Published online Mar 24, 2024. doi: 10.5306/wjco.v15.i3.434
Peer-review started: October 11, 2023
First decision: December 7, 2023
Revised: December 27, 2023
Accepted: February 5, 2024
Article in press: February 5, 2024
Published online: March 24, 2024
Processing time: 163 Days and 5.6 Hours
The ubiquitin-proteasome pathway (UPP) has been proven to play important roles in cancer.
To investigate the prognostic significance of genes involved in the UPP and develop a predictive model for liver cancer based on the expression of these genes.
In this study, UPP-related E1, E2, E3, deubiquitylating enzyme, and proteasome gene sets were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, aiming to screen the prognostic genes using univariate and multivariate regression analysis and develop a prognosis predictive model based on the Cancer Genome Atlas liver cancer cases.
Five genes (including autophagy related 10, proteasome 20S subunit alpha 8, proteasome 20S subunit beta 2, ubiquitin specific peptidase 17 like family member 2, and ubiquitin specific peptidase 8) were proven significantly correlated with prognosis and used to develop a prognosis predictive model for liver cancer. Among training, validation, and Gene Expression Omnibus sets, the overall survival differed significantly between the high-risk and low-risk groups. The expression of the five genes was significantly associated with immunocyte infiltration, tumor stage, and postoperative recurrence. A total of 111 differentially expressed genes (DEGs) were identified between the high-risk and low-risk groups and they were enriched in 20 and 5 gene ontology and KEGG pathways. Cell division cycle 20, Kelch repeat and BTB domain containing 11, and DDB1 and CUL4 associated factor 4 like 2 were the DEGs in the E3 gene set that correlated with survival.
We have constructed a prognosis predictive model in patients with liver cancer, which contains five genes that associate with immunocyte infiltration, tumor stage, and postoperative recurrence.
Core Tip: This study unveils the crucial role of the ubiquitin-proteasome pathway (UPP) in liver cancer prognosis. Five key genes (autophagy related 10, proteasome 20S subunit alpha 8, proteasome 20S subunit beta 2, ubiquitin specific peptidase 17 like family member 2, and ubiquitin specific peptidase 8) identified from The Cancer Genome Atlas datasets constitute a robust prognostic model, accurately predicting liver cancer outcomes. Immunocyte infiltration analysis highlights associations of these genes with immune cell abundance, while clinical correlations link them to tumor stage and recurrence. Differential gene expression and pathway enrichment elucidate underlying biological processes. E3 analysis identifies specific ligases (cell division cycle 20, Kelch repeat and BTB domain containing 11, and DCAF4L2) with significant expression differences, further emphasizing the integral role of the UPP in liver cancer development and providing valuable insights for precision medicine and prognosis prediction.
- Citation: Li H, Ma YP, Wang HL, Tian CJ, Guo YX, Zhang HB, Liu XM, Liu PF. Establishment of a prognosis predictive model for liver cancer based on expression of genes involved in the ubiquitin-proteasome pathway. World J Clin Oncol 2024; 15(3): 434-446
- URL: https://www.wjgnet.com/2218-4333/full/v15/i3/434.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i3.434
The prevalence of liver cancer has been increasing, with an annual growth rate of up to 2%-3%[1] and survival rate of 18% in 2020[2]. A total of 336400 new liver cancer cases were detected in China in 2016[3], and the sharply elevated incidence (18.0 per 100000) of liver cancer caused by sugar-sweetened food must be given extra attention[4].
Hepatitis B/C virus (HBV or HCV) infection, addiction to alcohol, liver cirrhosis, fatty hepatitis, and eating aflatoxin contaminated food are the risk factors for liver cancer[5]. Imaging examinations for liver cancer include ultrasonography, dynamic contrast-enhanced computed tomography (CT), multimodal magnetic resonance imaging, 18F-fluorodeoxyglucose positron emission tomography/CT, and so on. Virtual liver biopsy sampling pipeline for eliminating sampling bias may be the potential diagnostic method to investigate the nature of the lesions and etiology[6]. In recent years, using statistical models combined with machine learning techniques to elevate the diagnostic accuracy of serum biomarkers such as α-fetoprotein and cell-free DNA or RNA has been widely applied to the early diagnosis of hepatocellular carcinoma[7]. Additionally, surgical resection, transplantation, ablation, chemotherapy, and immunotherapy are common treatment options for liver cancer patients[8]. However, effective surveillance and prediction of the prognosis of liver cancer still face multiple challenges due to the high heterogeneity of this malignancy.
The ubiquitin-proteasome pathway (UPP) is one of the key pathways of protein selective degradation in organisms[9], which is related to cell cycle, proliferation, differentiation, apoptosis, transcription, signal transduction, immune response, stress response, and extracellular effectors[10]. The malfunction of the UPP is linked to various diseases, such as carcinogenesis, infection, autoimmunity, and inflammation. Based on The Cancer Genome Atlas (TCGA) datasets and 961 ubiquitin-proteasome system genes (UPSGs), Liu et al[11] found that DDB1 and CUL4 associated factor 13 (DCAF13), cell division cycle 20 (CDC20), and proteasome 20S subunit beta 5 (PSMB5) have excellent performance to predict the survival of liver cancer patients. Zhang et al[12] identified a seven-UPSG prognostic signature, of which autophagy related 10
The UPP-related gene set included 857 genes from the UPP-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways[13], among which 10 was related to E1, 38 related to E2, 651 to E3, 112 to deubiquitylating enzyme (DUB), and 46 to the proteasome.
The expression data of 424 samples related to liver cancer were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Three recurrent samples, 50 normal tissue samples, and one sample without overall survival (OS) data were deleted and the remaining 370 samples were randomly divided into a training group (n = 296) and a validation group (n = 74) in a ratio of 4:1. Another validation set (GSE54236) was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). This data set included 162 samples, including 81 tumor samples.
Univariate and multivariate regression analyses were performed to screen the prognostic genes in the E1, E2, DUB, and proteasome gene sets using the Survival (version 3.2-3) and Glmnet (version 4.0-2) packages in R. The threshold of univariate analysis was P < 0.1, and stepwise multivariate regression analysis was used to screen genes associated with OS. The risk score of the screened genes was calculated to construct a prognosis predictive model, and the prognostic ability was assessed using the receiver operating characteristic (ROC) curve drawn with Proc (version 1.16.2) package. According to the risk score, the patients were divided into either a high-risk or a low-risk group. The Maxstat (version 0.7-25) package in R was used to calculate the optimal cut-off value. The log-rank method was used to compare the difference in OS between the two groups, and the Survival (version 3.2-3) and Survminer (version 0.4.8) packages in R were used to draw the survival curve. In the validation group, the same method was used to verify the model.
The abundance of 40 types of immune cells in each sample was analyzed using GVSA (version 1.32.0). The correlation analysis between the screened genes and immune-related indicators was performed using the Psych (version 2.0.8) and Corrplot (version 0.84) packages in R.
The clinical parameters were compared between the high-risk and the low-risk groups using an independent sample t-test or two-sample Wilcoxon test, and Spearman correlation analysis was used to determine whether the gene expression and risk scores were statistically related to clinical parameters. The Psych (version 2.0.8) and Corrplot (version 0.84) packages in R were used for plotting. Univariate Cox regression analysis was used to determine the relationship between OS and clinical parameters, as well as the relationship between the gene expression and postoperative recurrence.
The Limma package (version 3.40.2) was used to screen DEGs between the high-risk and low-risk groups, with the threshold set at P < 0.05 and |log2FC| > 1. Then the functional enrichment analysis was carried out using the database for annotation, visualization, and integrated discovery (DAVID, https://david.ncifcrf.gov/) to identify the enriched gene ontology (GO) terms and KEGG pathways of the DEGs.
As a specific substrate recognition element, E3 plays an important role in the ubiquitin-mediated proteolytic cascade[14]. Because of its specificity, the relationship between the expression of genes in the E3 set and prognostic risk was analyzed separately. Similar to the screening method for DEGs, the Limma package was used to screen the DEGs in the E3 gene set between the high-risk and low-risk groups, and the screening threshold was P < 0.05 and |log2FC| > 1.
IBM SPSS Statistics 21 and R (version 3.6.2) were used for statistical analyses. The Shapiro-Wilk test was used for normality test, and the independent sample t test or the two-sample Wilcoxon test were used to analyze the differences in variables between two groups. The chi-square test or Fisher's test was used for analysis of categorical variables. The log-rank method was used to test the significance of survival data.
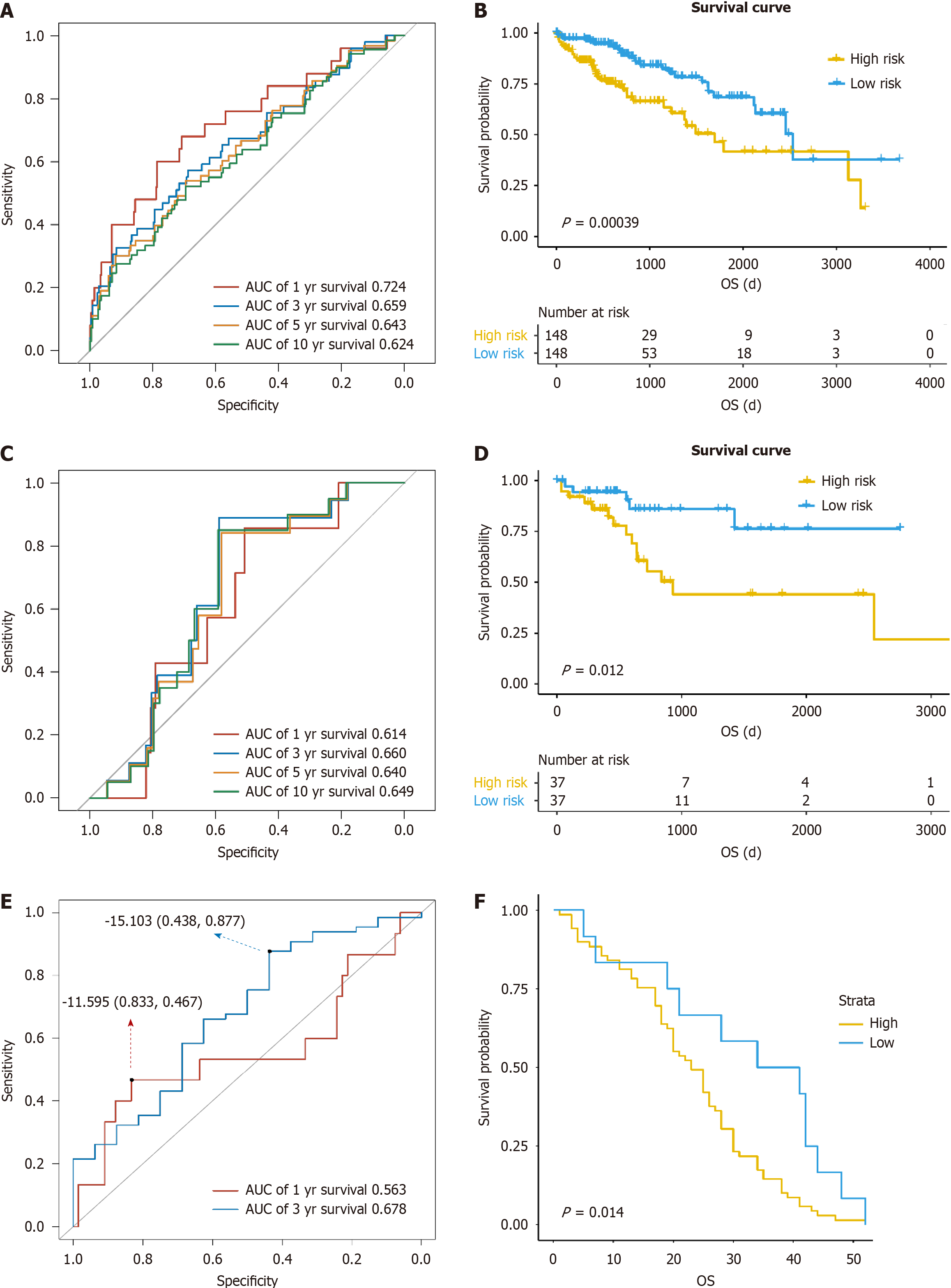
The clinical parameters of the whole samples, cases in the training group, and those in the validation group are listed in Table 1, with P value referring to the statistical results between the training group and the validation group. A total of five genes significantly related to prognosis were screened to construct a prognosis predictive model, including ATG10, proteasome 20S subunit alpha 8 (PSMA8), proteasome 20S subunit beta 2 (PSMB2), ubiquitin specific peptidase 17 like family member 2 (USP17L2), and ubiquitin specific peptidase 8 (USP8) (Table 2). In the training group, the area under the curve (AUC) values of the model for predicting 1-, 3-, 5-, and 10-year survival were 0.724, 0.659, 0.643, and 0.624, respectively (Figure 1A). All patients were classified into either a high-risk or low-risk group according to the score of risk. There was a significant difference in survival time between the high-risk and low-risk groups (P < 0.001, Figure 1B). In the validation group, the AUC values of the model for predicting 1-, 3-, 5-, and 10-year survival were 0.614, 0.66, 0.64, and 0.649, respectively, and the low-risk group exhibited a higher survival probability than the high-risk group (P = 0.012, Figure 1C and D), suggesting that the model can well predict the prognosis in liver cancer patients. In the GSE54236 data set, the AUC values of the model for predicting 1- and 3-year survival were 0.563 and 0.678, respectively, and the cases could be divided into a high-risk group and low-risk group by the risk score. There was a significantly difference in survival time between the high-risk group and low-risk group (P = 0.014, Figure 1E and F).
| Parameter | Category | TCGA (n = 370) | Training group (n = 296) | Validation group (n = 74) |
| Age | 59.441 ± 13.517 | 59.53 ± 13.71 | 59.081 ± 12.796 | |
| P value | 0.799 | |||
| Gender | Female | 121 (32.7) | 101 (34.1) | 20 (27) |
| Male | 249 (67.3) | 195 (65.9) | 54 (73) | |
| P value | 0.27 | |||
| Height | 167.34 ± 10.7 | 167.32 ± 11.368 | 167.43 ± 7.622 | |
| P value | 0.92 | |||
| Weight | 72.85 ± 19.468 | 73.05 ± 20.571 | 72.07 ± 14.478 | |
| P value | 0.646 | |||
| BMI | 26.13 ± 8.453 | 26.25 ± 9.145 | 25.66 ± 4.909 | |
| P value | 0.609 | |||
| Histological type | FLC | 3 (0.8) | 2 (0.7) | 1 (1.4) |
| HCC | 360 (97.3) | 287 (97) | 73 (98.6) | |
| FLHCC | 7 (1.9) | 7 (2.4) | 0 (0) | |
| P value | 0.35 | |||
| Stage | I/II | 256 (69.2) | 209 (69.6) | 47 (63.5) |
| III/IV | 90 (24.4) | 69 (23.3) | 21 (28.4) | |
| Not available | 24 (6.5) | 18 (6.1) | 6 (8.1) | |
| P value | 0.454 | |||
| Grade | G1/G2 | 232 (62.7) | 180 (60.8) | 52 (70.3) |
| G3/G4 | 133 (35.9) | 112 (37.9) | 21 (28.4) | |
| Not available | 5 (1.4) | 4 (1.4) | 1 (1.4) | |
| P value | 0.241 | |||
| Gene | Gene set | Coef | Hazard ratio | Z | P value |
| ATG10 | Ubiquitin-conjugating enzyme (E2) | 0.48387 | 1.62234 | 2.364 | 0.0181 |
| PSMA8 | Proteasome | 0.20721 | 1.23024 | 1.962 | 0.0497 |
| PSMB2 | Proteasome | 0.66763 | 1.94962 | 2.483 | 0.013 |
| USP17L2 | Deubiquitinating enzyme (DUB) | -2.8057 | 0.06046 | -3.048 | 0.0023 |
| USP8 | Deubiquitinating enzyme (DUB) | -0.46594 | 0.62755 | -1.701 | 0.0889 |
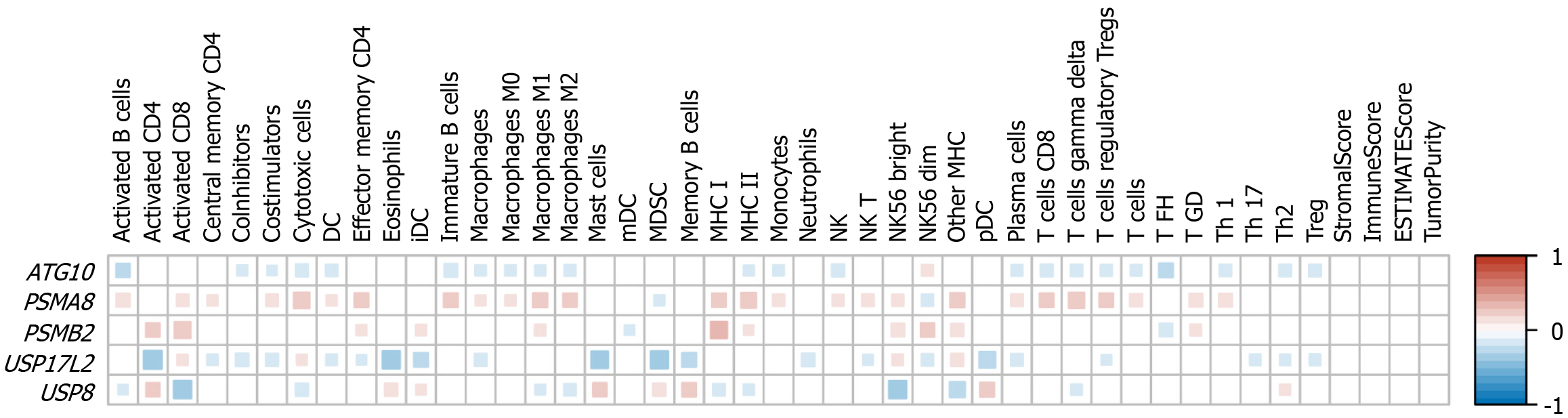
Through the above analysis, we found that the expression levels of the five genes can predict the prognosis of liver cancer. Because immunocyte infiltration is commonly affected by gene expression, we then studied the correlation of the expression levels of the five genes with the abundance of 40 types of immune cells. PSMA8 was associated with the abundance of the most immune cells, and the abundance of 28 immune cell types was significantly correlated with PSMA8 expression levels. This was followed by USP17L2, ATG10, USP8, and PSMB2, with 25, 23, 18, and 13 types of immune cells that were related to the expression levels of these genes, respectively (Figure 2). The abundance of most cells was negatively correlated with the expression levels of ATG10, USP17L2, and USP8, while PSMA8 and PSMB2 expression levels were positively correlated with the abundance of most cell types (Figure 2).
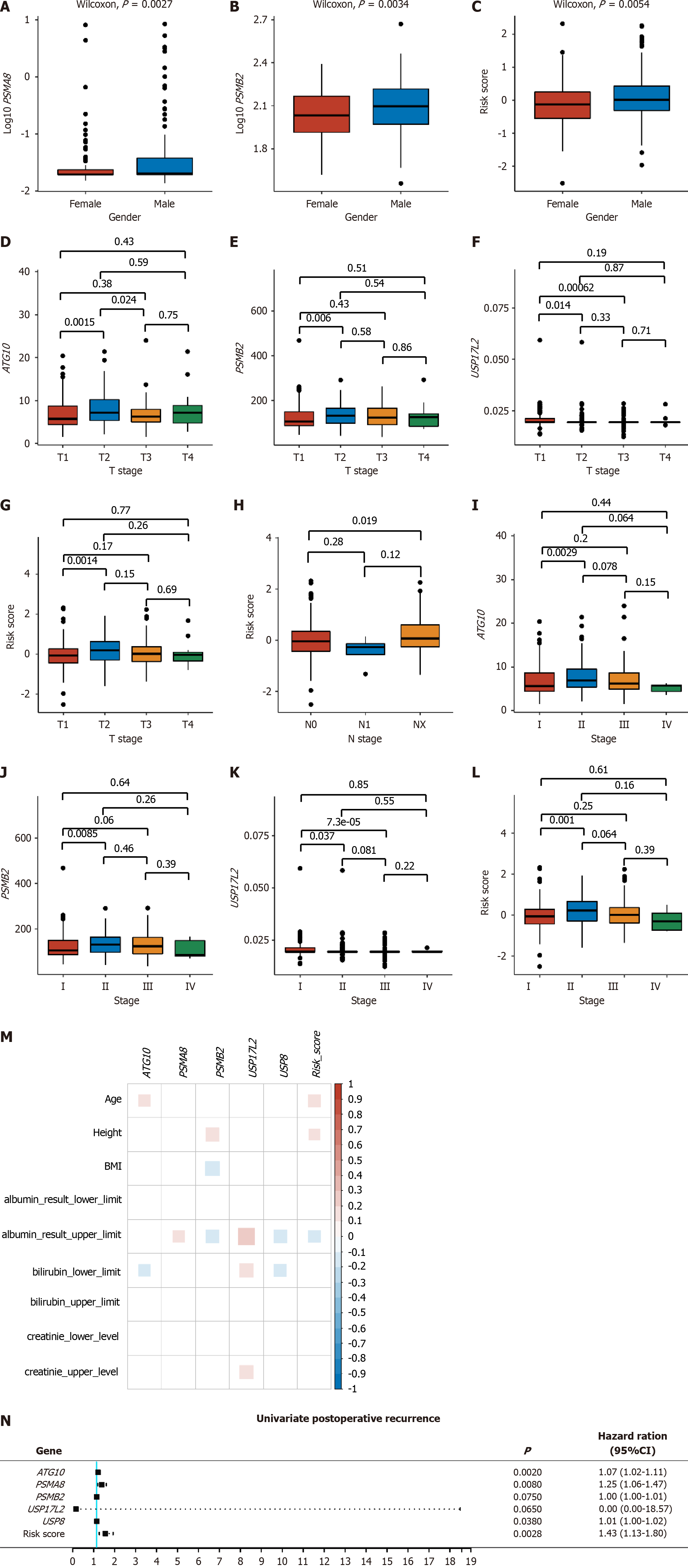
PSMA8 and PSMB2 expression and the risk score were significantly different between males and females (Figure 3A-C). For pathological and clinical stages, ATG10 expression and the risk score were significantly different between T2 and T3 stages, PSMB2 and USP17L2 expression was significantly different between T1 and T3 stages (Figure 3D-G), the risk score was statistically lower in N0 stage than in NX stage (Figure 3H), and the expression levels of ATG10, PSMB2, and USP17L2 and the risk score were significantly different between stage I and stage II (Figure 3I-L). Moreover, PSMA8, PSMB2, USP17L2, and USP8 expression was all correlated with the upper limit of albumin results, among which PSMA8 and USP17L2 were positively correlated, and PSMB2 and USP8 were negatively correlated with albumin results (Figure 3M). There was also a negative correlation between the risk score and the upper limit of albumin results, indicating that as the risk value increased, the albumin levels decreased, leading to an elevated prognosis risk for patients (Figure 3M).
Postoperative recurrence included extrahepatic recurrence, local recurrence, intrahepatic recurrence, and new primary tumor. After univariate Cox analysis, ATG10, PSMA8, and USP8, as well as the risk score, were found significantly correlated with postoperative recurrence (P < 0.05, Figure 3N).
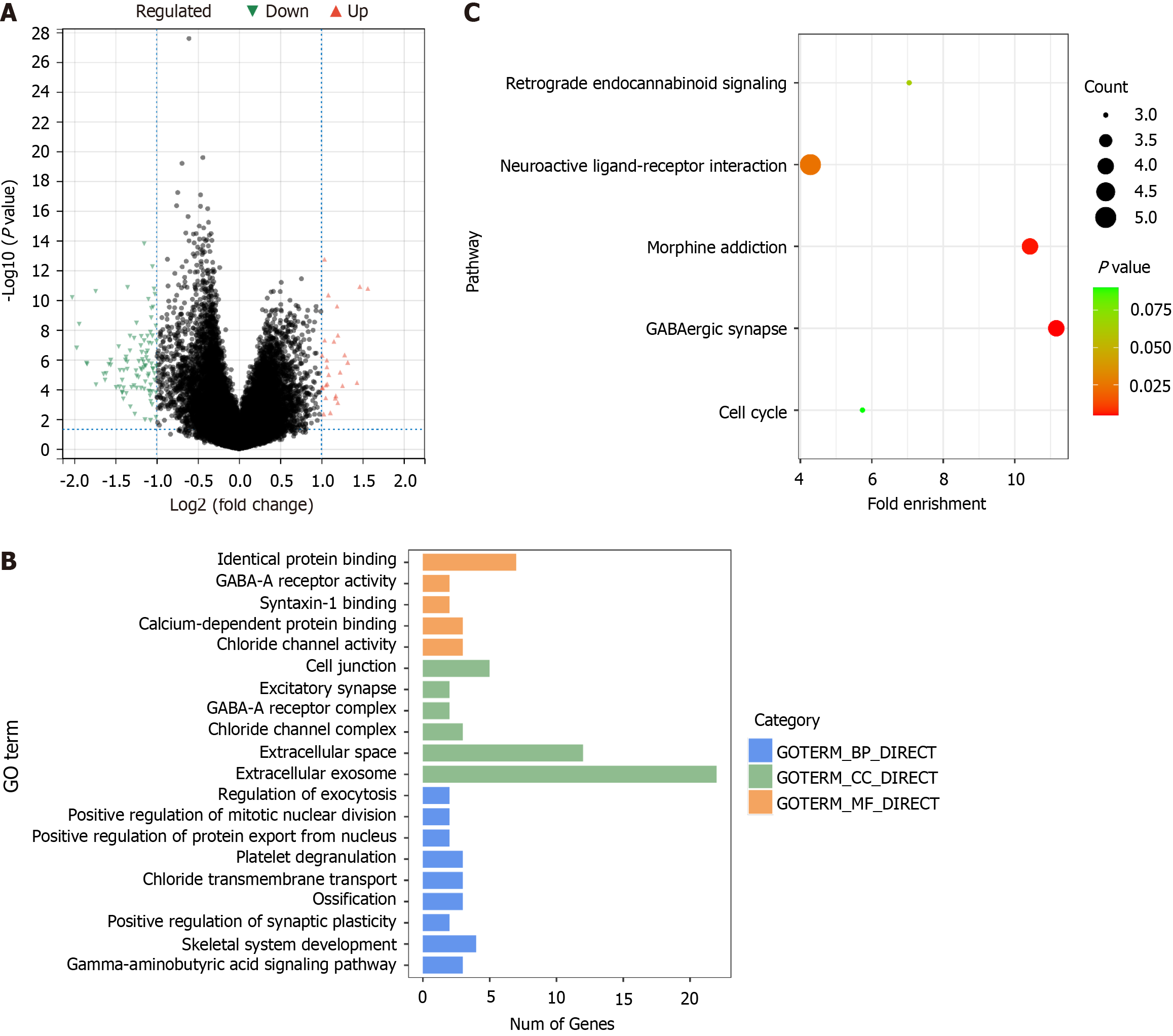
A total of 111 DEGs were screened out between the high-risk group and low-risk group, among which 27 were up-regulated and 84 down-regulated (Figure 4A). These DEGs were associated with 20 GO terms, comprising 9 biological processes, 6 cellular components, and 5 molecular functions (Figure 4B). Five KEGG pathways enriched were GABAergic synapse, morphine addiction, neuroactive ligand-receptor interaction, retrograde endocannabinoid signaling, and cell cycle (Figure 4C).
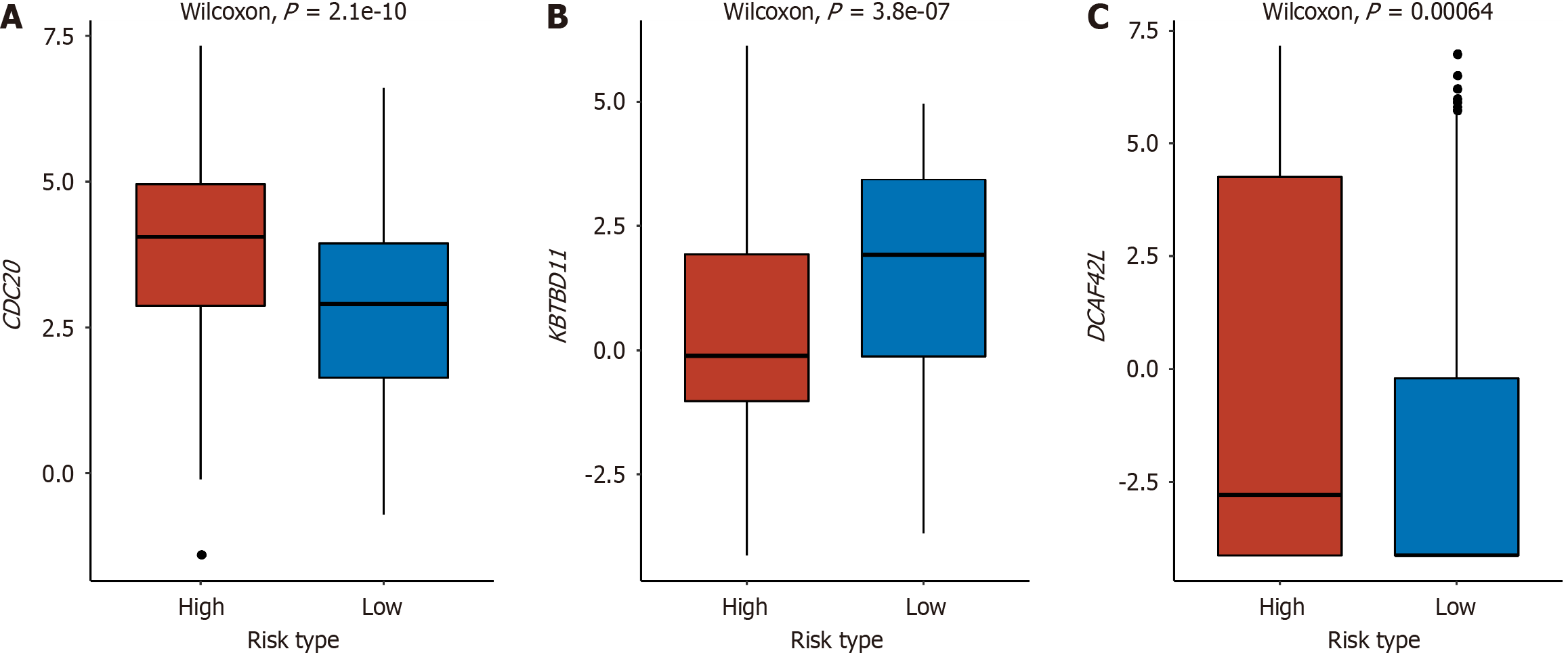
Between the high-risk and low-risk groups, significant differences were observed in three genes within the E3 gene set: CDC20, Kelch repeat and BTB domain containing 11 (KBTBD11), and DDB1 and CUL4 associated factor 4 like 2 (DCAF4L2). In the high-risk group, CDC20 and DCAF4L2 exhibited elevated expression levels, whereas KBTBD11 showed higher expression in the low-risk group. This suggested a negative correlation between the expression of CDC20 and DCAF4L2 and survival, while KBTBD11 displayed a positive correlation with the prognosis of liver cancer (Figure 5).
The key factor to cell survival lies in the balance of protein synthesis and decomposition. The UPP is an ATP-dependent non-lysosomal protein degradation pathway, which is important for the body to regulate the level and function of intracellular proteins, thus efficiently and selectively degrading intracellular proteins. This study showed that the expression of the UPP genes ATG10, PSMA8, PSMB2, USP17L2, and USP8 was significantly correlated with the prognosis of liver cancer. The prognosis model constructed based on these five genes could accurately predict the prognosis of patients (P < 0.001 and P = 0.012 in training and validation groups, respectively, Figure 1). These genes were statistically correlated with different clinical parameters and immune cell abundance (Figures 2 and 3). The model categorized all patients into either a high-risk group or a low-risk group, and a total of 111 DEGs were screened between the two groups, which were enriched in GO terms related to protein binding, GABA-A receptor, synapse, etc., and KEGG pathways of retrograde endocannabinoid signaling, neuroactive ligand-receptor interaction, morphine addiction, GABAergic synapse, and cell cycle (Figure 4).
Those five genes were found to promote the development of many malignant tumors, including liver cancer[15-22]. Our results showed that the increased expression of ATG10, PSMA8, and PSMB2 increased the risk of death (P = 0.018, 0.049, and 0.013, respectively), while the increased expression of USP17L2 and USP8 decreased the risk of death (P = 0.002 and 0.089, respectively). According to previous studies, the overexpression of ATG10 and PSMB2 in tumors promoted the invasion or metastasis of tumor cells[16,18], and USP8 showed the opposite effect[21,22]. Besides, PSMA8 could affect the progression and prognosis of colorectal cancer due to its strong association with PSMB2[23]. Interestingly, higher PSMA8 expression levels were correlated with good prognoses for breast cancer through epigenetic regulation[24]. In our study, it was found that PSMA8 was positively correlated with the prognosis of patients with liver cancer. On the contrary, USP17L2 has been found to be overexpressed in a variety of tumors[19,20], which is similar to our results. However, recent studies have found that up-regulation of USP17L2 causes chemotherapy resistance in colorectal cancer, and knockdown of USP17L2 could overcome bromodomain and extra-terminal domain inhibitor resistance in prostate cancer cells[25,26]. Hence, the role of USP17L2 in liver cancer still requires further exploration.
The global immune system functions pose great technical challenges to the research of tumor-immune interaction[27,28]. Because immune infiltration plays a key role in the development of liver cancer[27], we conducted a thorough correlation analysis to identify the immune cell types associated with the prognosis model. Minor alterations in the distribution of immune cells could potentially exert diverse impacts on the progression of tumors[29]. In this study, myeloid dendritic cells were the immune cell type with a significant difference in abundance only between groups with high and low expression of the PSMB2 gene, as well as neutrophils and Th17 cells between groups with different expression of the USP17L2 gene (Figure 2). However, no significant correlation was found between tumor-infiltrating immune cells and gene expression, and it is imperative to conduct additional confirmation and validation in an independent cohort. Furthermore, exploring the connection between the expression levels of some checkpoints and immune infiltration, as well as the tumor microenvironment, will be a hotspot for future research.
Moreover, the expression of one or more of the five genes and the risk score were different among different T, N, and clinical stages (Figure 3). It is widely known that tumor stage is a key prognostic factor for malignant tumors[30]. In addition, all genes except ATG10 and the risk score were correlated with the upper limit of albumin results (Figure 3). The risk score was not only statistically significant in different stages, but also negatively correlated with the upper limit of albumin results and postoperative recurrence, which proves that the model developed in this study has appreciated value in clinical prediction of recurrence and prognosis.
E3 is the key factor in the UPP, which can specifically recognize different substrates and show high selectivity in protein degradation. Therefore, we analyzed the E3 gene set independently of E1, E2, DUB, and proteasome-related genes. Finally, the expression levels of CDC20, KBTBD11, and DCAF4L2 were identified as significantly different between the high-risk and low-risk groups, which were also included in the above 111 DEGs. CDC20 plays a vital role in chromosome segregation and mitosis[31]. It regulates the stability of phosphorylated mitotic centromere-associated kinesin in metaphase-anaphase transition[32], which may play a role as a cancer protein to promote the development and progression of liver cancer. In the study of Zheng et al[33], CDC20, proliferating cell nuclear antigen, and minichromosome maintenance complex component 6 synergistically affect the regulation of the cell cycle and may be potential prognostic factors for liver cancer. Shi et al[34] found that CDC20 serves as a crucial factor in the development of hepatocellular carcinoma (HCC) by controlling the prolyl-4-hydroxylase domain 3 protein. By analyzing four expression profiles from the GEO database, it was found that the up-regulation of CDC20 in HCC tissues indicates poor OS and disease-free survival[35]. Recently, KBTBD11 was identified as a newly discovered adipogenesis-related gene[36]. In diverse cancer types, such as colorectal cancer, HCC, and head and neck squamous cell carcinoma, the expression of KBTBD11 was significantly decreased in tumor tissues as compared to normal tissues[37]. This is consistent with our result that patients in the high-risk group had lower KBTBD11 gene expression levels. DCAF4L2 is a member of the E3 complex, which is usually used as a mediator of protein-protein interaction and negatively regulates NF-κB signal transduction. Overexpression of DCAF4L2 has been observed in human colon cancer[38]. In a study of HCC, overexpression of DCAF4L2 is a common feature of nonalcoholic steatohepatitis-associated HCC and viral hepatitis-associated HCC, which can be used as a candidate therapeutic target for HCC[39]. We also found overexpression of DCAF4L2 in high-risk patients, which suggested a poor prognosis in patients with liver cancer.
One of the main shortcomings of this study is the lack of clinical cases. All the data were from TCGA and GEO, resulting in the lack of clinical data for some patients, and it was unable to validate the expression of the five genes and comprehensively analyze their correlation with clinical and prognostic indicators. This is a preliminary study, and the results reported are exploratory. We intend to validate these results and the detailed mechanisms in future studies.
In conclusion, we have used gene expression data in TCGA to screen genes involved in the UPP pathway that significantly correlate with the prognosis of liver cancer. Our findings indicate that the UPP plays an important role in the development of liver cancer, which provides new insights into the early prediction of prognosis and precision medicine in liver cancer.
The ubiquitin-proteasome pathway (UPP) is crucial for selective protein degradation, and its dysfunction is linked to various diseases, including cancer. Proteasome inhibitors are emerging as potential anti-tumor drugs. This study explored the association between UPP gene expression and liver cancer prognosis, aiming to identify key genes and develop a predictive model. By doing so, the research seeks to offer novel insights into the role and potential mechanisms of the UPP in liver cancer development, contributing to the ongoing exploration of effective therapeutic strategies for liver cancer.
Due to the high tumor heterogeneity, effective surveillance and predication of the prognosis of liver cancer still face multiple challenges. This study was performed to analyze the relationship between the expression of genes in the UPP and the prognosis of liver cancer and construct a prognosis predictive model for this malignancy.
The study aimed to investigate the prognostic significance of genes in the UPP in liver cancer. Using gene expression data from The Cancer Genome Atlas (TCGA) and gene expression comprehensive (GEO) databases, the study identified key genes involved in the UPP, constructed a prognostic predictive model for liver cancer, and explored the associations of the model with immune cell infiltration and clinical parameters, in order to enhance liver cancer prognosis prediction and provide insights into the role and potential mechanisms of the UPP in liver cancer development, contributing valuable information for precision medicine in the context of liver cancer management.
The research employed diverse methodologies, utilizing UPP-related gene sets and patient data from TCGA and GEO databases. A prognostic model was constructed using univariate and multivariate regression analyses, involving five key genes (ATG10, PSMA8, PSMB2, USP17L2, and USP8). The model demonstrated robust predictive abilities for liver cancer prognosis. Immunocyte infiltration analysis and correlation studies with clinical parameters provided additional insights. Differentially expressed genes and enrichment analyses shed light on relevant pathways. The study's comprehensive approach contributes a nuanced understanding of UPP gene implications in liver cancer prognosis.
This study investigated the role of the UPP in liver cancer, identifying five key genes (ATG10, PSMA8, PSMB2, USP17L2, and USP8) associated with prognosis. A predictive model was constructed and validated using TCGA and GEO datasets. The study highlighted differential gene expression between the high- and low-risk groups and enriched relevant pathways. Additionally, differentially expressed genes in the E3 gene set (CDC20, KBTBD11, and DCAF4L2) were identified as significant. The findings provide valuable insights into liver cancer prognosis, immunology, and potential therapeutic targets.
We have used gene expression data in TCGA to screen genes in the UPP that significantly correlated with the prognosis of liver cancer. Our findings indicate that the UPP plays an important role in the development of liver cancer, which provides new insights into the early prediction of prognosis and precision medicine in liver cancer.
This is a preliminary study, and the results reported are exploratory. We intend to validate these results and the detailed mechanisms in future studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Rodriguez JC, Spain S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Zheng XM
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64562] [Article Influence: 16140.5] [Reference Citation Analysis (176)] |
| 2. | Cui Y, Li H, Zhan H, Han T, Dong Y, Tian C, Guo Y, Yan F, Dai D, Liu P. Identification of Potential Biomarkers for Liver Cancer Through Gene Mutation and Clinical Characteristics. Front Oncol. 2021;11:733478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | Maomao C, He L, Dianqin S, Siyi H, Xinxin Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P, Wanqing C. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19:1121-1138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 161] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 4. | Zhao L, Zhang X, Coday M, Garcia DO, Li X, Mossavar-Rahmani Y, Naughton MJ, Lopez-Pentecost M, Saquib N, Shadyab AH, Simon MS, Snetselaar LG, Tabung FK, Tobias DK, VoPham T, McGlynn KA, Sesso HD, Giovannucci E, Manson JE, Hu FB, Tinker LF. Sugar-Sweetened and Artificially Sweetened Beverages and Risk of Liver Cancer and Chronic Liver Disease Mortality. JAMA. 2023;330:537-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 42] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 5. | Zhou J, Sun H, Wang Z, Cong W, Wang J, Zeng M, Zhou W, Bie P, Liu L, Wen T, Han G, Wang M, Liu R, Lu L, Ren Z, Chen M, Zeng Z, Liang P, Liang C, Yan F, Wang W, Ji Y, Yun J, Cai D, Chen Y, Cheng W, Cheng S, Dai C, Guo W, Hua B, Huang X, Jia W, Li Y, Liang J, Liu T, Lv G, Mao Y, Peng T, Ren W, Shi H, Shi G, Tao K, Wang X, Xiang B, Xing B, Xu J, Yang J, Yang Y, Ye S, Yin Z, Zhang B, Zhang L, Zhang S, Zhang T, Zhao Y, Zheng H, Zhu J, Zhu K, Shi Y, Xiao Y, Dai Z, Teng G, Cai J, Cai X, Li Q, Shen F, Qin S, Dong J, Fan J. Guidelines for the Diagnosis and Treatment of Hepatocellular Carcinoma (2019 Edition). Liver Cancer. 2020;9:682-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 573] [Article Influence: 114.6] [Reference Citation Analysis (1)] |
| 6. | Li Q, Wang F, Chen Y, Chen H, Wu S, Farris AB, Jiang Y, Kong J. Virtual liver needle biopsy from reconstructed three-dimensional histopathological images: Quantification of sampling error. Comput Biol Med. 2022;147:105764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19:670-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 165] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 8. | Li Y, Zhang R, Xu Z, Wang Z. Advances in Nanoliposomes for the Diagnosis and Treatment of Liver Cancer. Int J Nanomedicine. 2022;17:909-925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 9. | Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 241] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 10. | Staszczak M. [Ubiquitin-proteasome pathway as a target for therapeutic strategies]. Postepy Biochem. 2017;63:287-303. [PubMed] |
| 11. | Liu ZY, Li YH, Zhang QK, Li BW, Xin L. Development and validation of a ubiquitin-proteasome system gene signature for prognostic prediction and immune microenvironment evaluation in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2023;149:13363-13382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 12. | Zhang J, Liu L, Wang Z, Hou M, Dong Z, Yu J, Sun R, Cui G. Ubiquitin-proteasome system-based signature to predict the prognosis and drug sensitivity of hepatocellular carcinoma. Front Pharmacol. 2023;14:1172908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 13. | Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545-D551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1156] [Cited by in RCA: 2320] [Article Influence: 580.0] [Reference Citation Analysis (0)] |
| 14. | Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 158] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Sun W, Li J, Zhou L, Han J, Liu R, Zhang H, Ning T, Gao Z, Liu B, Chen X, Ba Y. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics. 2020;10:1981-1996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 16. | Liu P, Ma C, Wu Q, Zhang W, Wang C, Yuan L, Xi X. MiR-369-3p participates in endometrioid adenocarcinoma via the regulation of autophagy. Cancer Cell Int. 2019;19:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Jiao X, Liu W, Mahdessian H, Bryant P, Ringdahl J, Timofeeva M, Farrington SM, Dunlop M, Lindblom A. Recurrent, low-frequency coding variants contributing to colorectal cancer in the Swedish population. PLoS One. 2018;13:e0193547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Tan S, Li H, Zhang W, Shao Y, Liu Y, Guan H, Wu J, Kang Y, Zhao J, Yu Q, Gu Y, Ding K, Zhang M, Qian W, Zhu Y, Cai H, Chen C, Lobie PE, Zhao X, Sun J, Zhu T. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37:4887-4900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 19. | Hu B, Deng T, Ma H, Liu Y, Feng P, Wei D, Ling N, Li L, Qiu S, Zhang L, Peng B, Liu J, Ye M. Deubiquitinase DUB3 Regulates Cell Cycle Progression via Stabilizing Cyclin A for Proliferation of Non-Small Cell Lung Cancer Cells. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 20. | Baohai X, Shi F, Yongqi F. Inhibition of ubiquitin specific protease 17 restrains prostate cancer proliferation by regulation of epithelial-to-mesenchymal transition (EMT) via ROS production. Biomed Pharmacother. 2019;118:108946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Zhu Y, Xu J, Hu W, Wang F, Zhou Y, Gong W, Xu W. Inhibiting USP8 overcomes hepatocellular carcinoma resistance via suppressing receptor tyrosine kinases. Aging (Albany NY). 2021;13:14999-15012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Rong Z, Zhu Z, Cai S, Zhang B. Knockdown of USP8 Inhibits the Growth of Lung Cancer Cells. Cancer Manag Res. 2020;12:12415-12422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 23. | Wang Z, Huang C, Wu J, Zhang H, Shao Y, Fu Z. Analysis of the Prognostic Significance and Immune Infiltration of the Amino Acid Metabolism-Related Genes in Colon Adenocarcinoma. Front Genet. 2022;13:951461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 24. | Chiao CC, Liu YH, Phan NN, An Ton NT, Ta HDK, Anuraga G, Minh Xuan DT, Fitriani F, Putri Hermanto EM, Athoillah M, Andriani V, Ajiningrum PS, Wu YF, Lee KH, Chuang JY, Wang CY, Kao TJ. Prognostic and Genomic Analysis of Proteasome 20S Subunit Alpha (PSMA) Family Members in Breast Cancer. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 25. | Zhang Q, Zhang ZY, Du H, Li SZ, Tu R, Jia YF, Zheng Z, Song XM, Du RL, Zhang XD. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26:2300-2313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 26. | Jin X, Yan Y, Wang D, Ding D, Ma T, Ye Z, Jimenez R, Wang L, Wu H, Huang H. DUB3 Promotes BET Inhibitor Resistance and Cancer Progression by Deubiquitinating BRD4. Mol Cell. 2018;71:592-605.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 27. | Fu Y, Liu S, Zeng S, Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 301] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 28. | Quan Y, Liang F, Wu D, Yao X, Hu Z, Zhu Y, Chen Y, Wu A, Tang D, Huang B, Xu R, Lyu Z, Yan Q, Luo L, Ning Z, Li Y, Xiong J. Blood Cell DNA Methylation of Aging-Related Ubiquitination Gene DZIP3 Can Predict the Onset of Early Stage Colorectal Cancer. Front Oncol. 2020;10:544330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Pan S, Zhan Y, Chen X, Wu B, Liu B. Bladder Cancer Exhibiting High Immune Infiltration Shows the Lowest Response Rate to Immune Checkpoint Inhibitors. Front Oncol. 2019;9:1101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 328] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 31. | Kapanidou M, Curtis NL, Bolanos-Garcia VM. Cdc20: At the Crossroads between Chromosome Segregation and Mitotic Exit. Trends Biochem Sci. 2017;42:193-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 32. | Sanhaji M, Ritter A, Belsham HR, Friel CT, Roth S, Louwen F, Yuan J. Polo-like kinase 1 regulates the stability of the mitotic centromere-associated kinesin in mitosis. Oncotarget. 2014;5:3130-3144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Zheng Y, Shi Y, Yu S, Han Y, Kang K, Xu H, Gu H, Sang X, Chen Y, Wang J. GTSE1, CDC20, PCNA, and MCM6 Synergistically Affect Regulations in Cell Cycle and Indicate Poor Prognosis in Liver Cancer. Anal Cell Pathol (Amst). 2019;2019:1038069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Shi M, Dai WQ, Jia RR, Zhang QH, Wei J, Wang YG, Xiang SH, Liu B, Xu L. APC(CDC20)-mediated degradation of PHD3 stabilizes HIF-1a and promotes tumorigenesis in hepatocellular carcinoma. Cancer Lett. 2021;496:144-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 35. | Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in Tumor Tissues Predicted Worse Overall Survival and Disease-Free Survival in Hepatocellular Carcinoma Patients. Biomed Res Int. 2018;2018:7897346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 36. | Watanabe K, Yoshida K, Iwamoto S. Kbtbd11 gene expression in adipose tissue increases in response to feeding and affects adipocyte differentiation. J Diabetes Investig. 2019;10:925-932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Gong J, Tian J, Lou J, Wang X, Ke J, Li J, Yang Y, Gong Y, Zhu Y, Zou D, Peng X, Yang N, Mei S, Zhong R, Chang J, Miao X. A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann Oncol. 2018;29:632-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 38. | Wang H, Chen Y, Han J, Meng Q, Xi Q, Wu G, Zhang B. DCAF4L2 promotes colorectal cancer invasion and metastasis via mediating degradation of NFκb negative regulator PPM1B. Am J Transl Res. 2016;8:405-418. [PubMed] |
| 39. | Tian Y, Arai E, Makiuchi S, Tsuda N, Kuramoto J, Ohara K, Takahashi Y, Ito N, Ojima H, Hiraoka N, Gotoh M, Yoshida T, Kanai Y. Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas. J Cancer Res Clin Oncol. 2020;146:2461-2477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |