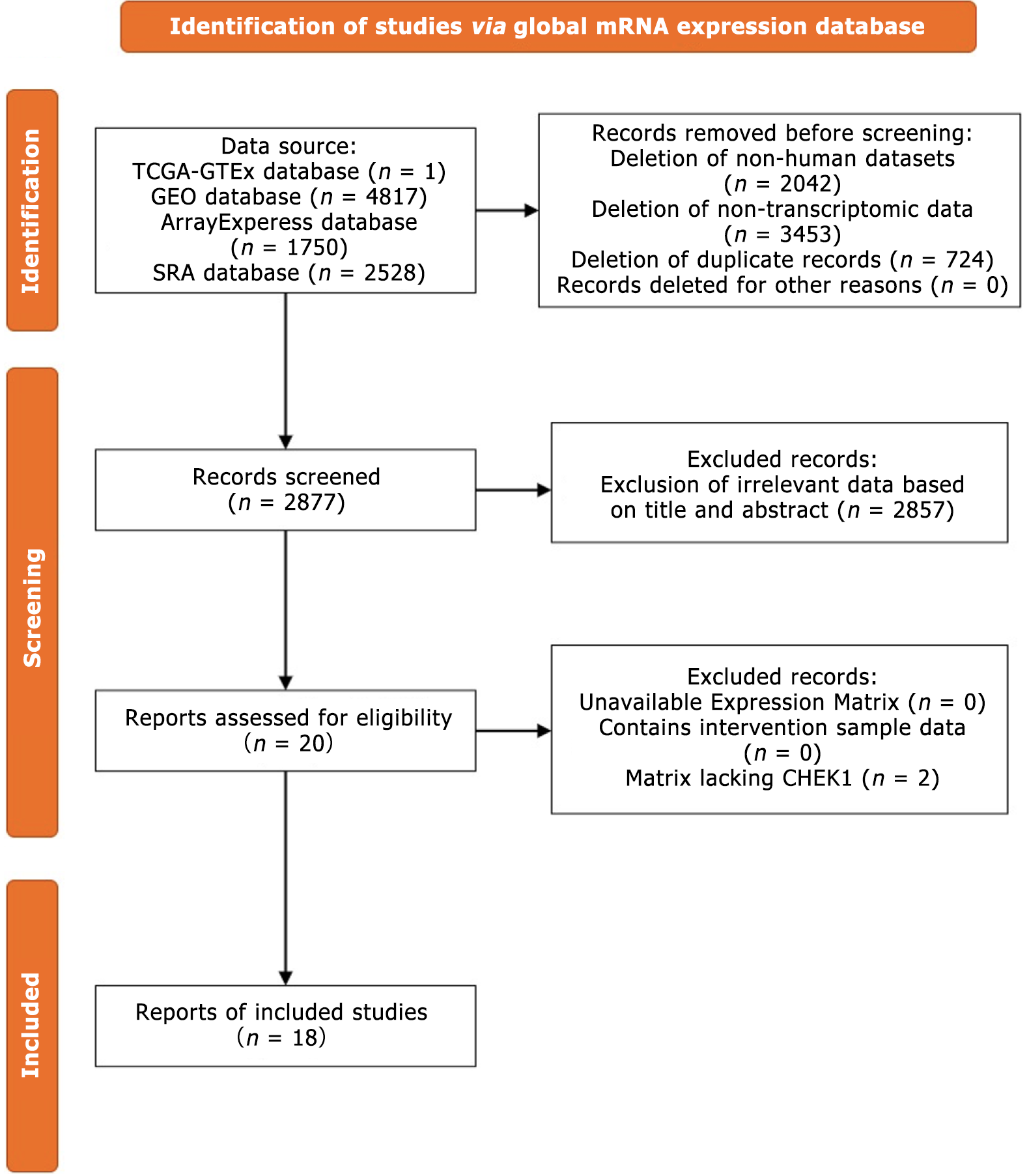
The mRNA expression datasets from CRC and noncancerous intestinal tissues were collected from public high-throughput databases, including Gene Expression Omnibus, Array Express, Sequence Read Archive, the Cancer Genome Atlas, and Genotype-Tissue Expression. The inclusion criteria included: (1) Datasets sourced from human tissues or cells; and (2) Datasets containing both experimental and control groups, including no fewer than three samples in each group. The exclusion criteria were: (1) Duplicate datasets; (2) Nontranscriptomic datasets; and (3) Datasets lacking an expression matrix. The collected datasets underwent log transformation, and batch effects were eliminated using the limma and sva packages after merging the platform matrices.
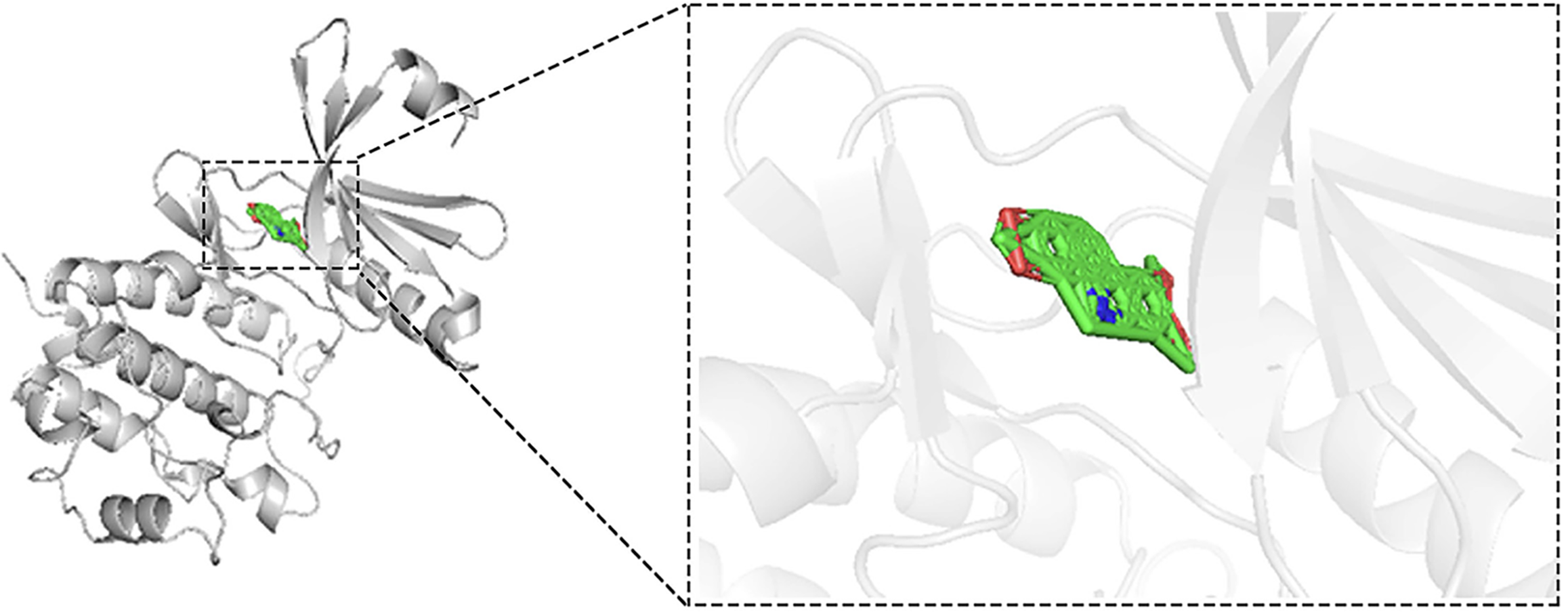
The crystal structure of CHEK1 was obtained from the research collaboratory for structural bioinformatics protein data bank, with subsequent removal of water molecules, metal bonds, and co-crystallized ligands accomplished using PyMOL version 2.4. The structure of nitidine chloride (NC) was retrieved from the PubChem database. Auto Dock Tools was employed to process the structures of CHEK1 and NC for molecular docking analysis. Finally, molecular docking experiments were conducted using Auto Dock Vina 1.5.7, and the results were visualized with PyMOL 2.4.
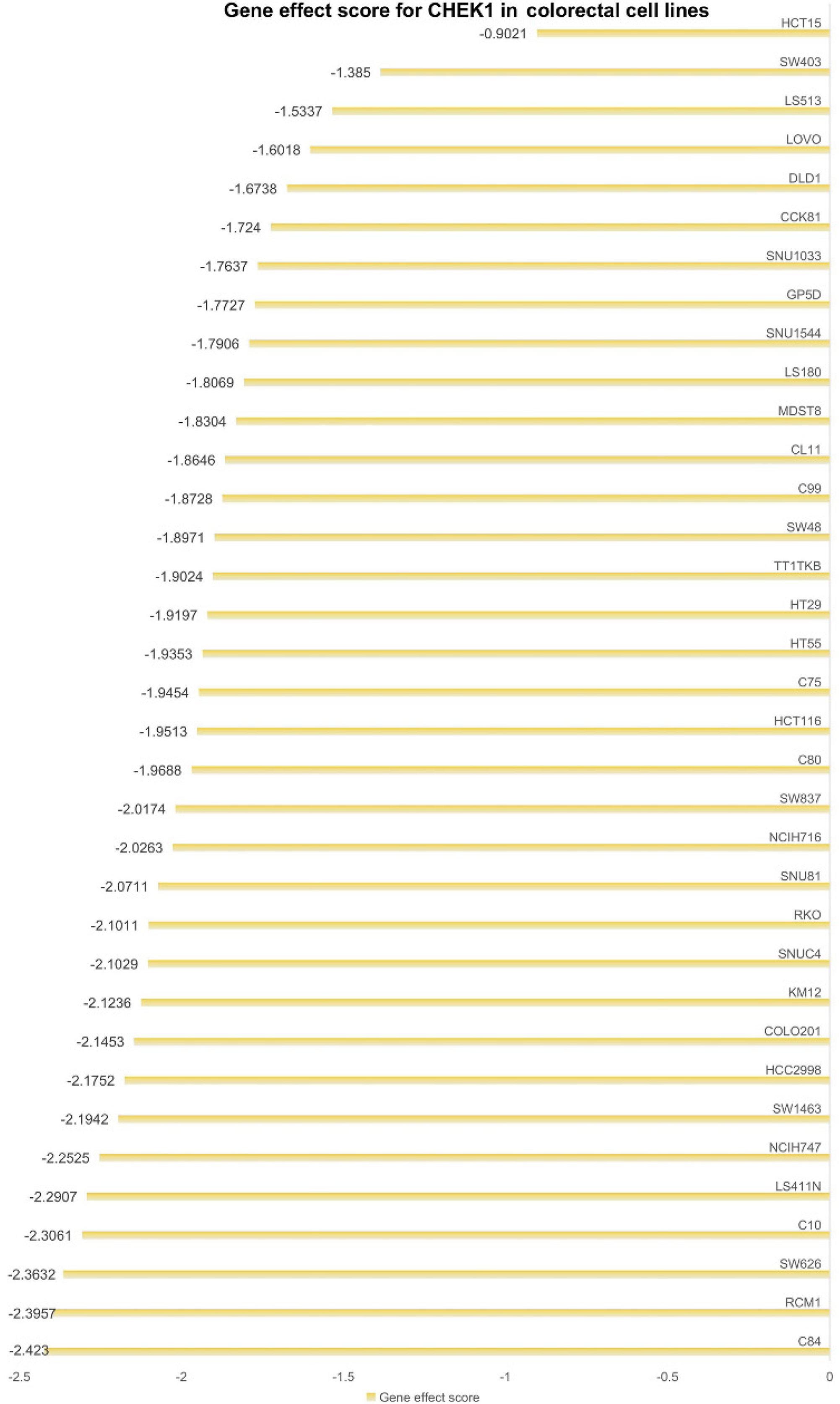
To clarify the role of CHEK1 in CRC cell proliferation, a clustered regularly interspaced short palindromic repeats (CRISPR) knockout screening technique was utilized to disrupt the CHEK1 gene in CRC cell lines. CERES is a computational method for CRISPR-Cas9 screening in cancer cell lines to accurately estimate gene dependency and reduce false-positive results[18]. The impact of CHEK1 knockout was assessed using the CERES algorithm to calculate gene effect scores, determining the inhibitory effect on CRC cell proliferation upon CHEK1 gene knockout. A negative score indicates impaired proliferation following CHEK1 knockout.
Statistical analyses were performed using R version 4.3.2 and Stata version 16. Differences in CHEK1 mRNA expression levels between CRC and control groups were evaluated using the Wilcoxon test. In the calculation of the standard mean difference (SMD) using Stata 16.0, a random-effects model was utilized when I2 exceeded 50% or P values were less than 0.05; otherwise, a fixed-effects model was applied for the integration of CHEK1 mRNA expression data. Publication bias in the SMD model was assessed using Begg’s and Egger’s tests. Summary receiver operating characteristic (SROC) curves were constructed to further verify expression differences of CHEK1 mRNA across various groups, with calculations of sensitivity, specificity, negative likelihood ratio, and positive likelihood ratio.
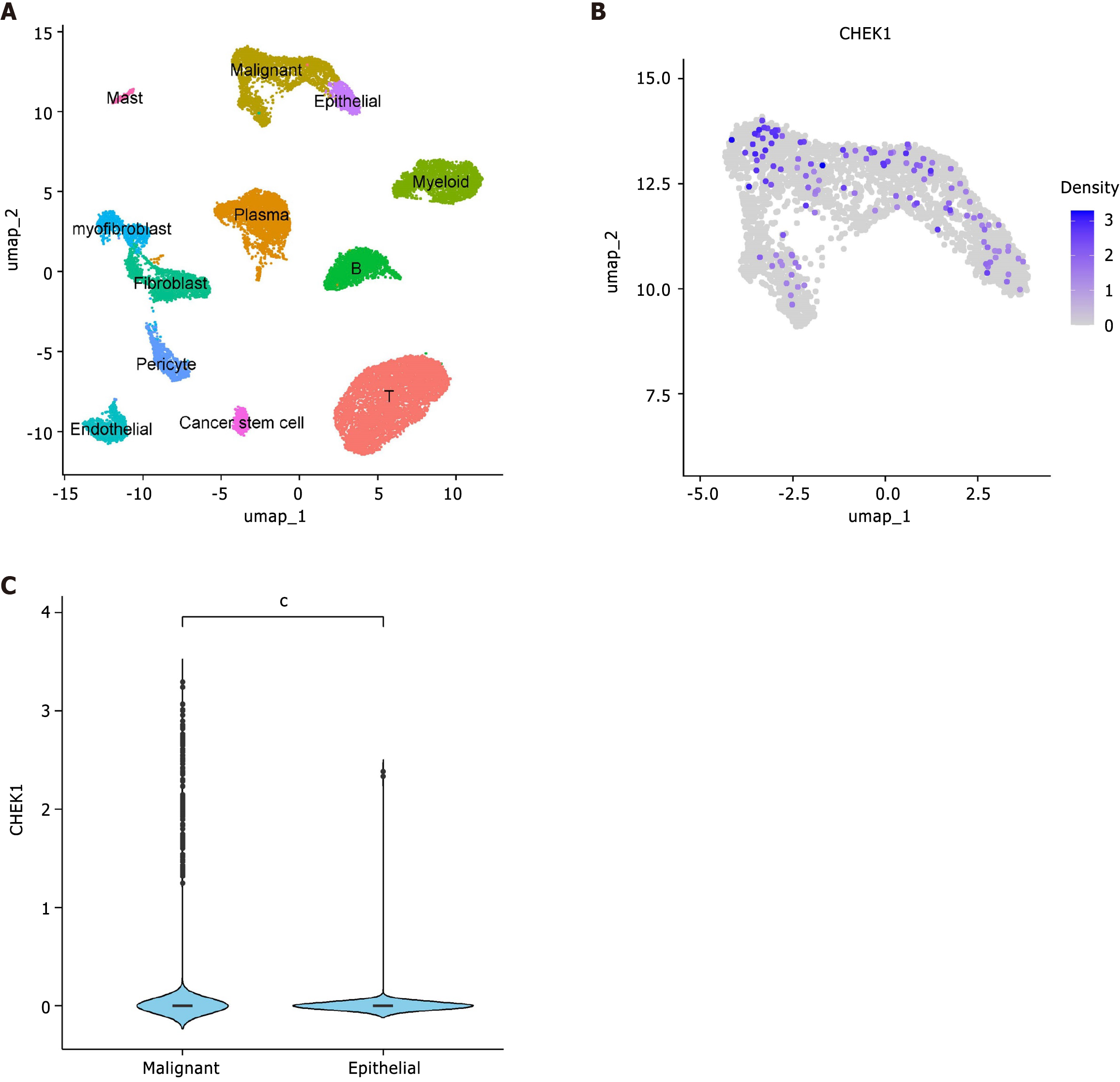
Upregulation of CHEK1 in CRC through single-cell analysis: The main structure of the article is shown in the flowchart (Figure 1). The workflow for the incorporation and filtration of single-cell sequencing data is presented in Supplementary Figure 1. After comprehensive data filtering and preprocessing, 12 distinct cell types were annotated, including mast cells, malignant epithelial cells, normal epithelial cells, myeloid cells, myofibroblasts, fibroblasts, plasma cells, B cells, pericytes, endothelial cells, cancer stem cells, and T cells (Figure 2A). Further evaluation of CHEK1 expression within epithelial cells revealed a comparatively higher expression level in malignant epithelial cells compared to normal epithelial cells, indicating an upregulation of CHEK1 in CRC with statistical significance (P < 0.05) (Figure 2B and C).
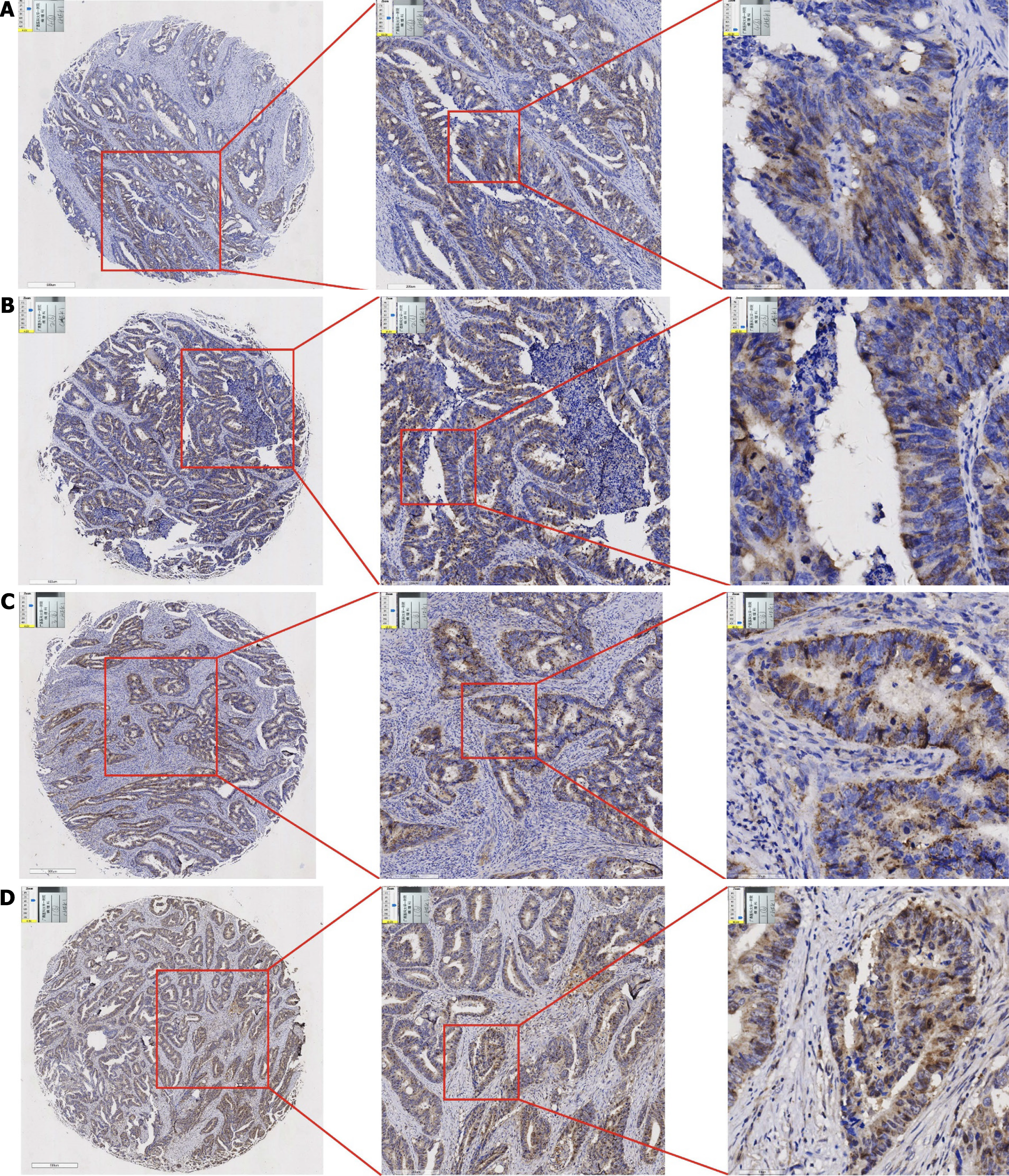
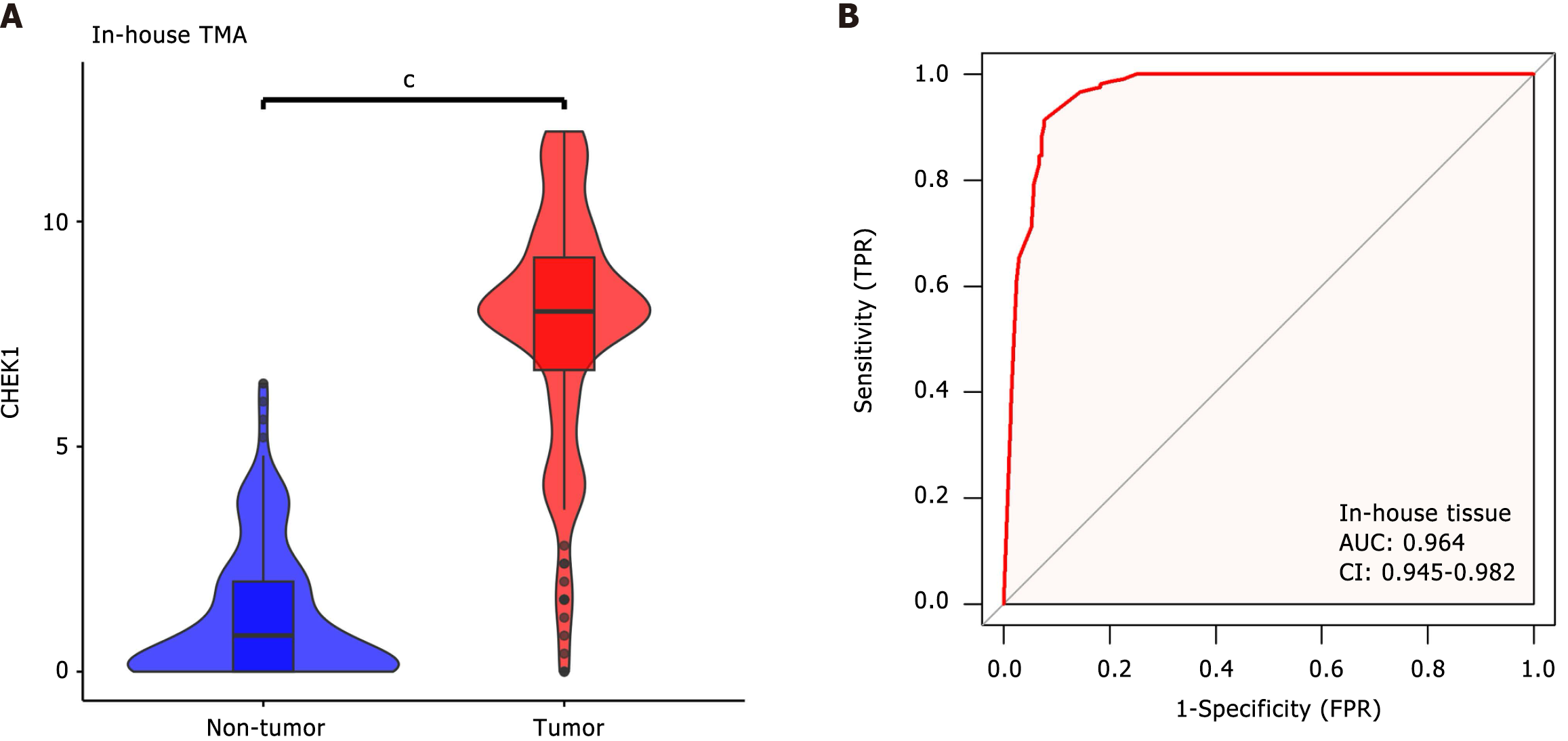
Comparison of CHEK1 protein expression in CRC and non-cancerous adjacent tissues: Immunohistochemistry was utilized to investigate the expression levels of CHEK1 at the protein level in non-cancerous adjacent tissues compared to CRC. Analysis of four pairs of clinical samples revealed that CHEK1 protein staining (Figure 3) in non-cancerous tissues was markedly lighter than that in the corresponding CRC tissues (Figure 4). Furthermore, evaluation with an internal tissue microarray indicated the overexpression of CHEK1 in CRC compared to non-cancerous adjacent tissues, with statistical significance (P < 0.001) (Figure 5A). The ROC analysis resulted in an area under the curve (AUC) of 0.964 (95%CI: 0.945–0.982) (Figure 5B).
Integration and assessment of global microarray and sequencing datasets: The flowchart for the selection of CHEK1 mRNA data from global datasets is illustrated in Figure 6. A total of 18 datasets were curated, encompassing 2498 CRC cases and 1330 non-CRC cases. The inclusion of a substantial number of study samples provides a robust basis for evaluating CHEK1 expression in CRC.
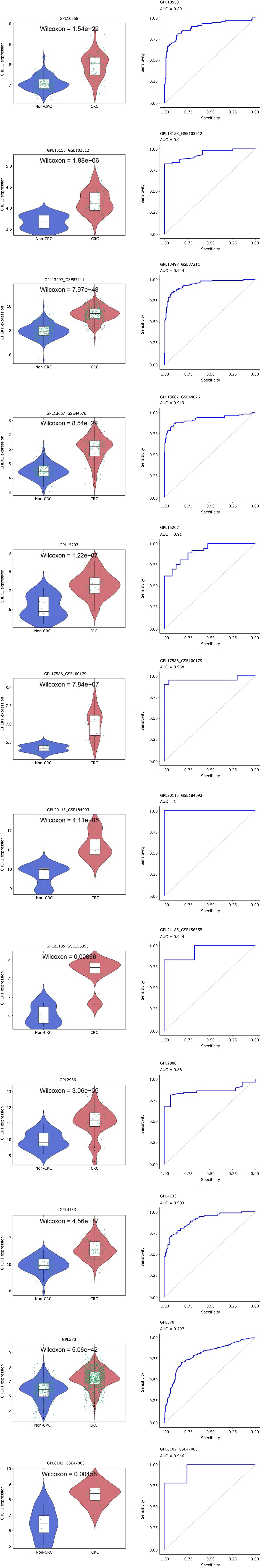
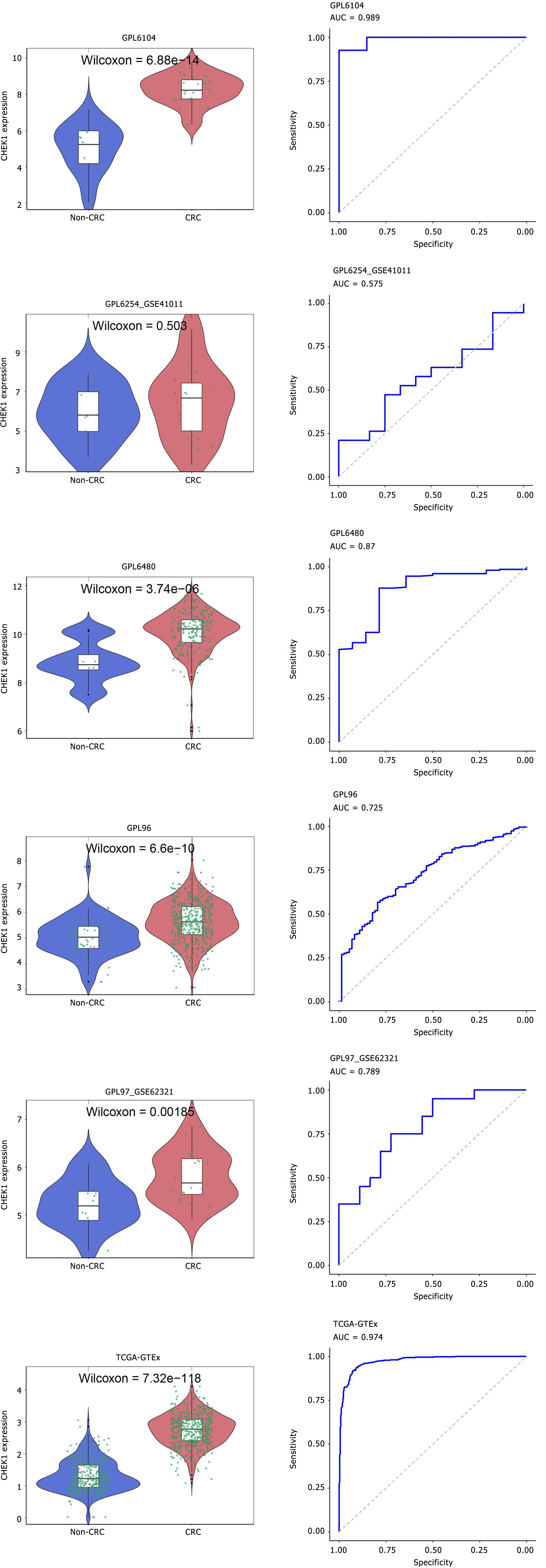
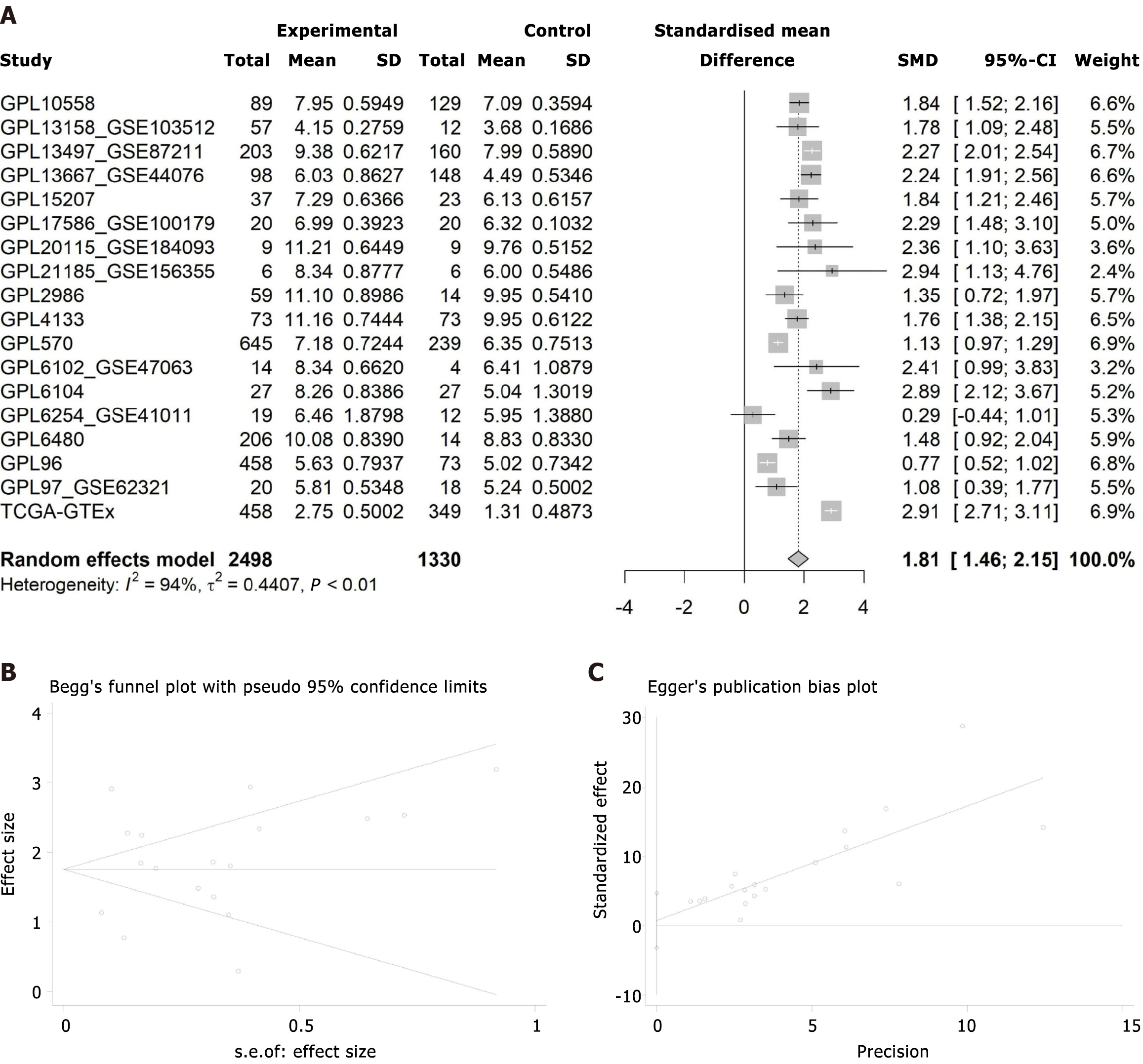
Overexpression of CHEK1 mRNA in CRC: To compare the expression levels of CHEK1 in CRC and non-CRC samples, the Wilcoxon test was employed across multiple datasets. The results indicated that CHEK1 mRNA is overexpressed in CRC cases across 17 platforms, with statistical significance (P < 0.05, AUC ≥ 0.7) as shown in Figure 7 and Figure 8. Comprehensive analysis using a random-effects model revealed a significant overexpression of CHEK1 mRNA in CRC, with a SMD of 1.81, a 95%CI: 1.46-2.15, and I2 = 94%, P < 0.01 (Figure 9A). Furthermore, the results from the Begg and Egger tests indicated the absence of publication bias (P = 0.705 and 0.703) (Figure 9B and C).
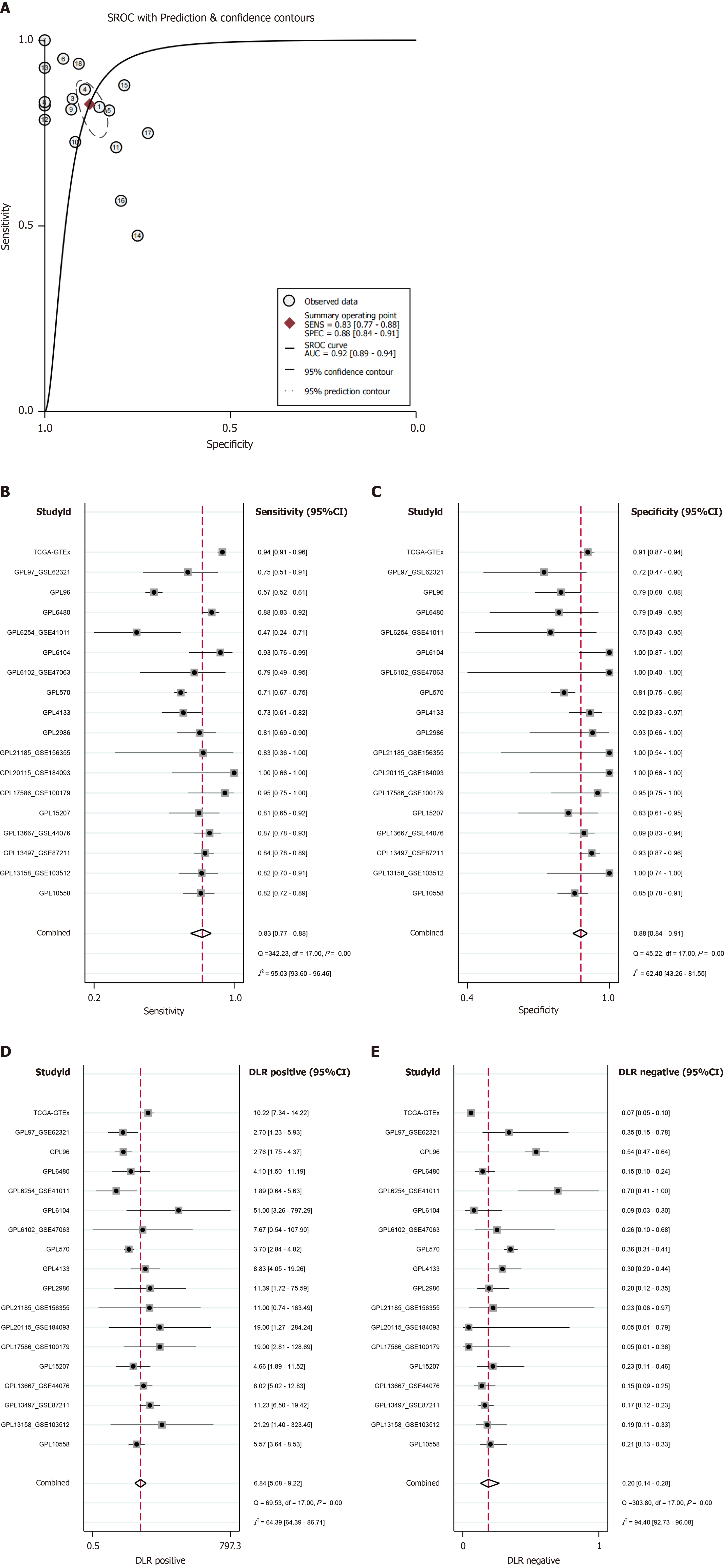
Quantitative characteristics of the relationship between CHEK1 and CRC: The differences in CHEK1 expression between CRC and non-CRC tissues were validated using SROC, which also assessed the quantitative characteristics across different groups. The AUC was found to be 0.92 (95%CI: 0.89–0.94) (Figure 10A), with sensitivity and specificity reported at 0.83 (95%CI: 0.77–0.88) and 0.88 (95%CI: 0.84–0.91), respectively (Figure 10B and C). The positive diagnostic likelihood ratio (DLR) was calculated as 6.84 (95%CI: 5.08–9.22, I2 = 64.39%), while the negative DLR was 0.20 (95%CI: 0.14–0.28, I2 = 94.40%) (Figure 10D and E).
NC specifically targets CHEK1 in CRC: Through molecular docking experiments, we investigated the interaction between NC and CHEK1. The findings suggest that NC can act as a specific inhibitor of CHEK1, with an optimal binding energy of -11.8 kcal/mol. Visualization confirmed the absence of hydrogen bonds between NC and CHEK1 (Figure 11).
Utilizing CRISPR knockout screening technology, we examined the impact of CHEK1 on the proliferation of CRC cell lines. The results demonstrated that following the knockout of the CHEK1 gene, the gene effect scores for 33 CRC cell lines were below 0 (Figure 12), resulting in a reduced rate of cell proliferation. This suggests that CHEK1 plays a role in promoting CRC cell proliferation.
CRC has emerged as one of the leading cancers globally, with increasing incidence and mortality rates[19]. Although early-stage CRC demonstrates high preventability and curability, the diagnosis and treatment of metastatic CRC continue to pose significant challenges. Current therapeutic approaches, including immunotherapy and targeted therapy, have shown limited efficacy in most patients[20]. Consequently, there is an urgent need to discover novel biomarkers and therapeutic targets aimed at improving patient prognoses in CRC. The integration of single-cell RNA sequencing and tissue microarray gene chip technologies in recent years has provided new insights into cancer research, particularly in exploring the tumor microenvironment and transcriptomic profiles.
In CRC research, CHEK1 has gradually gained recognition as a crucial cell cycle checkpoint kinase, with roles identified across various cancer types. Its critical role in the DNA damage response and the regulation of the cell cycle positions it as a potential therapeutic target[21]. Previous studies have indicated that aberrant CHEK1 expression in cancers such as breast cancer[22] and nonsmall cell lung cancer[23], correlates with disease progression. However, research on CHEK1 in CRC remains insufficient, particularly regarding its expression characteristics at both the single-cell and tissue levels.
This study systematically evaluated CHEK1 expression in CRC by integrating single-cell RNA sequencing data and tissue microarray gene analysis. The single-cell RNA sequencing results demonstrated that CHEK1 expression was comparatively higher in the malignant epithelial cells of CRC than in normal epithelial cells. This finding aligns with the established role of CHEK1 in tumor progression. Immunohistochemical analysis further substantiated this result, revealing remarkably elevated CHEK1 expression in CRC tissue compared to adjacent noncancerous tissues. These findings suggest that increased CHEK1 expression may be closely associated with CRC occurrence and development, implying its potential as a diagnostic biomarker.
In the integrated analysis of global expression datasets, a consistent elevation of CHEK1 in CRC tissues was observed, corroborating previous reports of CHEK1 overexpression in solid tumors[24]. Notably, in studies across 17 platforms, the differential expression of CHEK1 mRNA in CRC was found to be statistically significant. This outcome further supports the potential of CHEK1 as a biomarker for CRC. SROC analysis indicated that CHEK1 expression levels exhibited good sensitivity and specificity for distinguishing CRC, reinforcing its validity as a biomarker. Because of its richness in polyphenols, aronia berry extracts demonstrated significant anticancer effects in a variety of malignant tumors. Some researchers treated CRC cells and observed downregulation of CHEK1, which was significantly associated with cell cycle arrest and enhanced DNA damage[25]. This suggests that the upregulation of CHEK1 in CRC may be a response mechanism of tumor cells to cell cycle regulation and DNA damage. Combined with the anticancer activity of aronia berry extracts, inhibition of CHEK1 function may be a potential strategy to improve the therapeutic efficacy of CRC.
In the context of mitigating CRC cell proliferation, NC was shown to inhibit cell proliferation and induce apoptosis by blocking the extracellular signal-regulated kinase signaling pathway[26]. Moreover, our molecular docking studies suggested that NC could specifically target CHEK1 with a favorable binding affinity. This opens new avenues for targeted therapies against CHEK1 and lays the groundwork for the future development of anti-cancer drugs targeting CHEK1. Additionally, CRISPR knockout experiments further confirmed the role of CHEK1 in promoting CRC cell proliferation, underscoring its potential as a therapeutic target in CRC. CHEK1 regulates cell cycle and DNA damage response mechanisms and plays a key role in the Ataxia telangiectasia and Rad3-related (ATR) signaling pathway; studies have shown that the ATR inhibitor AZD6738 can be used in combination with trifluridine to efficiently induce cell cycle arrest at the G2/M checkpoint, as well as CHEK1 phosphorylation and enhanced apoptosis[27]. This finding, together with the results found in this study using CRISPR knockdown screening, reflects the importance of CHEK1 in CRC cell proliferation.
This study systematically explored CHEK1’s expression characteristics and clinical implications in CRC by combining single-cell RNA sequencing and tissue chip technologies. The high expression of CHEK1 in CRC is intimately linked to the initiation and progression of tumor cells, holding significant diagnostic and therapeutic value. Future investigations should delve into the specific mechanistic roles of CHEK1 in CRC and assess its clinical efficacy as a therapeutic target. Furthermore, the development of additional specific drugs targeting CHEK1 and subsequent clinical trials to validate their effectiveness in CRC treatment are needed. Such efforts will contribute to improving early diagnosis and therapeutic outcomes for CRC patients, ultimately leading to effective treatment strategies.
CHEK1 is comparatively overexpressed in CRC and is associated with cell proliferation in CRC, highlighting its potential as a biomarker for CRC. However, the limitations of this study include its reliance on specific datasets, which may not encompass the full spectrum of CRC heterogeneity, and the need for further in vivo validation to confirm these findings. Future research should focus on elucidating the precise mechanisms by which CHEK1 influences CRC progression and on exploring the therapeutic significance of targeting CHEK1 to develop effective treatment strategies for CRC patients.
| 1. | Zeng W, Liu H, Mao Y, Jiang S, Yi H, Zhang Z, Wang M, Zong Z. Myeloidderived suppressor cells: Key immunosuppressive regulators and therapeutic targets in colorectal cancer (Review). Int J Oncol. 2024;65. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Klimeck L, Heisser T, Hoffmeister M, Brenner H. Colorectal cancer: A health and economic problem. Best Pract Res Clin Gastroenterol. 2023;66:101839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 65] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 3. | Andrei P, Battuello P, Grasso G, Rovera E, Tesio N, Bardelli A. Integrated approaches for precision oncology in colorectal cancer: The more you know, the better. Semin Cancer Biol. 2022;84:199-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 4. | Kong WS, Li JJ, Deng YQ, Ju HQ, Xu RH. Immunomodulatory molecules in colorectal cancer liver metastasis. Cancer Lett. 2024;598:217113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 5. | Wang Y, Wu S, Song Z, Yang Y, Li Y, Li J. Unveiling the pathological functions of SOCS in colorectal cancer: Current concepts and future perspectives. Pathol Res Pract. 2024;262:155564. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 6. | Wang Y, Wei Y, Huang J, Li X, You D, Wang L, Ma X. Prognostic value of matrix metalloproteinase-2 protein and matrix metalloproteinase-9 protein in colorectal cancer: a meta-analysis. BMC Cancer. 2024;24:1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 7. | Ikumawoyi VO, Lynch KD, Iverson DT, Call MR, Yue GE, Prasad B, Clarke JD. Microcystin-LR activates serine/threonine kinases and alters the phosphoproteome in human HepaRG cells. Toxicon. 2024;249:108072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Chen B, Guo J, Wang T, Lee Q, Ming J, Ding F, Li H, Zhang Z, Li L, Cao Y, Na J. Maternal heterozygous mutation in CHEK1 leads to mitotic arrest in human zygotes. Protein Cell. 2022;13:148-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Xu H, Gitto SB, Ho GY, Medvedev S, Shield-Artin K, Kim H, Beard S, Kinose Y, Wang X, Barker HE, Ratnayake G, Hwang WT; Australian Ovarian Cancer Study, Hansen RJ, Strouse B, Milutinovic S, Hassig C, Wakefield MJ, Vandenberg CJ, Scott CL, Simpkins F. CHK1 inhibitor SRA737 is active in PARP inhibitor resistant and CCNE1 amplified ovarian cancer. iScience. 2024;27:109978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 10. | Yu D, Liu S, Chen Y, Yang L. Integrative Bioinformatics Analysis Reveals CHEK1 and UBE2C as Luminal A Breast Cancer Subtype Biomarkers. Front Genet. 2022;13:944259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 11. | Li C, Liao J, Wang X, Chen FX, Guo X, Chen X. Combined Aurora Kinase A and CHK1 Inhibition Enhances Radiosensitivity of Triple-Negative Breast Cancer Through Induction of Apoptosis and Mitotic Catastrophe Associated With Excessive DNA Damage. Int J Radiat Oncol Biol Phys. 2023;117:1241-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 12. | Wei TW, Shan TK, Wang H, Chen JW, Yang TT, Zhou LH, Zhao D, Sun JT, Wang SB, Gu LF, Du C, Jiang QQ, Sun R, Wang QM, Kong XQ, Lu XH, Sun HL, Xu Y, Xie LP, Gu AH, Chen F, Ji Y, Guo XJ, Wang LS. Checkpoint Kinase 1 Stimulates Endogenous Cardiomyocyte Renewal and Cardiac Repair by Binding to Pyruvate Kinase Isoform M2 C-Domain and Activating Cardiac Metabolic Reprogramming in a Porcine Model of Myocardial Ischemia/Reperfusion Injury. J Am Heart Assoc. 2024;13:e034805. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 13. | Fan J, Du X, Chen M, Xu Y, Xu J, Lu L, Zhou S, Kong X, Xu K, Zhang H. Critical role of checkpoint kinase 1 in spinal cord injury-induced motor dysfunction in mice. Int Immunopharmacol. 2024;138:112521. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 14. | Chen S, Zhu H, Lin L, Lu L, Chen L, Zeng L, Yue W, Kong X, Zhang H. Apelin-13 improves pulmonary epithelial barrier function in a mouse model of LPS-induced acute lung injury by inhibiting Chk1-mediated DNA damage. Biochem Pharmacol. 2024;226:116297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 15. | Manic G, Signore M, Sistigu A, Russo G, Corradi F, Siteni S, Musella M, Vitale S, De Angelis ML, Pallocca M, Amoreo CA, Sperati F, Di Franco S, Barresi S, Policicchio E, De Luca G, De Nicola F, Mottolese M, Zeuner A, Fanciulli M, Stassi G, Maugeri-Saccà M, Baiocchi M, Tartaglia M, Vitale I, De Maria R. CHK1-targeted therapy to deplete DNA replication-stressed, p53-deficient, hyperdiploid colorectal cancer stem cells. Gut. 2018;67:903-917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 16. | Stawinska M, Cygankiewicz A, Trzcinski R, Mik M, Dziki A, Krajewska WM. Alterations of Chk1 and Chk2 expression in colon cancer. Int J Colorectal Dis. 2008;23:1243-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Hu C, Li T, Xu Y, Zhang X, Li F, Bai J, Chen J, Jiang W, Yang K, Ou Q, Li X, Wang P, Zhang Y. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023;51:D870-D876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 493] [Reference Citation Analysis (0)] |
| 18. | Chen W, Lin Y, Jiang M, Wang Q, Shu Q. Identification of LARS as an essential gene for osteosarcoma proliferation through large-Scale CRISPR-Cas9 screening database and experimental verification. J Transl Med. 2022;20:355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 2022;7:262-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 379] [Article Influence: 126.3] [Reference Citation Analysis (6)] |
| 20. | Mangas-Sanjuan C, Jover R. Familial colorectal cancer. Best Pract Res Clin Gastroenterol. 2022;58-59:101798. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 21. | McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 22. | Lee S, Jee HY, Lee YG, Shin JI, Jeon YJ, Kim JB, Seo HE, Lee JY, Lee K. PKR-Mediated Phosphorylation of eIF2a and CHK1 Is Associated with Doxorubicin-Mediated Apoptosis in HCC1143 Triple-Negative Breast Cancer Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 23. | Jin M, Zhang F, Li Q, Xu R, Liu Y, Zhang Y. Circ_0011292 knockdown mitigates progression and drug resistance in PTX-resistant non-small-cell lung cancer cells by regulating miR-433-3p/CHEK1 axis. Thorac Cancer. 2022;13:1276-1288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 24. | Fadaka AO, Bakare OO, Sibuyi NRS, Klein A. Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Asahi Y, Xu C, Okuno K, Taketomi A, Goel A. The anticancer effects of Aronia berry extract are mediated by Chk1 and p53 in colorectal cancer. Phytomedicine. 2024;135:156086. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 26. | Zhai H, Hu S, Liu T, Wang F, Wang X, Wu G, Zhang Y, Sui M, Liu H, Jiang L. Nitidine chloride inhibits proliferation and induces apoptosis in colorectal cancer cells by suppressing the ERK signaling pathway. Mol Med Rep. 2016;13:2536-2542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Harata S, Suzuki T, Takahashi H, Hirokawa T, Kato A, Watanabe K, Yanagita T, Ushigome H, Shiga K, Ogawa R, Mitsui A, Kimura M, Matsuo Y, Takiguchi S. AZD6738 promotes the tumor suppressive effects of trifluridine in colorectal cancer cells. Oncol Rep. 2023;49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |