Published online Jan 24, 2025. doi: 10.5306/wjco.v16.i1.94813
Revised: May 17, 2024
Accepted: June 5, 2024
Published online: January 24, 2025
Processing time: 218 Days and 5.5 Hours
Mitochondrial genes are involved in tumor metabolism in ovarian cancer (OC) and affect immune cell infiltration and treatment responses.
To predict prognosis and immunotherapy response in patients diagnosed with OC using mitochondrial genes and neural networks.
Prognosis, immunotherapy efficacy, and next-generation sequencing data of patients with OC were downloaded from The Cancer Genome Atlas and Gene Expression Omnibus. Mitochondrial genes were sourced from the MitoCarta3.0 database. The discovery cohort for model construction was created from 70% of the patients, whereas the remaining 30% constituted the validation cohort. Using the expression of mitochondrial genes as the predictor variable and based on neural network algorithm, the overall survival time and immunotherapy efficacy (complete or partial response) of patients were predicted.
In total, 375 patients with OC were included to construct the prognostic model, and 26 patients were included to construct the immune efficacy model. The average area under the receiver operating characteristic curve of the prognostic model was 0.7268 [95% confidence interval (CI): 0.7258-0.7278] in the discovery cohort and 0.6475 (95%CI: 0.6466-0.6484) in the validation cohort. The average area under the receiver operating characteristic curve of the immunotherapy efficacy model was 0.9444 (95%CI: 0.8333-1.0000) in the discovery cohort and 0.9167 (95%CI: 0.6667-1.0000) in the validation cohort.
The application of mitochondrial genes and neural networks has the potential to predict prognosis and immunotherapy response in patients with OC, providing valuable insights into personalized treatment strategies.
Core Tip: In this study, we found that mitochondrial genes and neural networks can be used to predict ovarian cancer prognosis and immunotherapy response. These models were evaluated in detail. The average area under the receiver operating characteristic curve of the prognostic model was 0.7268 and 0.6475 for the discovery and validation cohorts, respectively. The average area under the receiver operating characteristic curve of the immunotherapy efficacy model was 0.9444 and 0.9167 for the discovery and validation cohorts, respectively. The Hosmer-Lemeshow goodness of fit test showed that the model had a good calibration performance.
- Citation: Tang ZJ, Pan YM, Li W, Ma RQ, Wang JL. Unlocking the future: Mitochondrial genes and neural networks in predicting ovarian cancer prognosis and immunotherapy response. World J Clin Oncol 2025; 16(1): 94813
- URL: https://www.wjgnet.com/2218-4333/full/v16/i1/94813.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i1.94813
Ovarian cancer (OC) is one of the most common gynecological cancers worldwide, with over 230000 new cases and 150000 deaths annually[1,2]. According to studies from the United States and the United Kingdom registries, 1 in 6 females dies within 90 days of diagnosis[1,2]. Genetic factors, gene mutations (such as in BRCA1 and BRCA2), nulliparity, infertility, endometriosis, obesity, and age are associated with the incidence of OC, whereas pregnancy, oral contraceptives, and nonsteroidal anti-inflammatory drugs are potential protective factors[1,3]. High-grade serous OC is the most common subtype of OC, accounting for over 70% of cases[4]. Treatment of OC primarily involves surgery, che
The traditional view holds that anaerobic glycolysis in the Warburg effect is the main energy source for tumor growth[6]. However, increasing evidence has shown that macromolecular synthesis in tumor cells depends on mitochondrial metabolism, and strategies targeting the mitochondrial oxidative respiratory chain can be developed for cancer treatment[7]. For example, the MYC pathway, one of the most common pathways in tumor synthetic metabolism and associated with increased mitochondrial and oxygen consumption, has been found to be more highly expressed in OC[8,9]. Changes in the MYC-CDK2/4-RB1 signaling pathway can be observed in 75% of ovarian clear cell carcinomas, and high MYC expression is associated with platinum resistance and poor prognosis. Moreover, MYC siRNA inhibits the growth of OC tumor cells[10-12].
This study collected data from The Cancer Genome Atlas Program (TCGA) and the Gene Expression Omnibus (GEO), sorted mitochondria-located genes, and used neural network technology to establish models to predict the prognosis and immunotherapy response of patients with OC.
TCGA data were downloaded from the UCSC-XENA website (https://xena.ucsc.edu/), the OC dataset (GDC TCGA OC) was selected, and samples with both prognostic and next-generation sequencing (NGS) data were retained for subsequent analysis. The OC immunotherapy dataset was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) (accession number: GSE188249). Both TCGA and GEO data were analyzed using fragments per kilobase million (FPKM_ sequencing results, and gene symbol conversion was performed using clusterProfiler. Mitochondria-localized genes were obtained from the MitoCarta3.0 database (https://www.broadinstitute.org/mitocarta).
The study was conducted in accordance with the Declaration of Helsinki (revised in 2013). Given that all data were retrieved from the publicly available TCGA and GEO databases and that patients’ private information has been concealed and is not traceable, this retrospective cohort was exempt from ethical considerations and written consent.
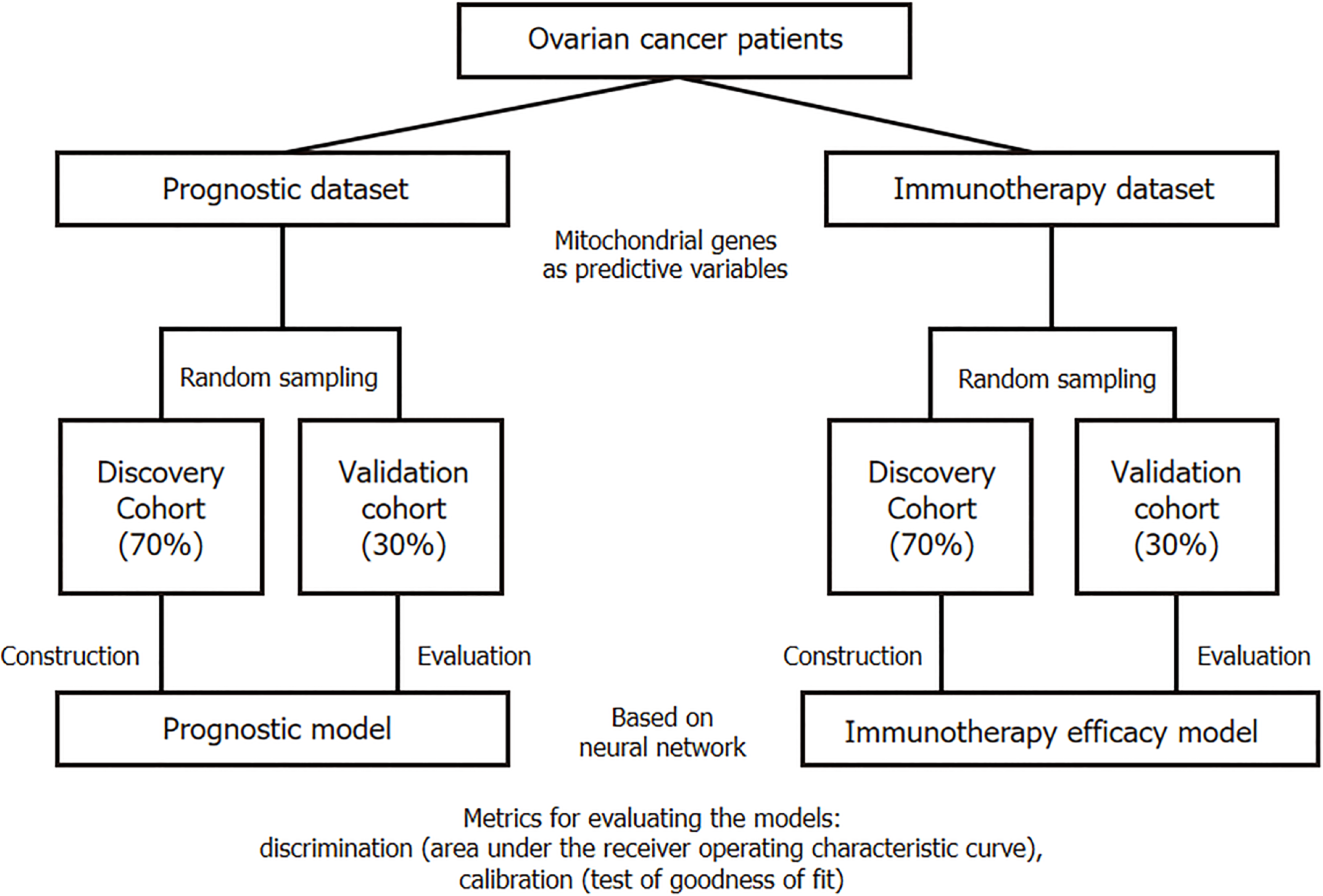
This retrospective study was designed for diagnostic testing. The main outcome was the overall survival of patients with OC, with the secondary endpoint being immunotherapy response. Sensitivity to immunotherapy was defined according to the data source authors’ definition, which meant that patients with sensitivity to OC showed partial response or complete response after the use of ICIs according to the Response Evaluation Criteria in Solid Tumors 1.1. TCGA data were used to construct an OC prognostic model, with patients randomly sampled into a discovery cohort (70% of the total) or a validation cohort (remaining 30% of the total). The discovery cohort was used to build the prognostic model (based on neural network technology), and the model performance was checked in the validation cohort. Similarly, GEO data were used to construct an OC immunotherapy efficacy model, with 70% of the GEO data randomly selected as the discovery cohort (for model construction) and the remaining 30% as the validation cohort (Figure 1).
The discovery cohort served as the basis for model training, whereas the validation cohort was used to further evaluate model performance. The model was constructed based on the neural network theory, and to enhance its performance, batch normalization layers and batch training functions were adopted. To prevent overfitting, dropout layers and early stopping functions were employed (which automatically terminated training when there was no significant improvement in model performance after several rounds). Adam was selected as the optimizer, with the learning rate set between 0.01 and 0.05. To predict patient survival, considering that this is a time-to-event classification task rather than a traditional classification task, a neural network was built based on the DeepSurv theory of Katzman et al[13]. The model was built in Python 3.9 using PyTorch, torchtuples, pandas, matplotlib, and NumPy.
The model was evaluated from two perspectives: Discrimination and calibration. The primary metric for evaluating discrimination was the area under the receiver operating characteristic curve (AUC). Generally, an AUC closer to 1 indicates better model performance, whereas an AUC closer to 0.5, suggests that the predictions of the model are akin to random guessing. The model is performing well when the AUC is > 0.7. Other indicators included the sensitivity, specificity, accuracy, negative predictive value (NPV), and positive predictive value (PPV).
In addition, we used the Hosmer-Lemeshow goodness of fit test to judge the calibration ability of the models. A P value greater than 0.05 indicated that the model had good calibration ability.
We truncated the survival data at 1 year, 2 years, and 3 years and evaluated the predictive performance of the model in detail. Finally, we used the PySide6 software package to compress the entire model into a Windows executable program for ease of use by clinicians.
Statistical analyses were performed using the R software (version 4.2.0). Numerical data were compared using the Wilcoxon test, whereas categorical data were compared using the χ2 test or Fisher’s exact test. A two-sided P value of less than 0.05 was considered statistically significant.
For the prognostic model, 375 patients from the TCGA with OC were included in the analysis, with 262 and 113 randomly assigned to the discovery and validation cohorts, respectively. The median survival time was 1012 days (interquartile range: 547-1712 days) in the discovery cohort and 1032 days (interquartile range: 394-1562 days) in the validation cohort, with no significant difference between the two groups (P = 0.5446). In the discovery cohort, 104 patients (39.69%) survived, whereas in the validation cohort, 41 patients (36.28%) survived, with no statistically significant difference (P = 0.5337). In the immunotherapy efficacy model, 18 patients were assigned to the discovery cohort, of which 50.00% (9 patients) were sensitive to treatment. In the validation cohort, 25.00% (2 of 8 patients) were sensitive to immunotherapy. There was no significant difference between the two groups (P = 0.3945) (Table 1).
| Discovery cohort | Validation cohort | Statistical method | P value | |
| Prognostic model | ||||
| (n = 262) | (n = 113) | |||
| Survival time in days | Wilcoxon | 0.5446 | ||
| Median (IQR) | 1012 (547, 1712) | 1032 (394, 1562) | ||
| Alive | χ2 test | 0.5337 | ||
| Yes | 104 (39.69) | 41 (36.28) | ||
| No | 158 (60.31) | 72 (63.72) | ||
| Immunotherapy efficacy model | ||||
| (n = 18) | (n = 8) | |||
| Sensitivity to immunotherapy | Fisher’s exact | 0.3945 | ||
| Yes | 9 (50.00) | 2 (25.00) | ||
| No | 9 (50.00) | 6 (75.00) |
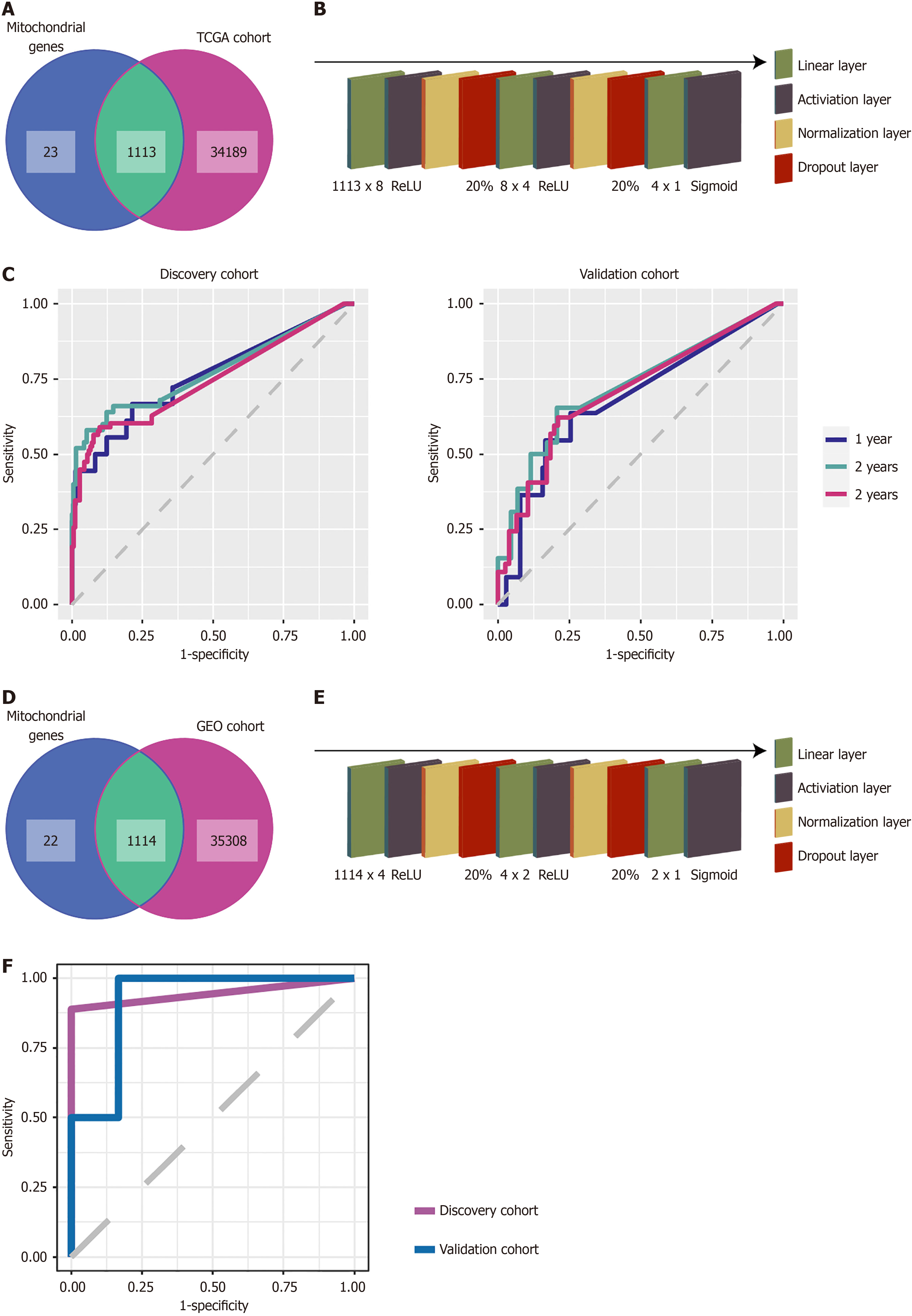
We intersected TCGA-sequenced genes with mitochondria-localized genes, and 1113 genes were simultaneously detected (Figure 2A). Using these 1113 genes as predictive variables and patient overall survival as the outcome variable, we built a neural network in Python. After 68 rounds of training, the early termination function automatically ended the training (Supplementary Figure 1A). The final OC prognostic model included 10 hidden layers: An input layer of 1113 × 8; a ReLU activation layer; a normalization layer; a 20% dropout layer; an 8 × 4 Linear layer; another ReLU activation layer; a normalization layer; a 20% dropout layer; a 4 × 1 Linear layer; and a sigmoid activation layer (Figure 2B). The average AUC of the prognostic model was 0.7268 [95% confidence interval (CI): 0.7258-0.7278] for the discovery cohort and 0.6475 (95%CI: 0.6466-0.6484) for the validation cohort (Table 2).
| Discovery cohort | Validation cohort | |
| AUC | 0.7268 | 0.6475 |
| AUC 95%CI | 0.7258-0.7278 | 0.6466-0.6484 |
We performed cutoffs at 1 year, 2 years, and 3 years to measure the performance of the OC prognostic model in detail. The receiver operating characteristic curves of the OC prognostic model at 1 year, 2 years, and 3 years are shown in Figure 2C. In the discovery cohort, at 1 year, the AUC was 0.7597 (95%CI: 0.6252-0.8942), specificity was 0.7869, sensitivity was 0.6667, accuracy was 0.7786, NPV was 0.9697, and PPV was 0.1875. At 2 years, the AUC was 0.7734 (95%CI: 0.6908-0.8560), specificity was 0.9481, sensitivity was 0.5800, accuracy was 0.8779, NPV was 0.9054, and PPV was 0.7250. At 3 years, the AUC was 0.7461 (95%CI: 0.6789-0.8134), specificity was 0.9022, sensitivity was 0.5897, accuracy was 0.8092, NPV was 0.8384, and PPV was 0.7188. In the validation cohort at 1 year, the AUC was 0.6827 (95%CI: 0.5114-0.8541), specificity was 0.7451, sensitivity was 0.6364, accuracy was 0.7345, NPV was 0.9500, and PPV was 0.2121. At 2 years, the AUC was 0.7250 (95%CI: 0.6128-0.8373), specificity was 0.7931, sensitivity was 0.6538, accuracy was 0.7611, NPV was 0.8846, and PPV was 0.4857. At 3 years, the AUC was 0.7087 (95%CI: 0.6126-0.8049), specificity was 0.7895, sensitivity was 0.6216, accuracy was 0.7345, NPV was 0.8108, and PPV was 0.5897.
The goodness-of-fit test results showed P values of 0.6699, 0.6304, and 0.4325 at 1 year, 2 years, and 3 years, respectively, in the discovery cohort. In the validation cohort, the P values were 0.0826, 0.1042, and 0.1084 at 1 year, 2 years, and 3 years, respectively (Table 3).
| Discovery cohort | Validation cohort | |||||
| 1 year | 2 years | 3 years | 1 year | 2 years | 3 years | |
| AUC | 0.7597 | 0.7734 | 0.7461 | 0.6827 | 0.725 | 0.7087 |
| AUC 95%CI | 0.6252-0.8942 | 0.6908-0.8560 | 0.6789-0.8134 | 0.5114-0.8541 | 0.6128-0.8373 | 0.6126-0.8049 |
| Specificity | 0.7869 | 0.9481 | 0.9022 | 0.7451 | 0.7931 | 0.7895 |
| Sensitivity | 0.6667 | 0.58 | 0.5897 | 0.6364 | 0.6538 | 0.6216 |
| Accuracy | 0.7786 | 0.8779 | 0.8092 | 0.7345 | 0.7611 | 0.7345 |
| NPV | 0.9697 | 0.9054 | 0.8384 | 0.95 | 0.8846 | 0.8108 |
| PPV | 0.1875 | 0.725 | 0.7188 | 0.2121 | 0.4857 | 0.5897 |
| P value, goodness of fit | 0.6699 | 0.6304 | 0.4325 | 0.0826 | 0.1042 | 0.1084 |
The GEO-sequenced genes were intersected with mitochondria-localized genes, and 1114 genes were simultaneously detected. These 1114 genes were used as predictive variables, with patient sensitivity to immunotherapy (complete response + partial response) as the outcome variable, to build a neural network in Python (Figure 2D). After 114 rounds of training, the early termination function automatically ended the training (Supplementary Figure 1B). The final OC immunotherapy efficacy model included 10 hidden layers: An input layer of 1114 × 4; a ReLU activation layer; a normalization layer; a 20% dropout layer; a 4 × 2 Linear layer; another ReLU activation layer; a normalization layer; a 20% dropout layer; a 2 × 1 Linear layer; and a sigmoid activation layer (Figure 2E).
The receiver operating characteristic curve of the OC immunotherapy efficacy model is shown in Figure 2F. In the discovery cohort, the model AUC was 0.9444 (95%CI: 0.8333-1.0000), specificity was 1.0000, sensitivity was 0.8889, accuracy was 0.9444, NPV was 0.9000, and PPV was 1.0000. In the validation cohort, the model AUC was 0.9167 (95%CI: 0.6667-1.0000), specificity was 0.8333, sensitivity was 1.0000, accuracy was 0.8750, NPV was 1.0000, and PPV was 0.6667.
The results of the goodness-of-fit test showed that the P value of the model was 0.3708 in the discovery cohort and 0.1175 in the validation cohort (Table 4). The model parameters are shown in Supplementary Figure 2.
| Discovery cohort | Validation cohort | |
| AUC | 0.9444 | 0.9167 |
| AUC 95%CI | 0.8333-1.0000 | 0.6667-1.0000 |
| Specificity | 1.0000 | 0.8333 |
| Sensitivity | 0.8889 | 1.0000 |
| Accuracy | 0.9444 | 0.8750 |
| NPV | 0.9000 | 1.0000 |
| PPV | 1.0000 | 0.6667 |
| P value, goodness of fit | 0.3708 | 0.1175 |
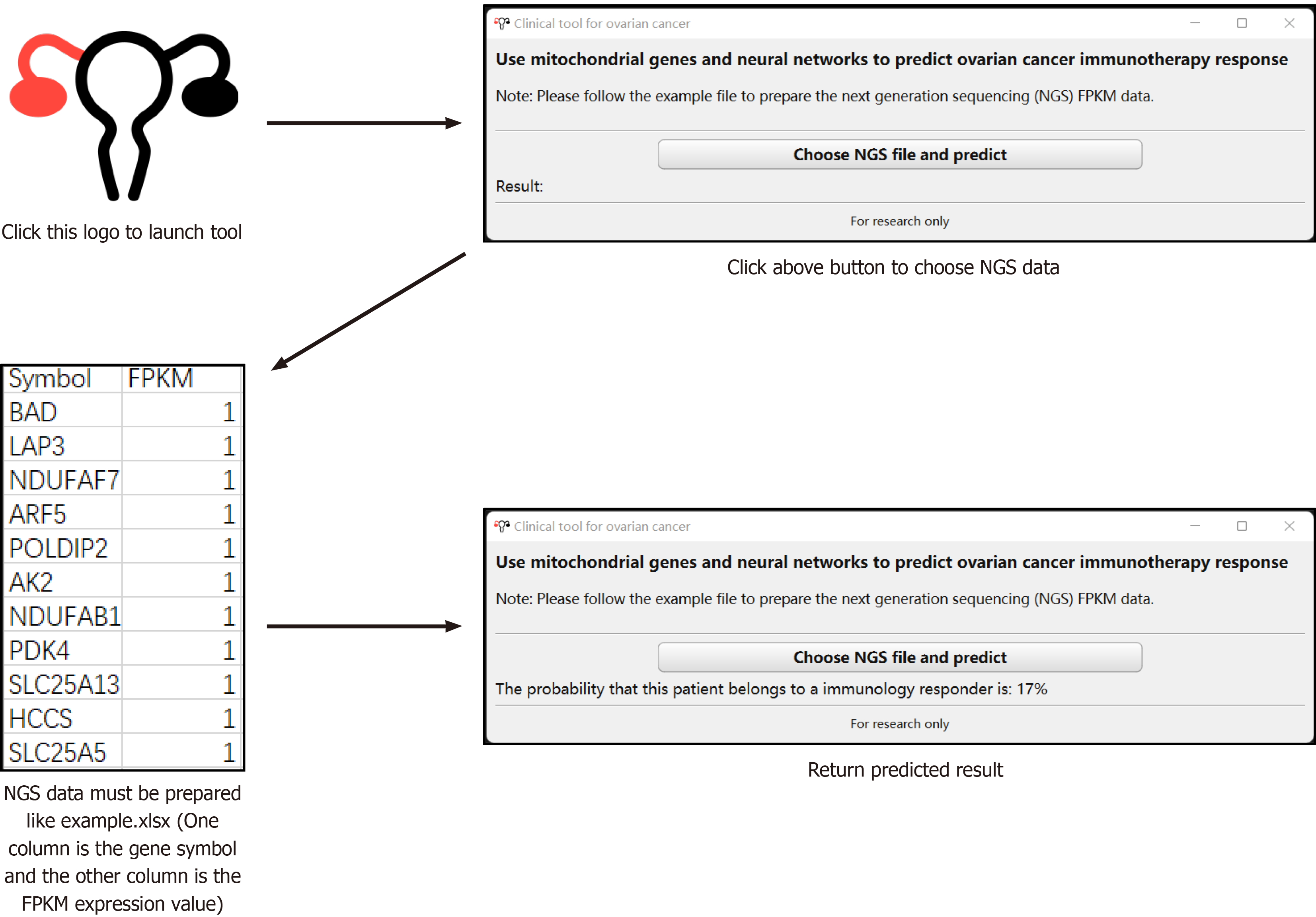
Considering the black-box characteristics of neural networks, the OC immunotherapy efficacy model was packaged into a Windows 64-bit executable program for ease of use by clinicians (Figure 3). After launching the tool, users can prepare the patient’s NGS FPKM file in the format of a built-in example file. By clicking “Choose NGS file and predict,” users can select the sequencing file to activate the built-in pretrained neural network. After the calculation is completed, the software will display the probability of the patient being sensitive to immunotherapy in the “Result” part.
As one of the most common lethal tumors in females, OC has a 5-year survival rate of approximately 45%[14]. The standard treatments for OC include surgery and chemotherapy. In recent years, poly (ADP-ribose) polymerase inhibitors and immunotherapies have emerged as new therapies attracting increasing attention[15-17]. Most immunotherapeutic drugs for OC are still undergoing clinical trials, and the objective response rate to ICIs remains limited at approximately 6%-15%[15-17].
There are no effective predictive factors for the efficacy of immunotherapy. The predictive value of classical indicators such as programmed death-ligand 1 and tumor mutational burden (TMB) remains limited. A study by Song et al[18] found that the AUC value for programmed death-ligand 1 in predicting immunotherapy efficacy was only 0.569, and the ability of TMB to predict the prognosis of patients receiving immunotherapy and its reproducibility across different samples have been questioned[18-21]. Therefore, there is an urgent need for a new indicator or algorithm to identify patients sensitive to immunotherapy and avoid unnecessary drug treatments and adverse events.
Traditional prognostic predictions often employ the Cox proportional hazards model. Based on linear assumptions, the model fits various risks and calculates the probability of a positive event. However, real-world situations are often complex and nonlinear, limiting the application of Cox regression. Machine learning algorithms, particularly deep learning neural networks, have gained increasing recognition from clinicians because of their superior predictive performance[22-24]. Our preliminary studies also found that neural networks have excellent predictive value for both prognosis and immunotherapy efficacy[25-27].
Increasing evidence suggests that mitochondria play a role in supplying energy to key metabolites in tumors. In
In this study, we integrated data from the TCGA and GEO databases to establish neural network models. These models used mitochondria-localized genes as variables to predict the prognosis and immunotherapy response of patients with OC. The average AUC of the OC prognostic model was 0.7268 and 0.6475 for the discovery and validation cohorts, respectively. For the OC immunotherapy efficacy model, the average AUC was 0.9444 for the discovery cohort and 0.9167 for the validation cohort. This study shows that the use of mitochondria-localized genes and neural networks can predict the prognosis and immunotherapy response of OC patients. Therefore, we packaged the models as a Windows executable tool for the convenience of clinicians, considering that there is a limited way to predict the immunotherapy response in patients with OC.
Yang et al[29] constructed an OC prognostic model based on the investigation of ferroptosis-related long noncoding RNAs, which had an AUC of 0.793 AUC in discovery data and 0.681 AUC in validation data. Li et al[30] developed prognostic models for OC based on ferroptosis and necroptosis, and the model had an AUC of 0.584-0.728. Jiang et al[31] built an OC prognostic model that used a combination of transcriptomic and proteomic data, which had an AUC of 0.596-0.749. Except for the Yang model, which had an AUC similar to ours, the other models performed poorly. This indicates both the feasibility of predicting OC prognosis at the genetic level and the complexity of OC prognosis. A multidimensional approach may be required to enhance the performance of the model further. Chen et al[32] found that autophagy-related genes could be used to predict OC survival and are related to the immunotherapy response. Wang et al[33] also observed a correlation between TMB-based genes and immune infiltration in OC. Unfortunately, these studies did not use these findings to create predictive models for assessing the efficacy of OC immunotherapy.
This study has some limitations. As this was a retrospective study, it may have suffered from selection and information biases. Further prospective studies with larger sample sizes can improve and validate the developed models.
The application of mitochondrial genes and neural networks has the potential to predict prognosis and immunotherapy response in patients with OC. This approach can provide valuable insights into personalized treatment strategies.
| 1. | Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 791] [Article Influence: 131.8] [Reference Citation Analysis (0)] |
| 2. | Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 636] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 3. | Werner H. BRCA1: An Endocrine and Metabolic Regulator. Front Endocrinol (Lausanne). 2022;13:844575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Prat J. Ovarian carcinomas: five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 404] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 5. | Morand S, Devanaboyina M, Staats H, Stanbery L, Nemunaitis J. Ovarian Cancer Immunotherapy and Personalized Medicine. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 227] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 6. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47133] [Article Influence: 3366.6] [Reference Citation Analysis (5)] |
| 7. | Vasan K, Werner M, Chandel NS. Mitochondrial Metabolism as a Target for Cancer Therapy. Cell Metab. 2020;32:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 458] [Article Influence: 91.6] [Reference Citation Analysis (0)] |
| 8. | Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225-6234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 483] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 9. | Emmings E, Mullany S, Chang Z, Landen CN Jr, Linder S, Bazzaro M. Targeting Mitochondria for Treatment of Chemoresistant Ovarian Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 10. | Reyes-González JM, Armaiz-Peña GN, Mangala LS, Valiyeva F, Ivan C, Pradeep S, Echevarría-Vargas IM, Rivera-Reyes A, Sood AK, Vivas-Mejía PE. Targeting c-MYC in Platinum-Resistant Ovarian Cancer. Mol Cancer Ther. 2015;14:2260-2269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 11. | Jung M, Russell AJ, Liu B, George J, Liu PY, Liu T, DeFazio A, Bowtell DD, Oberthuer A, London WB, Fletcher JI, Haber M, Norris MD, Henderson MJ. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res. 2017;77:971-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 12. | Murakami R, Matsumura N, Brown JB, Higasa K, Tsutsumi T, Kamada M, Abou-Taleb H, Hosoe Y, Kitamura S, Yamaguchi K, Abiko K, Hamanishi J, Baba T, Koshiyama M, Okuno Y, Yamada R, Matsuda F, Konishi I, Mandai M. Exome Sequencing Landscape Analysis in Ovarian Clear Cell Carcinoma Shed Light on Key Chromosomal Regions and Mutation Gene Networks. Am J Pathol. 2017;187:2246-2258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 13. | Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 732] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 14. | Chandra A, Pius C, Nabeel M, Nair M, Vishwanatha JK, Ahmad S, Basha R. Ovarian cancer: Current status and strategies for improving therapeutic outcomes. Cancer Med. 2019;8:7018-7031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 15. | Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. 2017;28:viii1-viii7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 291] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 16. | Yang C, Xia BR, Zhang ZC, Zhang YJ, Lou G, Jin WL. Immunotherapy for Ovarian Cancer: Adjuvant, Combination, and Neoadjuvant. Front Immunol. 2020;11:577869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 17. | O'Malley DM, Krivak TC, Kabil N, Munley J, Moore KN. PARP Inhibitors in Ovarian Cancer: A Review. Target Oncol. 2023;18:471-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 81] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 18. | Song P, Li W, Guo L, Ying J, Gao S, He J. Identification and Validation of a Novel Signature Based on NK Cell Marker Genes to Predict Prognosis and Immunotherapy Response in Lung Adenocarcinoma by Integrated Analysis of Single-Cell and Bulk RNA-Sequencing. Front Immunol. 2022;13:850745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 19. | Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR Jr, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA; CheckMate 026 Investigators. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2415-2426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1609] [Cited by in RCA: 2017] [Article Influence: 252.1] [Reference Citation Analysis (0)] |
| 20. | Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, Borghaei H, Ramalingam SS, Brahmer J, Reck M, O'Byrne KJ, Geese WJ, Green G, Chang H, Szustakowski J, Bhagavatheeswaran P, Healey D, Fu Y, Nathan F, Paz-Ares L. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018;378:2093-2104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1913] [Cited by in RCA: 2373] [Article Influence: 339.0] [Reference Citation Analysis (0)] |
| 21. | McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, Jonasch E, Khasraw M, Heimberger AB, Lim B, Ueno NT, Litton JK, Ferrarotto R, Chang JT, Moulder SL, Lin SY. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 754] [Article Influence: 188.5] [Reference Citation Analysis (0)] |
| 22. | van der Velden BHM, Kuijf HJ, Gilhuijs KGA, Viergever MA. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal. 2022;79:102470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 292] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 23. | Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun (Lond). 2020;40:154-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 240] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 24. | Chen Y, Wang J, Wang C, Liu M, Zou Q. Deep learning models for disease-associated circRNA prediction: a review. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 25. | Li S, Li W, Ma T, Fu S, Gao X, Qin N, Wu Y, Zhang X, Wang J, Pan Y, Liu Z. Assessing the efficacy of immunotherapy in lung squamous carcinoma using artificial intelligence neural network. Front Immunol. 2022;13:1024707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 26. | Li W, Zhang M, Cai S, Wu L, Li C, He Y, Yang G, Wang J, Pan Y. Neural network-based prognostic predictive tool for gastric cardiac cancer: the worldwide retrospective study. BioData Min. 2023;16:21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 27. | Li W, Lin S, He Y, Wang J, Pan Y. Deep learning survival model for colorectal cancer patients (DeepCRC) with Asian clinical data compared with different theories. Arch Med Sci. 2023;19:264-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 28. | Bajzikova M, Kovarova J, Coelho AR, Boukalova S, Oh S, Rohlenova K, Svec D, Hubackova S, Endaya B, Judasova K, Bezawork-Geleta A, Kluckova K, Chatre L, Zobalova R, Novakova A, Vanova K, Ezrova Z, Maghzal GJ, Magalhaes Novais S, Olsinova M, Krobova L, An YJ, Davidova E, Nahacka Z, Sobol M, Cunha-Oliveira T, Sandoval-Acuña C, Strnad H, Zhang T, Huynh T, Serafim TL, Hozak P, Sardao VA, Koopman WJH, Ricchetti M, Oliveira PJ, Kolar F, Kubista M, Truksa J, Dvorakova-Hortova K, Pacak K, Gurlich R, Stocker R, Zhou Y, Berridge MV, Park S, Dong L, Rohlena J, Neuzil J. Reactivation of Dihydroorotate Dehydrogenase-Driven Pyrimidine Biosynthesis Restores Tumor Growth of Respiration-Deficient Cancer Cells. Cell Metab. 2019;29:399-416.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 29. | Yang S, Ji J, Wang M, Nie J, Wang S. Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA. Biomolecules. 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 30. | Li Y, Gong X, Hu T, Chen Y. Two novel prognostic models for ovarian cancer respectively based on ferroptosis and necroptosis. BMC Cancer. 2022;22:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 31. | Jiang J, Chen Z, Wang H, Wang Y, Zheng J, Guo Y, Jiang Y, Mo Z. Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data. Biomolecules. 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Chen Y, Deng Z, Sun Y. Development of a risk model based on autophagy-related genes to predict survival and immunotherapy response in ovarian cancer. Hereditas. 2023;160:4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 33. | Wang H, Liu J, Yang J, Wang Z, Zhang Z, Peng J, Wang Y, Hong L. A novel tumor mutational burden-based risk model predicts prognosis and correlates with immune infiltration in ovarian cancer. Front Immunol. 2022;13:943389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |