Published online Mar 24, 2025. doi: 10.5306/wjco.v16.i3.100361
Revised: December 23, 2024
Accepted: January 9, 2025
Published online: March 24, 2025
Processing time: 159 Days and 6.6 Hours
This editorial comments on the review by Da Silva et al, published in the World Journal of Clinical Oncology which focuses on the molecular perspectives of lung cancer. With the rapid development of molecular technology, new diagnostic methods are constantly emerging, including liquid biopsy, the identification of gene mutations, and the monitoring biomarkers, thus providing precise in
Core Tip: The development of molecular biology techniques, including liquid biopsy techniques, the identification of gene mutations, and the application of immune checkpoint inhibitors can contribute to early and accurate diagnosis and personalized treatment strategies for lung cancer.
- Citation: Xiong Y, Cheng L, Zhou YJ, Ge WH, Qian M, Yang H. Diagnosis and treatment of lung cancer: A molecular perspective. World J Clin Oncol 2025; 16(3): 100361
- URL: https://www.wjgnet.com/2218-4333/full/v16/i3/100361.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i3.100361
Lung cancer remains the leading global cause of death by cancer[1] and contributes significantly to the global disease burden[2], accounting for 1 in 8 cancer diagnoses and 1 in 5 deaths worldwide. Lung cancer can be categorized into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for 80%–85% of all lung cancers, including adenocarcinoma, squamous lung cancer, large cell carcinoma, and adenosquamous carcinoma[3]. Traditional diagnostic methodology includes radiographic screening, sputum examination, bronchoscopy and lung tissue biopsy. Although radiographic screening is non-invasive, this screening method is associated with low levels of accuracy and sensitivity[4]. The examination of sputum is also associated with low sensitivity and is generally used only as an auxiliary form of diagnosis[5]. Bronchoscopy exhibits only limited specificity for distinguishing adenocarcinoma[6]. Furthermore, bronchoscopy and lung tissue biopsy are both invasive diagnostic tools. Surgical intervention, chemotherapy, and radiotherapy constitute the cornerstone of conventional therapeutics. However, radiotherapy and chemotherapy, as primary interventions for inoperable cancer, are associated with significant adverse effects, including bone marrow suppression and drug resistance, thus resulting in a low survival rate[7].
Consequently, improving the diagnostic efficacy and developing precision therapy for lung cancer remains a major challenge. With the ongoing development of sequencing and multi-omics analysis technologies, molecular biomarkers are frequently being identified and utilized in oncology[8,9]. Da Silva et al[10] reviewed the molecular perspectives of clinical oncology treatment and highlighted the importance of precision medicine by compiling data relating to molecular biomarkers and approaches. NSCLC has successfully adopted and utilized the concept of precision medicine by using molecular biomarkers to reflect tumor status and establish individualized therapy[9]. The editorial focuses on the current applications of emerging molecular technologies, as well as limitations and future prospects.
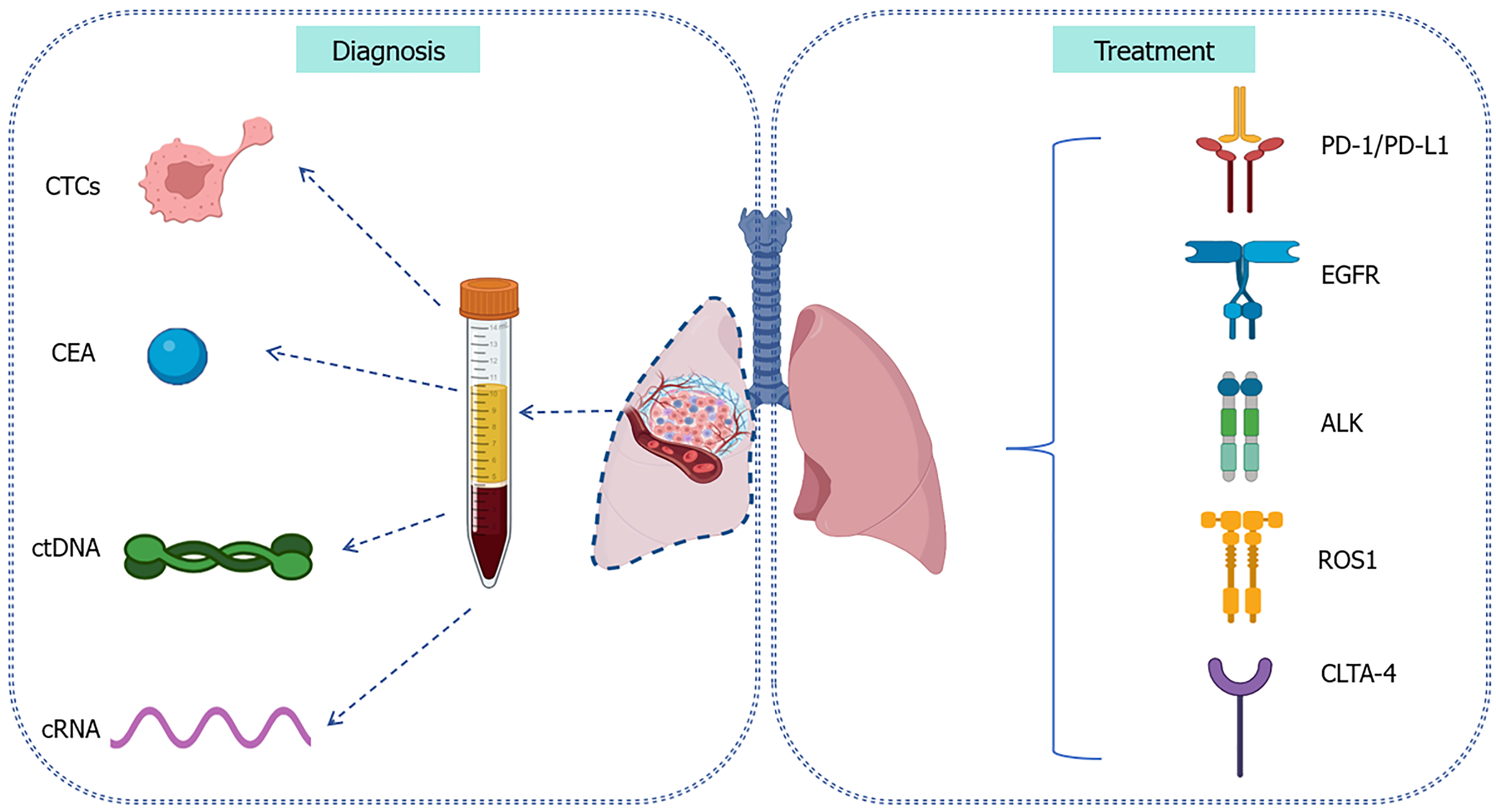
Accurate diagnosis is crucial if we are to delay the progression of disease and provide appropriate treatments for lung cancer during the initial stage. Histopathological biopsy is the definitive standard for diagnosing lung cancer. However, the invasive manner of this technique and associated constraints in facilitating early detection necessitate the identification and exploration of novel diagnostic modalities thereby enabling prompt therapeutic intervention[3]. Biomarkers occupy a critical position in identifying indeterminate pulmonary nodules found during traditional diagnosis and refining the clinical risk of lung cancer[11,12]. Detecting specific molecular biomarkers can minimize harm and cost without increasing the number of cancer deaths, such as carcinoembryonic antigen (CEA), complement fragments, circulating tumor DNA (ctDNA), circulating cell-free DNA (cfDNA) and blood protein profiling[11] (Figure 1).
CEA is one of the genetic cancer antigens found in the serum that is strongly associated with the progression of lung cancer[13]. The detection of CEA allows us to monitor the incidence of lung cancer and assess therapy efficacy[14]. The expression of CEA, cytokeratin19 fragment (CYFRA21-1) and neuron-specific enolase can provide useful information and facilitate treatment options. A previous meta-analysis reported that CEA levels could predict EGFR mutations in patients with NSCLC[15]. Furthermore, CEA is known to contribute to the regulation of fatty acid metabolism in NSCLC, along with induced tumor cell proliferation and migration[16]. Although CEA serves as a biomarker for various malignancies, its specificity for lung cancer is unsatisfactory, thus limiting its clinical utility[17]. Furthermore, the levels of CEA are low in vivo and susceptible to other biomarkers like cfDNA[17]. Thus, it is essential to enhance the detection limit and sensitivity of the equipment used to detect biomarkers.
Liquid biopsy, a non-traumatic diagnostic tool, is easier to acquire from a patient than a biopsy of lung tissue and can reflect the state of the tumor microenvironment in real-time. In addition, liquid biopsy allows for the dynamic monitoring of the response to treatment in patients with NSCLC. Liquid biopsy is predominantly used to analyze circulating tumor cells (CTCs), ctDNA, exosomes, and circulating RNA (cRNA)[18]. In addition, new technologies, such as next-generation sequencing (NGS) can enable the identification of diverse predictive biomarkers and the detection of oncogenic molecular targets with elevated levels of sensitivity and specificity[19]. NGS is more sensitive and specific than immunohistochemistry for the analysis of gene fusion mutations, which was particularly accurate for predicting the clinical benefit of crizotinib[20]. The testing of CTCs plays a pivotal role in the early recognition of lung cancer and can reflect tumor heterogeneity, a significant detail that are often overlooked by traditional biopsy methods[21,22]. The gene expression and mutation profile of CTCs can reflect the composition of primary and metastatic tumors, which can effectively distinguish between benign and malignant nodules[23]. The continuous monitoring of CTC concentrations and gene mutations can help us to assess the efficacy of treatment. ctDNA obtained from liquid biopsy can reflect the genomic profile and identify patients who might benefit from a particular form of targeted therapy[24,25]. Furthermore, the levels of ctDNA are known to be strongly correlated with tumor burden in patients and indicate a response to treatment earlier than conventional imaging modalities[23]. The sensitivity of ctDNA testing was found to be 82% for the detection of stage IV NSCLC while the specificity of ctDNA was 99.8% for the detection of gene mutations, thus highlighting the significant potential of ctDNA as a non-invasive diagnostic tool, although the screening of healthy individuals may result in false positives[26]. The specificities of CTCs, ctDNA and cRNA are notably higher than that of CEA (Table 1)[27-33]. Due to the heterogeneity of tumors, a single liquid biopsy may not fully reflect the genetic status, location and complexity of a given tumor.
| Types | Function | Specificity | Efficacy assessment |
| CTCs[27-29] | CTCs can identify all types of tumor cells and can be detected at early stages of lung cancer | High specificity | The number of CTCs is highly consistent with the therapy efficacy |
| ctDNA[26,30] | ctDNA serves as an independent molecular marker for NSCLC and allows for the optimization of clinical staging in the early to middle stages and monitor early metastasis | High specificity | ctDNA indicates the patient's response to treatment earlier than imaging examinations |
| CEA[15,16,31] | CEA is expressed at high levels in NSCLC and increases with the progression of clinical staging | Broad-spectrum but low specificity | Effective treatment can restore serum CEA concentration to baseline levels |
| cRNA[32,33] | An indicator of tumor proliferation and migration | High specificity | Up-regulation of cRNA is related to poor prognosis |
Simultaneously, molecular biology techniques have advanced our ability to treat cancer. Currently, the different subtypes of NSCLC can be identified by specific molecular characteristics based on a series of oncogenic mutations, which can also contribute to the development of targeted therapy[34]. Epidermal growth factor receptor (EGFR) mutations, including point mutations, deletions and insertions, is a common oncogenic driver event in patients with NSCLC[35]. The detection of EGFR alterations has been generally recommended for NSCLC patients in order to increase our understanding of the specific pathological features[3,36]. Tyrosine kinase inhibitors (TKIs) have been long regarded as the primary treatment for NSCLC patients with EGFR mutations[34]. These targeted therapies have been successfully used for the treatment of lung cancer by improving survival outcomes and reducing mortality[37]. Gefitinib is a first generation of EGFR-TKIs that can prolong survival when combined with chemotherapy[38]. For EGFR T790M-resistant mutations, third-generation TKIs, such as Osimertinib, can further improve progression-free survival (PFS) and intracerebral remission rate in patients with intracerebral metastasis[38,39]. In addition, anaplastic lymphoma kinase
| Types | Clinical relevance | Associated therapeutic strategies | Drugs |
| EGFR[43] | Lung adenocarcinoma has a substantially greater rate of EGFR mutation than lung squamous cell carcinoma | First-line treatment drugs for NSCLC patients with EGFR sensitive gene mutations | Osimertinib, Almonertinib, Furmonertinib, Befotertinib, Gefitinib |
| ALK[44] | ALK positive NSCLC patients are usually young, non-smoking, and EGFR non mutated lung adenocarcinoma populations | For advanced NSCLC patients accompanied by ALK gene fusion | Crizotinib, Alectinib, Ensartinib |
| ROS1[45] | ROS1-positive lung cancer is commonly found in young, non-smoking, or lightly smoking lung adenocarcinoma patients | For ROS1 positive NSCLC patients | Crizotinib, Entrectinib, Lorlatinib, Cabozantinib |
| PD-1/ PD-L1[46] | PD-1/PD-L1 inhibits T cell function via the TCR receptor signaling pathway, leading to immune escape in tumors | For driver gene negative advanced NSCLC patients | Nivolumab, Pembrolizumab, Camrelizumab, Tislelizumab, Sintilimab, Durvalumab, Atezolizumab |
| CLTA-4[47,48] | CLTA-4 facilitates tumor immune evasion | For advanced and metastatic NSCLC patients | Ipilimumab, Tremelimumab, Cadornilimab |
Immune checkpoint inhibitors have proven to be more effective than conventional chemotherapy[49], including programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein (CTLA-4). Both PD-1 and PD-L1 are known to be expressed at high levels in tumor cells and can exert negative effects on the immune system, particularly in terms of T cells[50,51]. The activation of PD-1 and PD-L1 can reduce the immune response of T cells and mediate the immune escape of tumor cells. Furthermore, the blockade of PD-1 and PD-L1 represents the most effective immunotherapeutic approach for NSCLC[52] and works by reversing the immunosuppressive status of the tumor microenvironment, thereby inhibiting tumor growth and eliminating tumor cells[49,51]. Pembrolizumab, a humanized antibody against PD-1, has been shown to exert significant anti-tumor activity in patients with advanced NSCLC, especially for tumor cells with high expression levels of PD-1[52,53]. The combination of pembrolizumab and chemotherapy as a first-line treatment significantly improved PFS and OS in NSCLC patients when compared to chemotherapy alone[54,55]. Despite the significant potential of PD-1 and PD-L1 inhibitors, their efficacy is constrained in tumor cells lacking inflammation, thus leading to a diminished response[56]. Furthermore, due to drug resistance, efficacy can be reduced even further. Appropriate combination therapy can enhance the antigen presenting functionality of T cells and reduce immunosuppressive resistance[57]. CTLA-4, an immunosuppressive molecule that is predominantly expressed in T cells, is a significant ‘hot-target’ for tumor-targeted immunotherapy and can facilitate tumor immune evasion by inhibiting the activation and proliferation of T cells[58]. Notably, the blockade of CTLA-4 remains controversial due to limited efficacy and high toxicity, although a corresponding CTLA-4 antibody (ipilimumab) has been used in the clinic[59].
The crux of precision medicine in lung cancer lies in the effective amalgamation of multi-omics data with artificial intelligence (AI) to identify specific molecular characteristics of tumors, thereby enhancing diagnostic precision and tailoring therapeutic strategies. Future studies should concentrate on harnessing and dissecting a multi-omics dataset, which will encompass a spectrum of biological insights, ranging from genomics to proteomics and metabolomics[60]. The incorporation of AI technology could significantly enhance early detection, pathological diagnosis, and classification, as well as facilitate prognostic evaluation and a more profound comprehension of the underlying molecular mechanisms[61]. However, multi-source, complex, and unstandardized datasets pose significant challenges to the establishment and application of AI models[61]. Therefore, a critical area for future investigation involves the effective integration of multi-omics approaches with AI technologies to enhance diagnostic accuracy and precision medicine.
Owing to our increasing comprehension of the molecular features of lung cancer, we are making significant progress regards to the diagnosis, molecular targeted therapy, and immunotherapy of this disease. The detection of biomarkers has become vital for the diagnosis of lung cancer and the development of personalized clinical management strategies. Furthermore, liquid biopsy will gradually be integrated with multi-omics information, AI and machine learning algorithms to facilitate the selection of targeted therapies for patients and allow for the dynamic surveillance of tumor progression. Concurrently, the establishment of standardized operating protocols are paramount if we are to enhance the precision and reliability of outcomes arising from liquid biopsy. Based on the complexity of medical conditions, further investigation is required for the development of precision oncology.
| 1. | Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20:624-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 517] [Article Influence: 258.5] [Reference Citation Analysis (0)] |
| 2. | GBD 2019 Respiratory Tract Cancers Collaborators. Global, regional, and national burden of respiratory tract cancers and associated risk factors from 1990 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Respir Med. 2021;9:1030-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 3. | Nooreldeen R, Bach H. Current and Future Development in Lung Cancer Diagnosis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 343] [Cited by in RCA: 434] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 4. | Feng J, Zhang P, Wang D, Li Y, Tan J. New strategies for lung cancer diagnosis and treatment: applications and advances in nanotechnology. Biomark Res. 2024;12:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 5. | Bederka LH, Sanchez JR, Rebeles J, Araujo PR, Grayson MH, Lai SC, DePalo LR, Habib SA, Hill DG, Lopez K, Patriquin L, Sussman R, Humphreys J, Reveles XT, Rebel VI. Sputum analysis by flow cytometry; an effective platform to analyze the lung environment. PLoS One. 2022;17:e0272069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 6. | Devi L, Verma Y, Kumar A, Khan F, Verma S, Kumar A. Understanding the Landscape of Bronchoscopy in Lung Cancer: Insights From Lesion Location, Gender, and Diagnostic Efficacy. Cureus. 2024;16:e53918. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Lee SM, Schulz C, Prabhash K, Kowalski D, Szczesna A, Han B, Rittmeyer A, Talbot T, Vicente D, Califano R, Cortinovis D, Le AT, Huang D, Liu G, Cappuzzo F, Reyes Contreras J, Reck M, Palmero R, Mak MP, Hu Y, Morris S, Höglander E, Connors M, Biggane AM, Vollan HK, Peters S. First-line atezolizumab monotherapy versus single-agent chemotherapy in patients with non-small-cell lung cancer ineligible for treatment with a platinum-containing regimen (IPSOS): a phase 3, global, multicentre, open-label, randomised controlled study. Lancet. 2023;402:451-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 8. | Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. 2023;24:695-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 411] [Article Influence: 205.5] [Reference Citation Analysis (0)] |
| 9. | Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 1434] [Article Influence: 358.5] [Reference Citation Analysis (0)] |
| 10. | Da Silva RCDS, Simon NA, Dos Santos AA, Olegário GM, Da Silva JF, Sousa NO, Corbacho MAT, de Melo FF. Personalized medicine: Clinical oncology on molecular view of treatment. World J Clin Oncol. 2024;15:992-1001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Seijo LM, Peled N, Ajona D, Boeri M, Field JK, Sozzi G, Pio R, Zulueta JJ, Spira A, Massion PP, Mazzone PJ, Montuenga LM. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J Thorac Oncol. 2019;14:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 349] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 12. | Kuwahara N, Honjo T, Sone N, Imanishi J, Nakayama K, Kamemura K, Iwahashi M, Ohta S, Kaihotsu K. Clinical impact of portal vein pulsatility on the prognosis of hospitalized patients with acute heart failure. World J Cardiol. 2023;15:599-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (2)] |
| 13. | Lung Cancer Cohort Consortium (LC3). The blood proteome of imminent lung cancer diagnosis. Nat Commun. 2023;14:3042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 31] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 14. | Yang Y, Xu M, Huang H, Jiang X, Gong K, Liu Y, Kuang X, Yang X. Serum carcinoembryonic antigen elevation in benign lung diseases. Sci Rep. 2021;11:19044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 15. | Gan T, An W, Long Y, Wang J, Zhang H, Liao M. Correlation between carcinoembryonic antigen (CEA) expression and EGFR mutations in non-small-cell lung cancer: a meta-analysis. Clin Transl Oncol. 2024;26:991-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 16. | Lei J, Wu L, Zhang N, Liu X, Zhang J, Kuang L, Chen J, Chen Y, Li D, Li Y. Carcinoembryonic antigen potentiates non-small cell lung cancer progression via PKA-PGC-1ɑ axis. Mol Biomed. 2024;5:19. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Ren F, Fei Q, Qiu K, Zhang Y, Zhang H, Sun L. Liquid biopsy techniques and lung cancer: diagnosis, monitoring and evaluation. J Exp Clin Cancer Res. 2024;43:96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 18. | Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W, Tang XM, Sun F, Lu HM, Deng J, Bai J, Li J, Wu CY, Lin QL, Lv ZW, Wang GR, Jiang GX, Ma YS, Fu D. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. 2022;21:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 241] [Article Influence: 80.3] [Reference Citation Analysis (0)] |
| 19. | Stanzione B, Del Conte A, Bertoli E, De Carlo E, Revelant A, Spina M, Bearz A. Therapeutical Options in ROS1-Rearranged Advanced Non Small Cell Lung Cancer. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 20. | Lin C, Shi X, Yang S, Zhao J, He Q, Jin Y, Yu X. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small-cell lung cancer. Lung Cancer. 2019;131:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 21. | Nagasaka M, Uddin MH, Al-Hallak MN, Rahman S, Balasubramanian S, Sukari A, Azmi AS. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol Cancer. 2021;20:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 22. | Huang L, Xu Y, Wang N, Yi K, Xi X, Si H, Zhang Q, Xiang M, Rong Y, Yuan Y, Wang F. Next-Generation Preclinical Functional Testing Models in Cancer Precision Medicine: CTC-Derived Organoids. Small Methods. 2024;8:e2301009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 23. | Shen H, Jin Y, Zhao H, Wu M, Zhang K, Wei Z, Wang X, Wang Z, Li Y, Yang F, Wang J, Chen K. Potential clinical utility of liquid biopsy in early-stage non-small cell lung cancer. BMC Med. 2022;20:480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 24. | Nakamura Y, Okamoto W, Kato T, Esaki T, Kato K, Komatsu Y, Yuki S, Masuishi T, Nishina T, Ebi H, Sawada K, Taniguchi H, Fuse N, Nomura S, Fukui M, Matsuda S, Sakamoto Y, Uchigata H, Kitajima K, Kuramoto N, Asakawa T, Olsen S, Odegaard JI, Sato A, Fujii S, Ohtsu A, Yoshino T. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nat Med. 2021;27:1899-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 159] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 25. | Pellini B, Chaudhuri AA. Circulating Tumor DNA Minimal Residual Disease Detection of Non-Small-Cell Lung Cancer Treated With Curative Intent. J Clin Oncol. 2022;40:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 130] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 26. | Li RY, Liang ZY. Circulating tumor DNA in lung cancer: real-time monitoring of disease evolution and treatment response. Chin Med J (Engl). 2020;133:2476-2485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Jiang JH, Gao J, Chen CY, Ao YQ, Li J, Lu Y, Fang W, Wang HK, de Castro DG, Santarpia M, Hashimoto M, Yuan YF, Ding JY. Circulating tumor cell methylation profiles reveal the classification and evolution of non-small cell lung cancer. Transl Lung Cancer Res. 2022;11:224-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Kapeleris J, Kulasinghe A, Warkiani ME, Vela I, Kenny L, O'Byrne K, Punyadeera C. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front Oncol. 2018;8:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 29. | Zhang Z, Ramnath N, Nagrath S. Current Status of CTCs as Liquid Biopsy in Lung Cancer and Future Directions. Front Oncol. 2015;5:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 30. | Zaman FY, Subramaniam A, Afroz A, Samoon Z, Gough D, Arulananda S, Alamgeer M. Circulating Tumour DNA (ctDNA) as a Predictor of Clinical Outcome in Non-Small Cell Lung Cancer Undergoing Targeted Therapies: A Systematic Review and Meta-Analysis. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 31. | Fu W, Yue Y, Song Y, Zhang S, Shi J, Zhao R, Wang Q, Zhang R. Comparable analysis of six immunoassays for carcinoembryonic antigen detection. Heliyon. 2024;10:e25158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 32. | Zhang C, Ma L, Niu Y, Wang Z, Xu X, Li Y, Yu Y. Circular RNA in Lung Cancer Research: Biogenesis, Functions, and Roles. Int J Biol Sci. 2020;16:803-814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 33. | Dou Y, Zhu Y, Ai J, Chen H, Liu H, Borgia JA, Li X, Yang F, Jiang B, Wang J, Deng Y. Plasma small ncRNA pair panels as novel biomarkers for early-stage lung adenocarcinoma screening. BMC Genomics. 2018;19:545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Tan AC, Tan DSW. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J Clin Oncol. 2022;40:611-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 436] [Article Influence: 145.3] [Reference Citation Analysis (0)] |
| 35. | Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol. 2020;61:167-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 396] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 36. | Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr K, Kwiatkowski DJ, Ladanyi M, Nowak JA, Sholl L, Temple-Smolkin R, Solomon B, Souter LH, Thunnissen E, Tsao MS, Ventura CB, Wynes MW, Yatabe Y. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 363] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 37. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9955] [Article Influence: 4977.5] [Reference Citation Analysis (2)] |
| 38. | Hou X, Li M, Wu G, Feng W, Su J, Jiang H, Jiang G, Chen J, Zhang B, You Z, Liu Q, Chen L. Gefitinib Plus Chemotherapy vs Gefitinib Alone in Untreated EGFR-Mutant Non-Small Cell Lung Cancer in Patients With Brain Metastases: The GAP BRAIN Open-Label, Randomized, Multicenter, Phase 3 Study. JAMA Netw Open. 2023;6:e2255050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 39. | Li Y, Yan B, He S. Advances and challenges in the treatment of lung cancer. Biomed Pharmacother. 2023;169:115891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 143] [Reference Citation Analysis (0)] |
| 40. | Schneider JL, Lin JJ, Shaw AT. ALK-positive lung cancer: a moving target. Nat Cancer. 2023;4:330-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 93] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 41. | Yu ZQ, Wang M, Zhou W, Mao MX, Chen YY, Li N, Peng XC, Cai J, Cai ZQ. ROS1-positive non-small cell lung cancer (NSCLC): biology, diagnostics, therapeutics and resistance. J Drug Target. 2022;30:845-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 42. | Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 681] [Article Influence: 113.5] [Reference Citation Analysis (0)] |
| 43. | Chen HY, Chen CH, Liao WC, Lin YC, Chen HJ, Hsia TC, Cheng WC, Tu CY. Optimal first-line treatment for EGFR-mutated NSCLC: a comparative analysis of osimertinib and second-generation EGFR-TKIs. BMC Pulm Med. 2024;24:517. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 44. | Barreira JV, Mendes JL, Parmanande A. Personalized medicine: paradigm shift in ALK positive non-small cell lung cancer: a case report. J Med Case Rep. 2023;17:374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 45. | Gendarme S, Bylicki O, Chouaid C, Guisier F. ROS-1 Fusions in Non-Small-Cell Lung Cancer: Evidence to Date. Curr Oncol. 2022;29:641-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 68] [Reference Citation Analysis (0)] |
| 46. | Chen QA, Ma K, Zhang L, Lin WH, Wu XX, Gao YB. Efficacy and Safety of Anti-Programmed Cell Death Protein 1/Programmed Death-Ligand 1 Antibodies Plus Chemotherapy as First-Line Treatment for NSCLC in the People's Republic of China: a Systematic Review and Meta-Analysis. JTO Clin Res Rep. 2024;5:100678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 47. | Chen B, Yao W, Li X, Lin G, Chu Q, Liu H, Du Y, Lin J, Duan H, Wang H, Xiao Z, Sun H, Liu L, Xu L, Xu Y, Xu F, Kong Y, Pu X, Li K, Wang Q, Li J, Li B, Xia Y, Wu L. A phase Ib/II study of cadonilimab (PD-1/CTLA-4 bispecific antibody) plus anlotinib as first-line treatment in patients with advanced non-small cell lung cancer. Br J Cancer. 2024;130:450-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 48. | Zhao Y, Ma Y, Fan Y, Zhou J, Yang N, Yu Q, Zhuang W, Song W, Wang ZM, Li B, Xia Y, Zhao H, Zhang L. A multicenter, open-label phase Ib/II study of cadonilimab (anti PD-1 and CTLA-4 bispecific antibody) monotherapy in previously treated advanced non-small-cell lung cancer (AK104-202 study). Lung Cancer. 2023;184:107355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 49. | Tang S, Qin C, Hu H, Liu T, He Y, Guo H, Yan H, Zhang J, Tang S, Zhou H. Immune Checkpoint Inhibitors in Non-Small Cell Lung Cancer: Progress, Challenges, and Prospects. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 123] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 50. | Tang Q, Chen Y, Li X, Long S, Shi Y, Yu Y, Wu W, Han L, Wang S. The role of PD-1/PD-L1 and application of immune-checkpoint inhibitors in human cancers. Front Immunol. 2022;13:964442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 294] [Reference Citation Analysis (0)] |
| 51. | Wu Q, Jiang L, Li SC, He QJ, Yang B, Cao J. Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway. Acta Pharmacol Sin. 2021;42:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 222] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 52. | Lahiri A, Maji A, Potdar PD, Singh N, Parikh P, Bisht B, Mukherjee A, Paul MK. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer. 2023;22:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 500] [Cited by in RCA: 526] [Article Influence: 263.0] [Reference Citation Analysis (1)] |
| 53. | Harrington KJ, Burtness B, Greil R, Soulières D, Tahara M, de Castro G Jr, Psyrri A, Brana I, Basté N, Neupane P, Bratland Å, Fuereder T, Hughes BGM, Mesia R, Ngamphaiboon N, Rordorf T, Wan Ishak WZ, Lin J, Gumuscu B, Swaby RF, Rischin D. Pembrolizumab With or Without Chemotherapy in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma: Updated Results of the Phase III KEYNOTE-048 Study. J Clin Oncol. 2023;41:790-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 252] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 54. | Hektoen HH, Tsuruda KM, Fjellbirkeland L, Nilssen Y, Brustugun OT, Andreassen BK. Real-world evidence for pembrolizumab in non-small cell lung cancer: a nationwide cohort study. Br J Cancer. 2024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Chen M, Xu Y, Zhao J, Liu X, Liu X, Zhang D, Shi Y, Zhang L, Zhong W, Wang M. Comparison of Chemotherapy Plus Pembrolizumab vs. Chemotherapy Alone in EGFR-Mutant Non-small-Cell Lung Cancer Patients. Clin Lung Cancer. 2023;24:278-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 56. | Yi M, Zheng X, Niu M, Zhu S, Ge H, Wu K. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer. 2022;21:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 526] [Cited by in RCA: 818] [Article Influence: 272.7] [Reference Citation Analysis (0)] |
| 57. | Wu M, Huang Q, Xie Y, Wu X, Ma H, Zhang Y, Xia Y. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J Hematol Oncol. 2022;15:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 271] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 58. | Babamohamadi M, Mohammadi N, Faryadi E, Haddadi M, Merati A, Ghobadinezhad F, Amirian R, Izadi Z, Hadjati J. Anti-CTLA-4 nanobody as a promising approach in cancer immunotherapy. Cell Death Dis. 2024;15:17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 59. | Liu Y, Zheng P. Preserving the CTLA-4 Checkpoint for Safer and More Effective Cancer Immunotherapy. Trends Pharmacol Sci. 2020;41:4-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 60. | Micheletti C, Dhuli K, Donato K, Gadler M, Benedetti S, Guerri G, Cristofoli F, Generali D, Donofrio CA, Cominetti M, Fioravanti A, Riccio L, Bernini A, Fulcheri E, Stuppia L, Stuppia L, Gatta V, Cristoni S, Cecchin S, Marceddu G, Bertelli M. Omics sciences and precision medicine in lung cancer. Clin Ter. 2023;174:37-45. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 61. | Huang S, Yang J, Shen N, Xu Q, Zhao Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin Cancer Biol. 2023;89:30-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 99] [Reference Citation Analysis (0)] |