Published online Aug 24, 2024. doi: 10.5306/wjco.v15.i8.1061
Revised: July 10, 2024
Accepted: July 25, 2024
Published online: August 24, 2024
Processing time: 109 Days and 16.4 Hours
Chemoresistance is the primary contributor to distant metastasis in the context of neoadjuvant chemoradiotherapy (nCRT) for rectal cancer. However, the under
To detect the differential expression profiles of plasma exosomal microRNAs (miRNAs) in poor and good responders and explore the potential mechanisms of chemoresistance.
In this study, the profiles of plasma exosomal miRNAs were compared in two di
Exosome hsa-miR-483-5p was up-regulated in good responders post-nCRT. Bioin
This study provides insights into the mechanism of chemoresistance in terms of exosomal miRNAs. However, further research is required within the framework of our established miRNA-mRNA network.
Core Tip: Neoadjuvant chemoradiotherapy (nCRT) is the standard of care for the management of locally advanced rectal cancer; however, chemoresistance remains a life-threatening problem in patients with colorectal cancer. In this study, we used RNA sequencing to identify the expression profiles of plasma exosomal microRNAs (miRNAs) in two dimensions, according to treatment response (poor/good responders) and treatment course (pre/post-nCRT). Exosome hsa-miR-483-5p was upregulated in good responders post-nCRT and was chosen for further analysis. A network of hsa-miR-483-5p/MA
- Citation: Li GB, Shi WK, Zhang X, Qiu XY, Lin GL. Hsa-miR-483-5p/mRNA network that regulates chemotherapy resistance in locally advanced rectal cancer identified through plasma exosome transcriptomics. World J Clin Oncol 2024; 15(8): 1061-1077
- URL: https://www.wjgnet.com/2218-4333/full/v15/i8/1061.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i8.1061
Neoadjuvant chemoradiotherapy (nCRT), followed by total mesorectal excision, is the gold standard treatment for locally advanced rectal cancer (LARC). This approach has significant benefits, including a marked improvement in local tumor control and a notable reduction in the local recurrence rare[1,2]. Nonetheless, the distant metastasis rate persists at approximately 30% and is the primary contributor to disease-associated mortality[2]. Good responders to nCRT, as re
Exosomes are small membrane vesicles released by various cell types, with diameters ranging from 30 to 100 nm[5,6]. They function as vehicles for intercellular communication and carry various molecules, including microRNAs (miRNAs)[7]. By transporting miRNAs, exosomes influence gene expression and regulate cellular functions in target cells. The intricate interaction between exosomes and miRNAs is pivotal for maintaining effective intercellular communication, governing gene expression, and influencing crucial biological processes (BP). Notably, exosomal miRNAs are closely associated with tumor pathogenesis and progression[8-10]. The exosome miR-208b has been identified as a promoter of oxaliplatin-related chemoresistance by inhibiting the expression of PDCD4[10]. Exosomal miR-423-3p is a potential pre
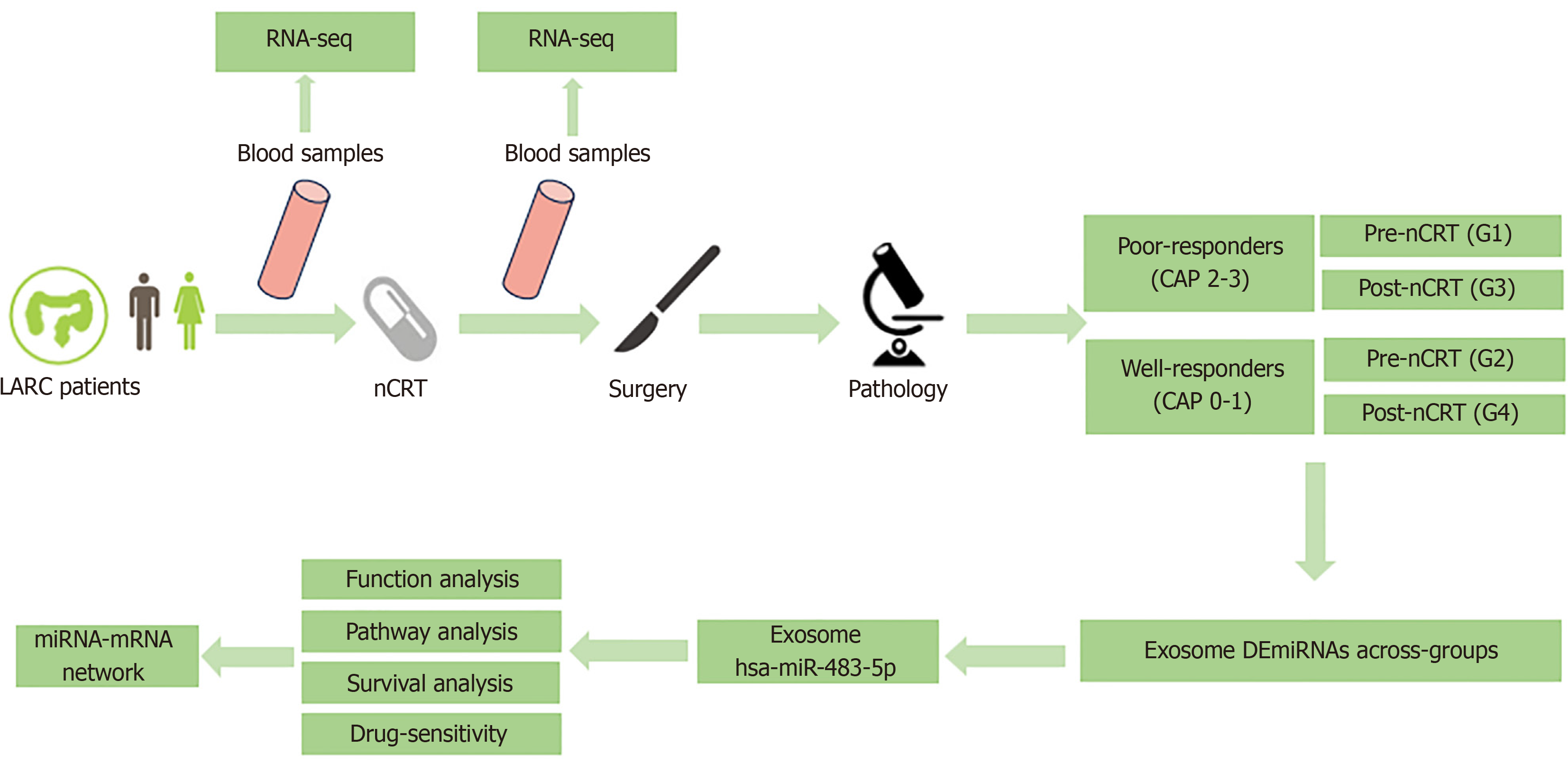
Hence, we designed this study to compare the differential expression profiles of plasma exosomal miRNAs between poor and good responders at pre- and post-nCRT time points using whole transcriptome RNA-seq. RNA-seq identified a set of dysregulated exosomal miRNAs, and several biological pathways and hub genes were identified using bioin
Fourteen patients diagnosed with rectal cancer were included in this study. Patients with pathologically confirmed adenocarcinoma (cT3-4 or cN1-2) who were scheduled to receive neoadjuvant long-course radiotherapy (45 Gy/25 fractions) concurrently with three cycles of oxaliplatin and capecitabine, followed by radical surgery, were enrolled. The degree of tumor response following nCRT was evaluated using the CAP standards (Table 1). Patients were categorized as responders (CAP 0-1) and poor responders (CAP 2-3).
| The criteria of CAP standards for the assessment of tumor response to nCRT | ||
| Good responder | CAP0 | Complete response with no viable tumor cells |
| CAP1 | Only small clusters or single cancer cells remained | |
| Poor-responder | CAP2 | Residual tumor cells remaining, but with predominant fibrosis |
| CAP3 | Poor response with extensive residual tumor cells | |
| The sub-group information of this study | ||
| Groups | Group 1 | Poor responders before nCRT |
| Group 2 | Good responders before nCRT | |
| Group 3 | Poor responders after nCRT | |
| Group 4 | Good responders after nCRT | |
Peripheral blood samples (20 mL) from these 14 patients at the pre- and post-nCRT time points were collected for RNA-seq. Subsequently, the samples were categorized into four groups based on the CAP grades and different time points: Poor responders pre-nCRT, good responders pre-nCRT, poor responders post-nCRT, and good responders post-nCRT. Detailed information on the study design is shown in Figure 1. The study protocol was approved by the Ethics Committee of Peking Union Medical College Hospital (No. JS-3209). Written informed consent was obtained from all the participants.
Peripheral blood samples were collected before and after nCRT using EDTA tubes. Following centrifugation at 3000 × g for 15 minutes at 4 °C, plasma was preserved at -80 °C until use. The thawed plasma supernatant underwent a series of steps, including dilution, filtration, and ultracentrifugation (100000 × g, 2 hours, 4 °C) to obtain exosome pellets. Exosomes were isolated via size exclusion chromatography[11]. In brief, 1 mL of 0.8 μm filtered plasma was 1.5-fold diluted with PBS, purified using Exosupur® columns, eluted with 0.1 M PBS, and concentrated to 200 μL. Small RNA libraries (1-500 ng) were prepared using the QIAseq miRNA Library Kit, and library quality was assessed using an Agilent Bioanalyzer 2100 and qPCR. Sequencing was performed using an Illumina HiSeq platform. The reads identified known and predicted new miRNAs by comparison with miRbase and the Human Genome (GRCh38).
Read counts and transcripts per million were calculated based on the mapping results. Differential analysis was performed using the DESeq2 R package v.1.10.1 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). RNAs discovered by RNA-seq with an adjusted P value < 0.05 and an absolute log2 fold change > 1 were considered differentially expressed (DE).
The differential expression profiles of exosomal miRNAs among groups were presented in the forms of a heatmap and a volcano plot using R packages “pheatmap” version 1.0.12 (https://CRAN.R-project.org/package=pheatmap). After exploratory visualization, a sophisticated least absolute shrinkage and selection operator (LASSO) regression analysis was performed using R version 4.3.1.
Target genes of the identified DE miRNAs were extracted from three databases: TargetScan (http://www.targetscan.org/), miRPath (https://mpd.bioinf.uni-sb.de/), and miRDB (http://mirdb.org/). The selected common target genes were visualized using Venn diagrams (https://omics.pnl.gov/software/venns). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of target genes were conducted using DAVID (https://david.ncifcrf.gov) and KOBAS (http://kobas.cbi.pku.edu.cn/kobas3), respectively. GO comprised three categories: BP, molecular functions, and cellular components. The SangerBox platform (http://sangerbox.com) was us11s used to identify the top 10 hub genes within the protein-protein interaction (PPI) network. Results were visualized using Cytoscape (v3.10.1).
To further explore the functions of the target genes, survival analysis was performed online using the TCGA database (https://kmplot.com/analysis/). The subgroup criteria were colorectal cancer (CRC), without distant metastasis, five-year recurrence-free survival (5y-RFS), and overall survival (OS). The study cohort included 625 patients with CRC. Based on the median expression profiles of the target genes, all patients were stratified into low- and high-expression groups, and then the 5y-RFS and OS between groups were compared using Kaplan-Meier curves and log-rank tests. Correlations between the expression profiles of the target genes and tumor stage in patients with CRC were also analyzed using the online TCGA database (http://sangerbox.com/home.html).
All the patients received an oxaliplatin-based chemotherapy regimen. To investigate the molecular mechanisms of DE miRNAs and their target genes in individuals with varying responses, we conducted a drug sensitivity analysis using the GDSC database (https://ngdc.cncb.ac.cn/). The expression profiles of the target genes and the oxaliplatin-associated IC50 values in six CRC cell lines (HCT116, HT-29, SW480, SW620, COLO-205, and HCT116) were extracted from the GDSC database. Subsequently, we categorized the target genes into low- and high-expression groups based on their median levels in the cell lines. Comparisons of IC50 values between these groups were performed to assess differences.
SPSS 25.0 (IBM Corp., Armonk, NY, United States) was used for statistical analysis, and GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, United States) was used to draw the figures. Normally distributed variables were reported as mean ± SD, and categorical data were reported as numbers and percentages (%). Between-group differences were compared using Student’s t-test. P values < 0.05 were considered statistically significant.
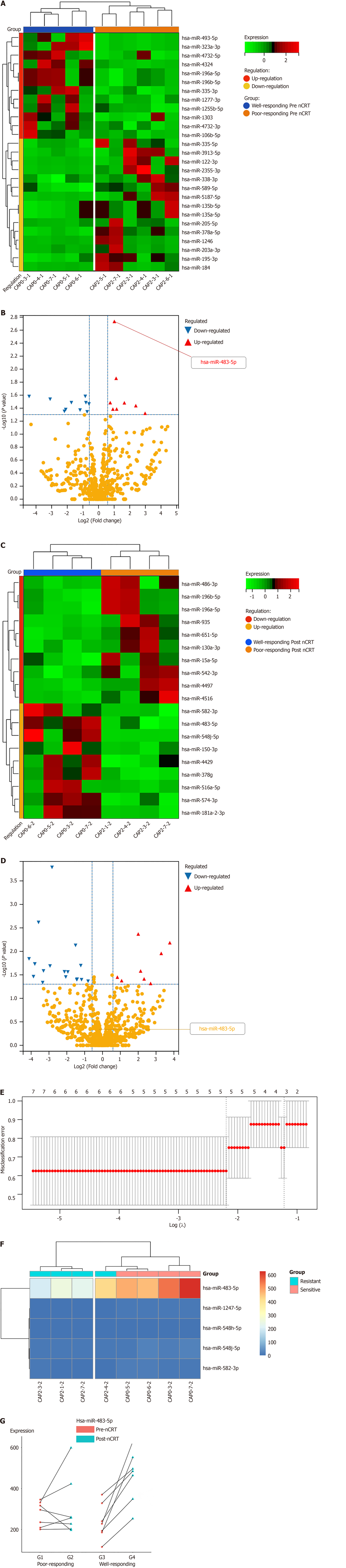
RNA-seq identified 1167 exosomal miRNAs, including 106 newly discovered miRNAs. Initially, the expression profiles of plasma exosomal miRNAs pre-nCRT were compared between poor and good responders, and 27 DEmiRNAs (19 downregulated and 8 upregulated) were identified (Figure 2A and B). The detection of candidate DEmiRNAs pre-nCRT may enable us to predict the treatment response to nCRT. In addition, compared to poor responders, approximately 19 exosomal DEmiRNAs (10 downregulated and 9 upregulated) post-nCRT were identified in good responders (Figure 2C and D). The top five upregulated and downregulated exosomal DEmiRNAs are listed in Table 2.
| Pre-nCRT (G1 vs G2) | Post-nCRT (G3 vs G4) | ||||||
| miRNAs | Log2FC | P value | Regulation | miRNAs | Log2FC | P value | Regulation |
| MiR-493-5p | 4.64 | 0.001 | Up | MiR-516a-5p | 4.74 | 0.041 | Up |
| MiR-332a-3p | 3.85 | 0.018 | Up | MiR-378 g | 2.98 | 0.047 | Up |
| MiR-4324 | 2.89 | 0.043 | Up | MiR-150-3p | 2.38 | 0.036 | Up |
| MiR-335-3p | 2.33 | 0.001 | Up | MiR-483-5p | 1.65 | 0.001 | Up |
| MiR-1303 | 2.25 | 0.048 | Up | MiR-582-3p | 1.16 | 0.032 | Up |
| MiR-2355-3p | -4.61 | 0.006 | Down | MiR-4497 | -4.44 | 0.026 | Down |
| MiR-184 | -4.37 | 0.027 | Down | MiR-935 | -3.09 | 0.029 | Down |
| MiR-5187-5p | -4.33 | 0.015 | Down | MiR-4516 | -2.18 | 0.044 | Down |
| MiR-122-3p | -4.37 | 0.027 | Down | MiR-486-3p | -2.09 | 0.041 | Down |
| MiR-378a-5p | -3.49 | 0.001 | Down | MiR-651-5p | -1.73 | 0.032 | Down |
The profiles of plasma exosomal miRNAs also exhibited differences at the pre- and post-nCRT time points. During the treatment course, 21 DEmiRNAs (10 downregulated and 11 upregulated) and 36 DEmiRNAs (21 downregulated and 15 upregulated) were identified in poor responders (Supplementary Figure 1A and B) and good responders (Supplementary Figure 1C and D), respectively. The top five upregulated and downregulated DE miRNAs are listed in Supplementary Table 1.
The identification of exosomal DE miRNAs pre-nCRT would enable the selection of potential responders to chemoradiation. Additionally, distinct DEmiRNAs profiles observed post-nCRT may serve as valuable indicators for predicting patient prognosis. LASSO regression analysis was used to select potential exosomal DEmiRNAs that might play a significant role in the response to nCRT. Thus, hsa-miR-483-5p was identified as a prognostic indicator for further analyses (Figure 2E and F).
Comparative analysis between groups revealed an upregulation of exosome hsa-miR-483-5p in good responders post-nCRT, in contrast to poor responders. Furthermore, within the subgroup of good responders, hsa-miR-483-5p exhibited increased expression post-nCRT compared with its pre-nCRT levels, as illustrated in Figure 2G. These results emphasize the potential utility of hsa-miR-483-5p as a predictive biomarker for the treatment response and prognosis of patients with LARC in the context of nCRT.
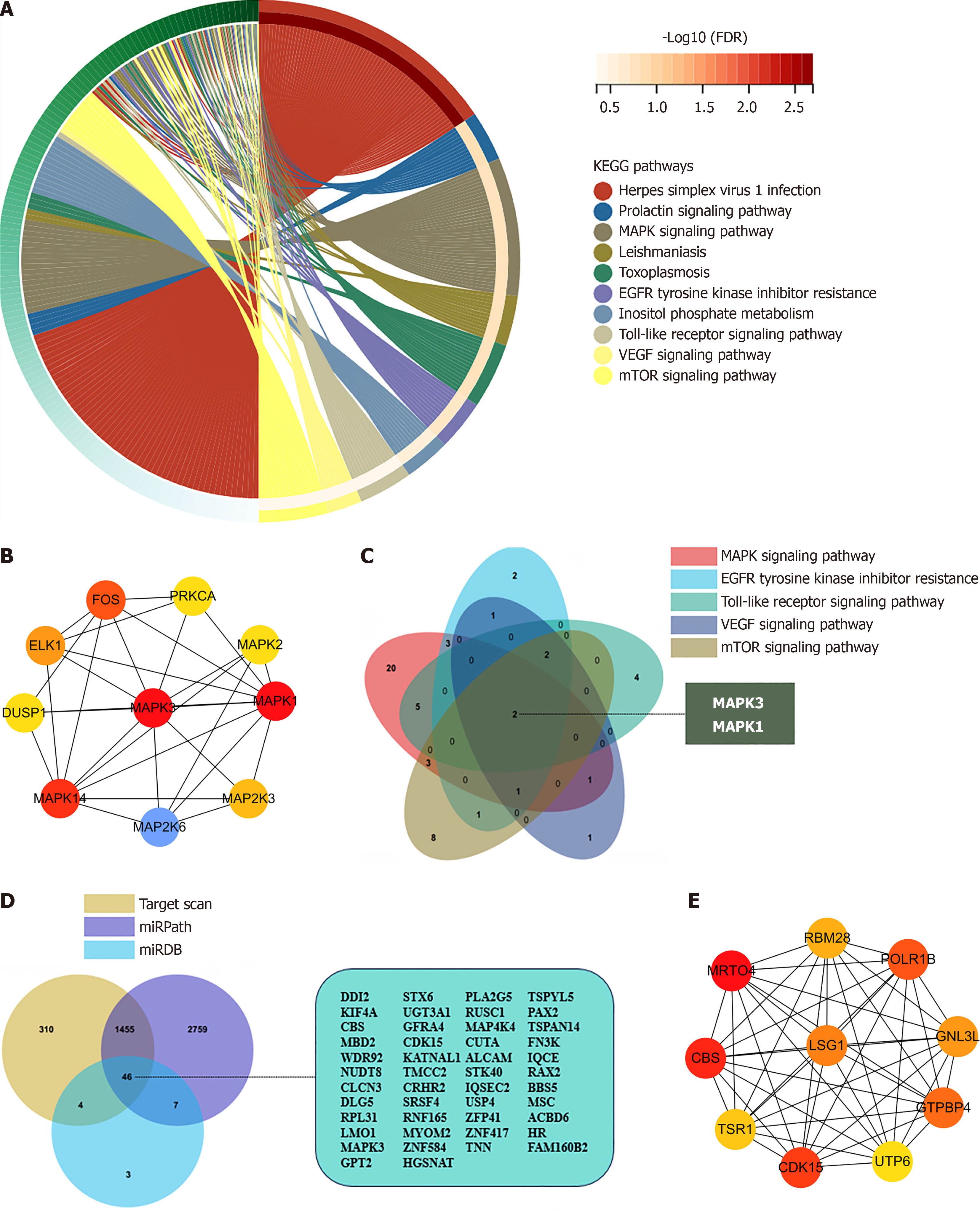
To further explore the potential functions of hsa-miR-483-5p and its target genes, a comprehensive bioinformatics an
The target genes of hsa-miR-483-5p were identified using online databases, with 1816 target genes identified in TargetScan, 4266 genes in miRPath, and 59 genes in miRDB. Subsequently, comprehensive analysis identified approximately 46 common target genes in the three databases (Figure 3D). PPI analysis of these 46 common target genes also identified ten hub genes, as shown in Figure 3E. This convergence of genes across multiple databases underscores their potential significance and serves as a robust foundation for further in-depth analyses and explorations.
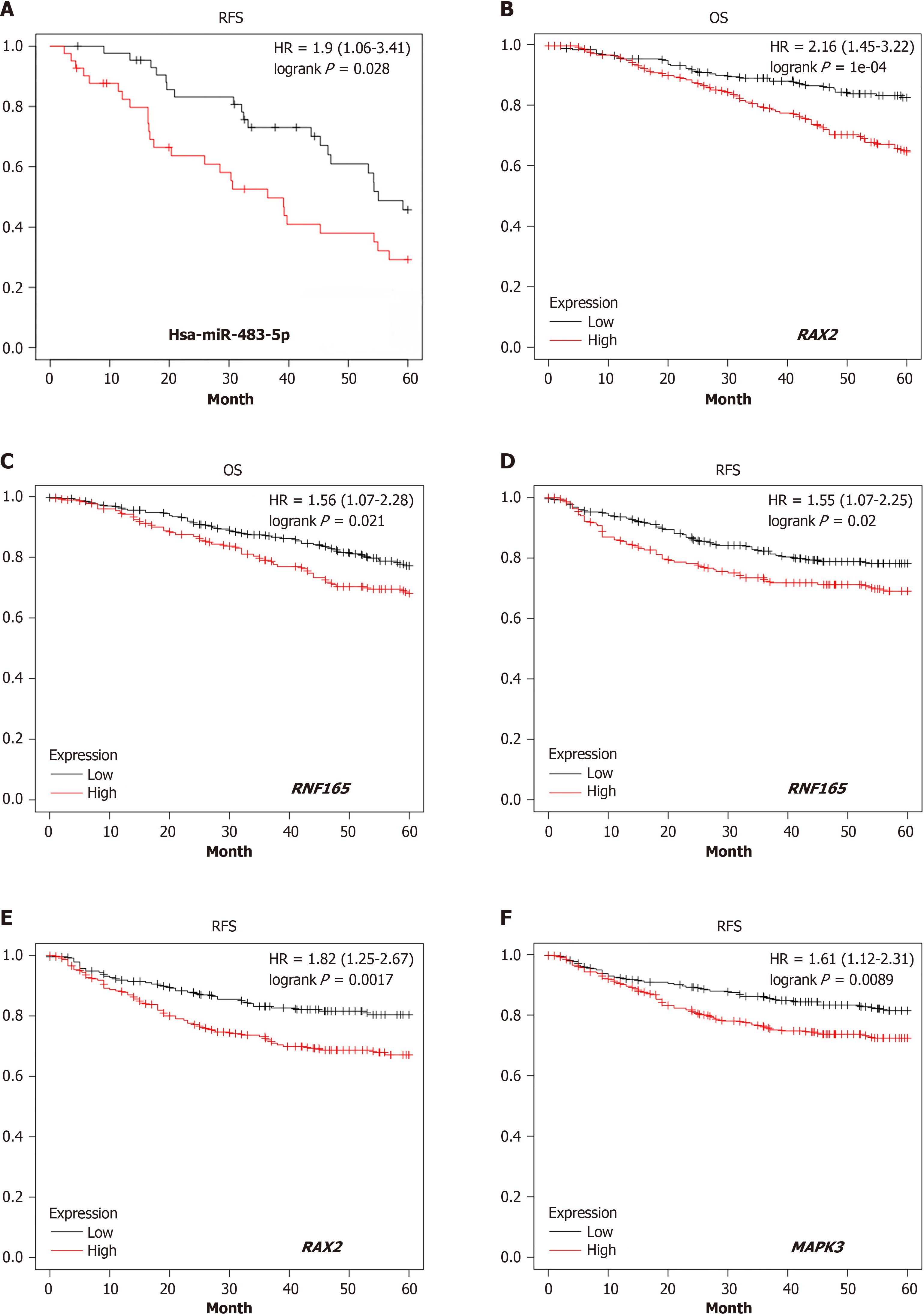
The TCGA database was used for survival analysis. The results indicated that upregulation of hsa-miR-483-5p correlated with worse RFS in breast cancer, demonstrating its potential role in the pathogenesis and progression of tumors (Figure 4A). Forty-six selected target genes were analyzed for RFS and OS. The results demonstrated that low expression of 22 target genes was correlated with superior RFS in patients with CRC (9 genes were positively correlated and 13 genes were negatively correlated) (Supplementary Figure 3A), and 21 target genes were correlated with superior OS (7 genes were positively correlated and 14 genes were negatively correlated) (Supplementary Figure 3B). We also discovered that the low expression of two target genes, RAX2 and RNF 165, was correlated with relatively better RFS and OS in patients with CRC (Figure 4B-E). In addition, low expression of MAPK3, selected from the above five KEGG pathways, was correlated with better RFS (Figure 4F).
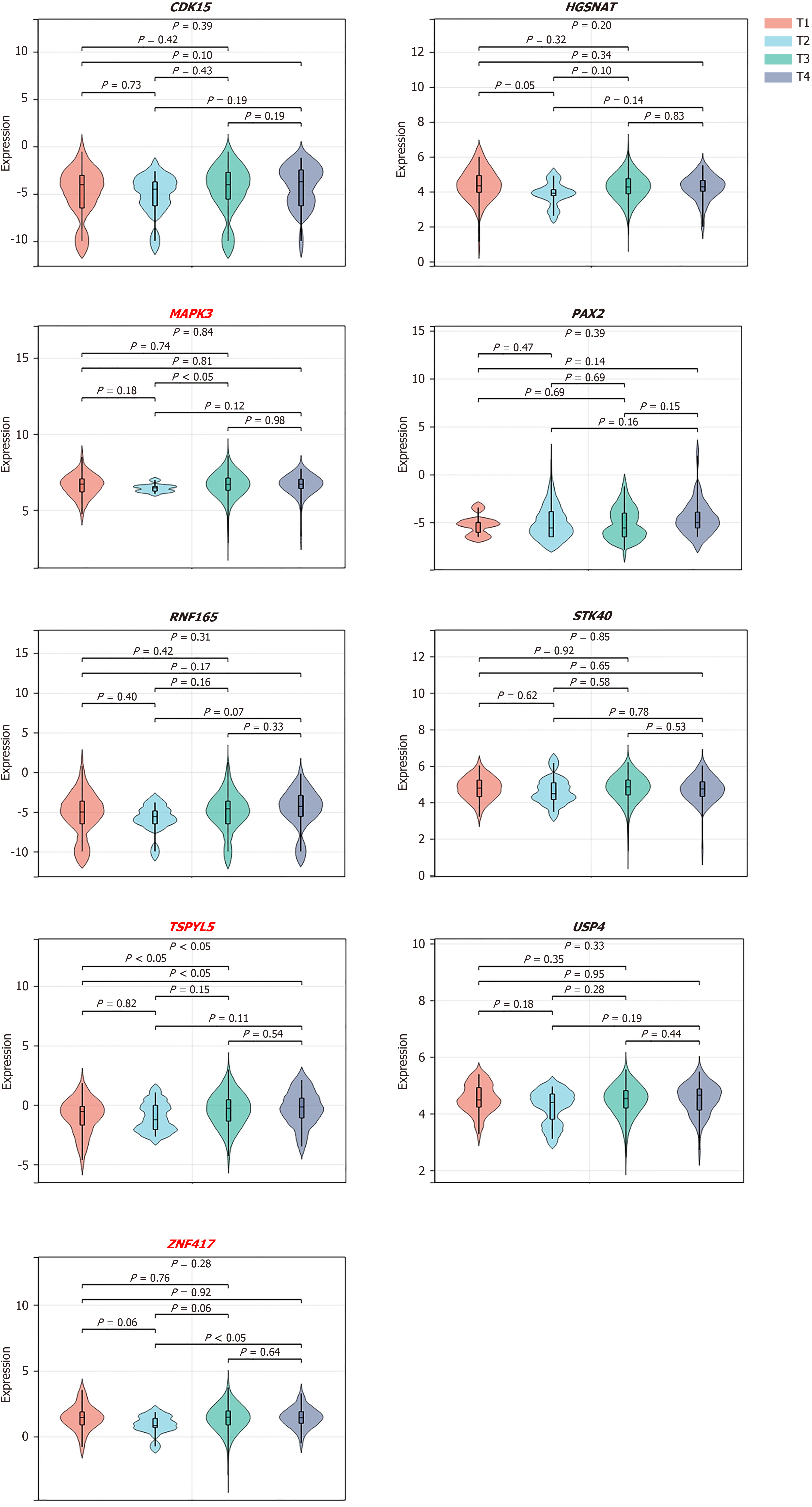
In addition, nine target genes that were positively correlated with RFS, including CDK15, HGSNAT, MAPK3, PAX2, RNF165, STK40, TSPYL5, USP4, and ZNF417, were selected to evaluate the relationships between their expression profiles and tumor stage (Figure 5). The results revealed that low expression of the target genes, MAPK3, TSPYL5, and ZNF417, was related to a relatively early tumor stage, further demonstrating its positive impact on survival outcomes.
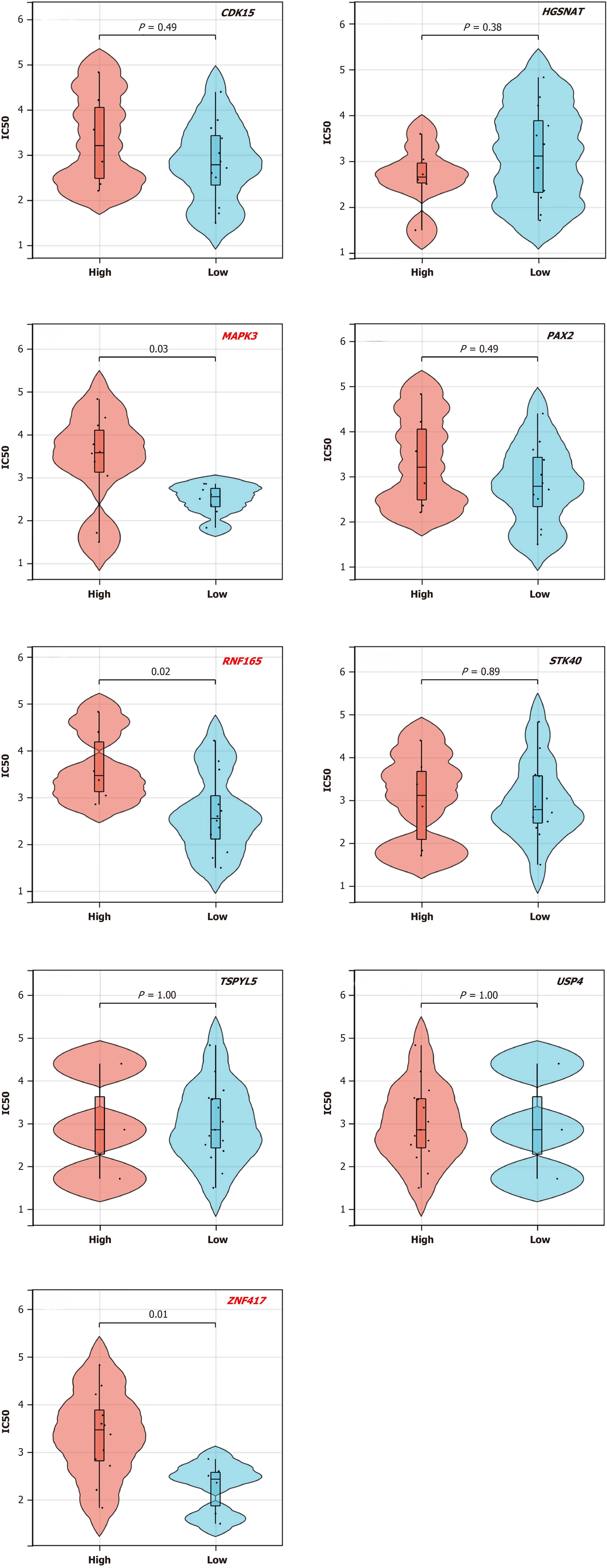
Oxaliplatin and capecitabine are the foundations of current chemotherapy regimens for patients with LARC, and sensitivity analysis of tumor cells to oxaliplatin could enable us to further elucidate the mechanisms of chemoresistance. Drug sensitivity analyses on the nine target genes selected from the survival analysis was performed using the GDSC database. The results indicated that low expression of MAPK3, RNF165, and ZNF417 was associated with decreased IC50 value (Figure 6). The lower the IC50 value, the better the tumor response to chemotherapy. Therefore, these three target genes may play important roles in chemotherapy sensitivity.
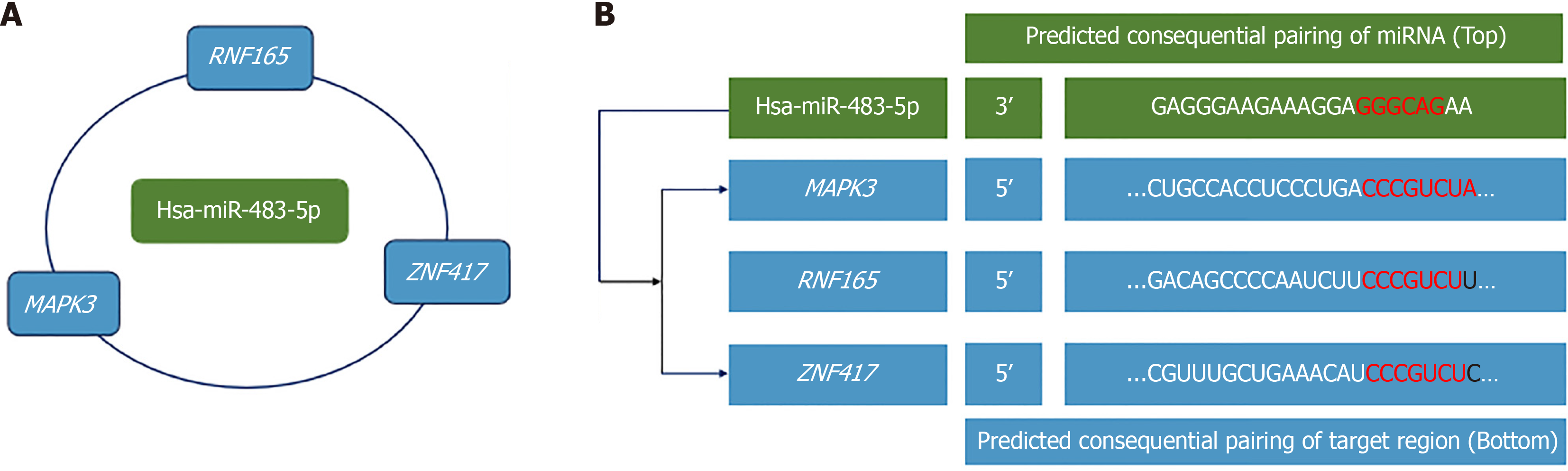
Based on the above analysis, the target genes of hsa-miR-483-5p, MAPK3, RNF 165, and ZNF417 were finally selected, and networks of hsa-miR-483-5p-mRNAs were constructed and are presented in Figure 7A. The potential binding sites of hsa-miR-483-5p and its target genes are also listed in Figure 7B.
Approximately one-third of patients with LARC do not benefit from the current standard nCRT, leading to distant metastasis. Chemoresistance may play a crucial role in tumor metastasis. Understanding the molecular alterations in poor and good responders during nCRT may enable us to explore the potential mechanisms of chemoresistance. RNA-seq identified several plasma exosomal DEmiRNAs in poor and good responders and revealed their potential functions by bioinformatic analysis. In addition, the expression profiles of exosomal DE miRNAs in LARC patients pre- and post-nCRT were determined.
miRNAs have been shown to be correlated with chemoresistance[12,13]. Chen et al[14] demonstrated that decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhi
In this study, the expression profiles of exosomal DEmiRNAs were compared in two dimensions according to the treatment response (poor responders vs good responders) and treatment courses (pre-nCRT vs post-nCRT). The results identified exosomal hsa-miR-483-5p, which was upregulated in good responders post-nCRT, as a candidate miRNA to further explore the mechanism of chemoresistance. Although no previous studies have demonstrated the functions of hsa-miR-483-5p in CRC, especially in chemoresistance, several studies have demonstrated that dysregulation of hsa-miR-483-5p is associated with relatively inferior survival outcomes in adrenocortical cancer[20], hepatocellular carcinoma[21], esophageal cancer[22], and breast cancer[23]. Accordingly, exosomal hsa-miR-483-5p might play an important role in the pathogenesis and progression of CRC, as well as in the process of chemoresistance, as indicated through the RNA-seq analysis.
The physiological roles of miRNAs depend on their capacity to post-transcriptionally regulate gene expression. Conse
To better explore the functions of hsa-miR-483-5p, bioinformatic analysis was conducted on its target genes, and as a result, several tumor-specific KEGG pathways were enriched. Fang and Richardson[24] demonstrated that MAPK sig
Upregulation of exosomal hsa-miR-483-5p in responders is associated with better survival outcomes. Moreover, hsa-miR-483-5p serves as a molecular sponge that inhibits the expression of its target genes. Accordingly, the selected target genes of hsa-miR-483-5p were crucial for exploring the mechanisms involved in chemoresistance.
In this study, the target genes MAPK3, RNF 165, and ZNF417 were identified in the miRNA-mRNA regulatory network. Several studies have illustrated the role of MAPK3 in the pathogenesis of CRC[30,31]. MAPK3, which encodes ERK1, is a crucial gene within the MAPK family that participates in several crucial cellular processes, including cell proliferation and differentiation, as well as the regulation of cell survival and apoptosis[30,31]. Over-activation may be associated with cancer development and treatment responses[24,32]. Although bioinformatics analysis identified that lower expression of RNF165 and ZNF417 was predicted to have a superior RFS, few studies have focused on the biological function of these two target genes in the setting of CRC, especially during nCRT. However, further molecular experiments are required to elucidate their exact functions.
These hub genes, particularly MAPK3, RNF165, and ZNF417, are pivotal players in the molecular networks associated with the identified pathways. Their consistent presence suggests their potential role in mediating biological responses related to tumor-specific signaling pathways. This comprehensive analysis enhances our understanding of the regulatory landscape of hsa-miR-483-5p and its implicated pathways.
In conclusion, the upregulation of exosomal hsa-miR-483-5p reflects a relatively better tumor response to nCRT and thus a superior survival outcome. An interactive network of hsa-miR-483-5p/MAPK3/RNF165/ZNF417 was constructed using bioinformatic analysis. Further research is required to elucidate the mechanisms of chemoresistance within the framework of our miRNA-mRNA network.
| 1. | Rödel C, Graeven U, Fietkau R, Hohenberger W, Hothorn T, Arnold D, Hofheinz RD, Ghadimi M, Wolff HA, Lang-Welzenbach M, Raab HR, Wittekind C, Ströbel P, Staib L, Wilhelm M, Grabenbauer GG, Hoffmanns H, Lindemann F, Schlenska-Lange A, Folprecht G, Sauer R, Liersch T; German Rectal Cancer Study Group. Oxaliplatin added to fluorouracil-based preoperative chemoradiotherapy and postoperative chemotherapy of locally advanced rectal cancer (the German CAO/ARO/AIO-04 study): final results of the multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2015;16:979-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 535] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 2. | Hong YS, Nam BH, Kim KP, Kim JE, Park SJ, Park YS, Park JO, Kim SY, Kim TY, Kim JH, Ahn JB, Lim SB, Yu CS, Kim JC, Yun SH, Kim JH, Park JH, Park HC, Jung KH, Kim TW. Oxaliplatin, fluorouracil, and leucovorin versus fluorouracil and leucovorin as adjuvant chemotherapy for locally advanced rectal cancer after preoperative chemoradiotherapy (ADORE): an open-label, multicentre, phase 2, randomised controlled trial. Lancet Oncol. 2014;15:1245-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 288] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 3. | Borg C, André T, Mantion G, Boudghène F, Mornex F, Maingon P, Adenis A, Azria D, Piutti M, Morsli O, Bosset JF. Pathological response and safety of two neoadjuvant strategies with bevacizumab in MRI-defined locally advanced T3 resectable rectal cancer: a randomized, noncomparative phase II study. Ann Oncol. 2014;25:2205-2210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Carlomagno C, Farella A, Bucci L, D'Armiento FP, Pesce G, Pepe S, Cannella L, Pacelli R, De Stefano A, Solla R, D'Armiento MR, De Placido S. Neo-adjuvant treatment of rectal cancer with capecitabine and oxaliplatin in combination with radiotherapy: a phase II study. Ann Oncol. 2009;20:906-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6517] [Article Influence: 1303.4] [Reference Citation Analysis (0)] |
| 6. | Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 1818] [Article Influence: 363.6] [Reference Citation Analysis (0)] |
| 7. | Tran N. Cancer Exosomes as miRNA Factories. Trends Cancer. 2016;2:329-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, Tan W, Nandy D, Bevan GH, Longenbach S, Sun Z, Lu Y, Wang T, Thibodeau SN; Boardman L; Kohli M, Wang L. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 507] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 9. | Guo T, Wang Y, Jia J, Mao X, Stankiewicz E, Scandura G, Burke E, Xu L, Marzec J, Davies CR, Lu JJ, Rajan P, Grey A, Tipples K, Hines J, Kudahetti S, Oliver T, Powles T, Alifrangis C, Kohli M, Shaw G, Wang W, Feng N, Shamash J, Berney D, Wang L, Lu YJ. The Identification of Plasma Exosomal miR-423-3p as a Potential Predictive Biomarker for Prostate Cancer Castration-Resistance Development by Plasma Exosomal miRNA Sequencing. Front Cell Dev Biol. 2020;8:602493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 10. | Ning T, Li J, He Y, Zhang H, Wang X, Deng T, Liu R, Li H, Bai M, Fan Q, Zhu K, Ying G, Ba Y. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol Ther. 2021;29:2723-2736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 11. | Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 854] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 12. | Al-Harbi S, Choudhary GS, Ebron JS, Hill BT, Vivekanathan N, Ting AH, Radivoyevitch T, Smith MR, Shukla GC, Almasan A. miR-377-dependent BCL-xL regulation drives chemotherapeutic resistance in B-cell lymphoid malignancies. Mol Cancer. 2015;14:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 14. | Chen W, Wang P, Lu Y, Jin T, Lei X, Liu M, Zhuang P, Liao J, Lin Z, Li B, Peng Y, Pan G, Lv X, Zhang H, Ou Z, Xie S, Lin X, Sun S, Ferrone S, Tannous BA, Ruan Y, Li J, Fan S. Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation. Theranostics. 2019;9:5739-5754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Gasiulė S, Dreize N, Kaupinis A, Ražanskas R, Čiupas L, Stankevičius V, Kapustina Ž, Laurinavičius A, Valius M, Vilkaitis G. Molecular Insights into miRNA-Driven Resistance to 5-Fluorouracil and Oxaliplatin Chemotherapy: miR-23b Modulates the Epithelial–Mesenchymal Transition of Colorectal Cancer Cells. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J, Thomas GJ, Packham G, Mirnezami AH. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73:6435-6447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 17. | Wang H, Wang X, Zhang H, Deng T, Liu R, Liu Y, Li H, Bai M, Ning T, Wang J, Ge S, Ba Y. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene. 2021;40:4695-4708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 18. | Wang L, Zhang X, Sheng L, Qiu C, Luo R. LINC00473 promotes the Taxol resistance via miR-15a in colorectal cancer. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Liu Z, Qin Y, Dong S, Chen X, Huo Z, Zhen Z. Overexpression of miR-106a enhances oxaliplatin sensitivity of colorectal cancer through regulation of FOXQ1. Oncol Lett. 2020;19:663-670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Soon PS, Tacon LJ, Gill AJ, Bambach CP, Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson BG, Sidhu SB. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin Cancer Res. 2009;15:7684-7692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 205] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 21. | Shen J, Wang A, Wang Q, Gurvich I, Siegel AB, Remotti H, Santella RM. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol Biomarkers Prev. 2013;22:2364-2373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Chen Y, Wang H, Zhu S, Lan X. miR-483-5p promotes esophageal cancer progression by targeting KCNQ1. Biochem Biophys Res Commun. 2020;531:615-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 23. | Ren J, Xu G, Sun H, Lin T, Xu S, Zhao Y. Inhibition of miR-483-5p improves the proliferation, invasion and inflammatory response of triple-negative breast cancer cells by targeting SOCS3. Exp Ther Med. 2021;22:1047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 859] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 25. | Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 26. | Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, Wang R, Peng Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 27. | Angius A, Pira G, Scanu AM, Uva P, Sotgiu G, Saderi L, Manca A, Serra C, Uleri E, Piu C, Caocci M, Ibba G, Zinellu A, Cesaraccio MR, Sanges F, Muroni MR, Dolei A, Cossu-Rocca P, De Miglio MR. MicroRNA-425-5p Expression Affects BRAF/RAS/MAPK Pathways In Colorectal Cancers. Int J Med Sci. 2019;16:1480-1491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Moradi-Marjaneh R, Hassanian SM, Fiuji H, Soleimanpour S, Ferns GA, Avan A, Khazaei M. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J Cell Physiol. 2018;233:5613-5622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Chen M, Zhong K, Tan J, Meng M, Liu CM, Chen B, Huang C, Wong HLX, Bian Z, Su T, Kwan HY. Baicalein is a novel TLR4-targeting therapeutics agent that inhibits TLR4/HIF-1α/VEGF signaling pathway in colorectal cancer. Clin Transl Med. 2021;11:e564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Li P, Zhou D, Chen D, Cheng Y, Chen Y, Lin Z, Zhang X, Huang Z, Cai J, Huang W, Lin Y, Ke H, Long J, Zou Y, Ye S, Lan P. Tumor-secreted IFI35 promotes proliferation and cytotoxic activity of CD8(+) T cells through PI3K/AKT/mTOR signaling pathway in colorectal cancer. J Biomed Sci. 2023;30:47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 31. | Forcet C, Stein E, Pays L, Corset V, Llambi F, Tessier-Lavigne M, Mehlen P. Netrin-1-mediated axon outgrowth requires deleted in colorectal cancer-dependent MAPK activation. Nature. 2002;417:443-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 205] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 32. | He L, Li H, Pan C, Hua Y, Peng J, Zhou Z, Zhao Y, Lin M. Squalene epoxidase promotes colorectal cancer cell proliferation through accumulating calcitriol and activating CYP24A1-mediated MAPK signaling. Cancer Commun (Lond). 2021;41:726-746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |