Published online Feb 24, 2024. doi: 10.5306/wjco.v15.i2.302
Peer-review started: September 15, 2023
First decision: October 17, 2023
Revised: October 20, 2023
Accepted: November 27, 2023
Article in press: November 27, 2023
Published online: February 24, 2024
Processing time: 157 Days and 22.8 Hours
Bladder cancer (BC) is the most common urological tumor. It has a high recur
To investigate the biological function and mechanism of TM9SF1 in BC.
Cells at 60%-80% confluence were transfected with lentiviral vectors for 48-72 h to achieve stable TM9SF1 overexpression or silencing in three BC cell lines (5637, T24, and UM-UC-3). The effect of TM9SF1 on the biological behavior of BC cells was then investigated through CCK8, wound-healing assay, transwell assay, and flow cytometry.
Overexpression of TM9SF1 increased the in vitro proliferation, migration, and invasion of BC cells by promoting the entry of BC cells into the G2/M phase. Silencing of TM9SF1 inhibited in vitro proliferation, migration, and invasion of BC cells and blocked BC cells in the G1 phase.
TM9SF1 may be an oncogene in BC.
Core Tip: This study was the first to attempt to construct a stable bladder cancer (BC) cell line to investigate the overexpression and silencing of transmembrane 9 superfamily member 1 (TM9SF1) using in vitro experiments for the purpose of exploring the pro-cancer effect of TM9SF1 in BC. We verified that overexpression of TM9SF1 could enhance the growth, migration, and invasion of BC cells and promote their entry into G2/M phase of the cell cycle. This information not only provides a new target in developing treatments for BC but is also a source of hope for BC patients.
- Citation: Wei L, Wang SS, Huang ZG, He RQ, Luo JY, Li B, Cheng JW, Wu KJ, Zhou YH, Liu S, Li SH, Chen G. TM9SF1 promotes bladder cancer cell growth and infiltration. World J Clin Oncol 2024; 15(2): 302-316
- URL: https://www.wjgnet.com/2218-4333/full/v15/i2/302.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i2.302
Bladder cancer (BC) is the most common urological tumor, ranking tenth among global malignant tumors and fourth among male malignant tumors. It has a high recurrence rate, displays tutor heterogeneity, and resists chemotherapy, imposing a huge cost burden on healthcare systems and affecting the prognosis and quality of survival in patients[1-4]. Age-standardized prevalence rates show considerable variation across geographic regions and are expected to continue to rise over the next two decades[5]. Several BC risk factors have been identified; in addition to geography and age, gender and exposure to a variety of carcinogens, of which smoking is the most prevalent, greatly influence the risk[6,7]. Moreover, age-standardized mortality rates have begun to decline in developed countries but are on the rise in low-income regions worldwide[8].
The most prominent symptom of BC is microscopic or gross hematuria. Seventy-five percent of bladder tumors are uroepithelial carcinomas confined to the mucosa, that is, non-muscle-invasive BC (NMIBC)[9]. Muscle-invasive BC (MIBC) is BC that has invaded the deeper layers of the bladder wall or has metastasized[10-13]. For patients with NMIBC, transurethral resection of the bladder tumor (TURBT) is the standard treatment, while radical cystectomy is indicated for patients with MIBC. To prevent BC recurrence or worsening, TURBT in selected patients is supplemented with an intravesical drip[10,14].
Despite some improvements in surgery and anesthesia techniques and the widespread adoption of perioperative chemotherapy, the long-term survival of BC patients has remained unchanged for decades[15]. However, advanced molecular studies have greatly increased the understanding of disease biology. To find better treatments for this disease, we investigated transmembrane 9 superfamily member 1 (TM9SF1) to determine its biological function and mechanism in BC and to assess whether it could be a therapeutic target.
TM9SF1, also known as MP70 and HMP70, was first identified in 1997[16]. TM9SF1 is localized on membranes and is an integral part of the membranes in which it is active, including autophagosome and lysosomal membranes and cytoplasmic vesicles. TM9SF1 is ubiquitously expressed in human tissues, widely expressed in yeast, plants, and mammals, and highly conserved[17]. Studies have identified TM9SF1 expression in BC, and through genome-wide microarray analyses of tissue sections, TM9SF1 has been determined to be a common differentially expressed gene in BC[18]. However, the biological function of TM9SF1 in BC cells is still unclear, and more in-depth exploration of its biological function is needed.
This study explored the effect of TM9SF1 on BC cells’ biological phenotype by constructing stable cell transfectants with overexpression or silencing of TM9SF1. The biological relationship between TM9SF1 and BC in vitro was validated, with an aim to provide a new direction for and new way of thinking about targeted BC therapy.
We purchased the uman BC cell line 5637, tool cells, and the human embryonic kidney cell line 293T from the cell bank at the Committee of Typical Culture Conservation at the Chinese Academy of Sciences as well as the human bladder transitional cell cancer cell line T24 from Guangzhou Genio Biotechnology Co., Ltd. and the human bladder transitional cell cancer cell line UM-UC-3 from Wuhan Punose Life Technology Co., Ltd. Cells were cultured in a 5% CO2 incubator at 37 °C.
The lentivirus used for constructing the stable TM9SF1-overexpressing cell strain was provided by Hanheng Biologicals; its vector system consisted of pSPAX2, pMD2G, and a shuttle plasmid carrying the target genes. The three plasmids were co-transfected into the 293T packaging cell via a transfection reagent. After lentiviral infection, quantitative real-time polymerase chain reaction (qRT-PCR), which is an experimental method for detecting RNA or DNA molecules and quantifying their content, was used to verify the efficiency of TM9SF1 overexpression in BC cells.
Lentiviral vectors are gene therapy vectors that have been developed based on human immunodeficiency virus-1. The TM9SF1 silencing plasmid used for constructing short hairpin RNA (shRNA) was provided by GeneCopoeia. The vector used is psi-LVRU6H, which has BamHI (5') and EcoRI (3') cloning sites, is 7451 bp in length, is ampicillin resistant, and has a thaumatin screening marker. The lentiviral infection was followed by qRT-PCR to validate the silencing efficiency of TM9SF1 in BC cells.
Cell Counting Kit-8 (CCK8) was used to detect cell proliferation. WST-8 (2-[2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5(2,4-disulfophenyl])-2H-tetrazole monosodium salt], the main component of the kit, reacts with intracellular mitochondria to generate an orange formazan dye in proportion to the number of viable cells. This indirectly detects the number of viable cells and shows cell proliferation. The optical density (OD) value was measured at 450 nm using a microplate reader; the higher the OD value, the more the live cells.
The migration of the stably transfected strains, which demonstrated silenced or overexpressed TM9SF1, was verified by cell wound-healing and transwell assays. First, the cells were incubated until they spread over the bottom of the plate. Next, a 1000 μL pipette tip was used to trace a line across the cell plane along a straight ruler held perpendicular to the bottom of the plate. The culture was then continued by adding medium containing the appropriate percentage of fetal bovine serum (FBS), which is the natural medium in the cell culture (5637: 1%; T24: 1%; UM-UC-3: 4%). Starting at 0 h, scratches at the same location were photographed under an inverted fluorescence microscope at intervals of 12 h or 48 h. The area of the scratches was calculated, and the healing rate was determined using ImageJ software.
For the transwell migration and invasion assays, 500 μL of complete medium was first added to the 24-well plates. Logarithmically growing 5637, T24, and UM-UC-3 BC cells were digested, centrifuged, and counted using cell resuspension with a culture medium containing 2% FBS. To measure invasion, 100 μL of suspended cells was added on top of the solidified matrix gel in the upper chamber of each transwell and incubated for 24 h. After which, the chambers were removed, fixed, and stained with methanol and crystal violet. All photography was performed using a Leica DMi8 inverted fluorescence microscope and calculated using ImageJ software.
The original medium was aspirated, and fresh serum-free medium was added based on starvation treatment in the incubator for 12 h. Then, the serum-free medium was aspirated, and a complete medium containing 10% FBS and 1% double antibody was added. Incubation continued for 12 h to synchronize the cell status of the experimental and control groups. Finally, 5 × 105 cells were removed and processed using a cell cycle staining kit (Lianke Bio). A flow cytometer was used to detect the cells.
Student’s t test in SPSS 25.0 was employed to analyze the statistical significance of the experiments, and P < 0.05 was defined as statistically significant.
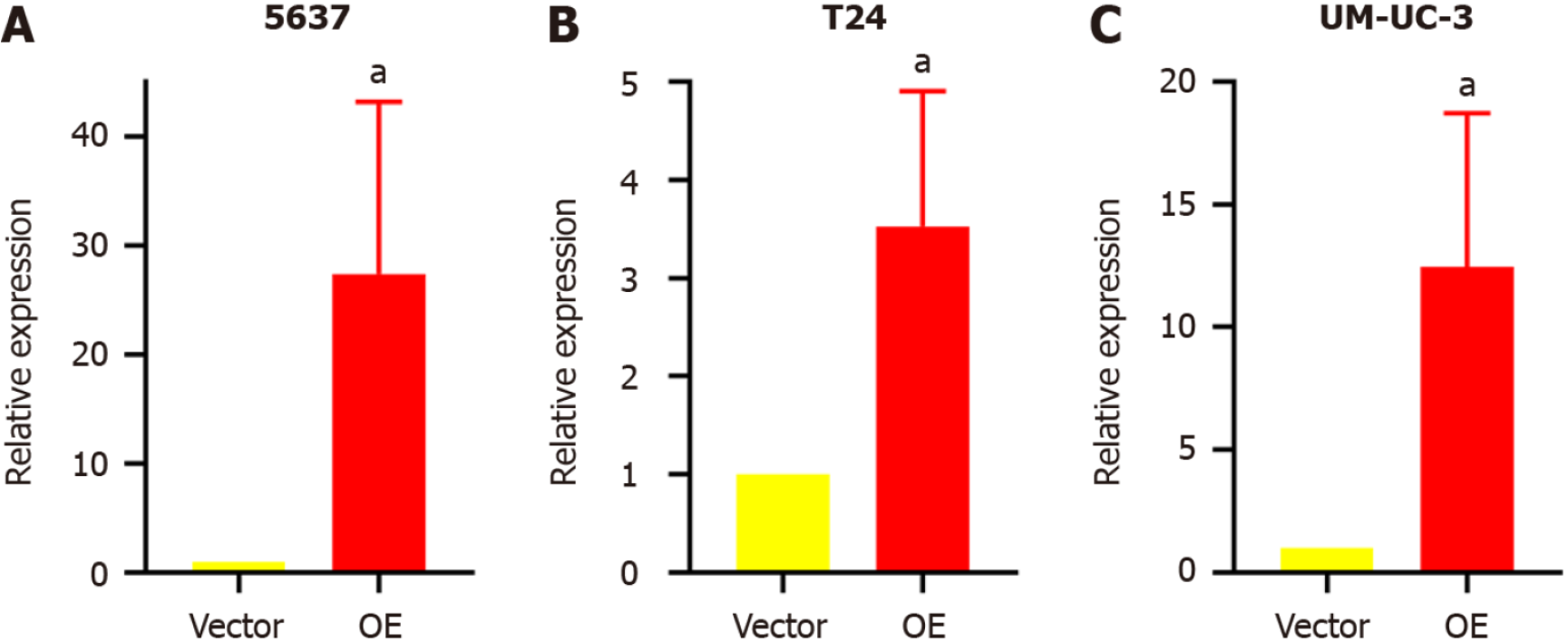
Stable transfectant construction for TM9SF1 overexpression: The relative expression of the TM9SF1 gene in BC cells transfected with TM9SF1-overexpressed lentiviral vector was detected by qRT-PCR in three cell lines (5637, T24, and UM-UC-3) using cells transfected with the empty lentiviral vector as a control group. The expression of TM9SF1 was 27.4-fold higher in the TM9SF1 overexpression group (OE group) of 5637 cells than in the empty vector group, 3.5-fold higher in TM9SF1-overexpressing T24 cells, and 12.5-fold higher in TM9SF1-overexpressing UM-UC-3 cells. All these values were statistically significant (P < 0.05), indicating that stable transfectants overexpressing TM9SF1 were successfully constructed in all three BC cell lines (Figure 1).
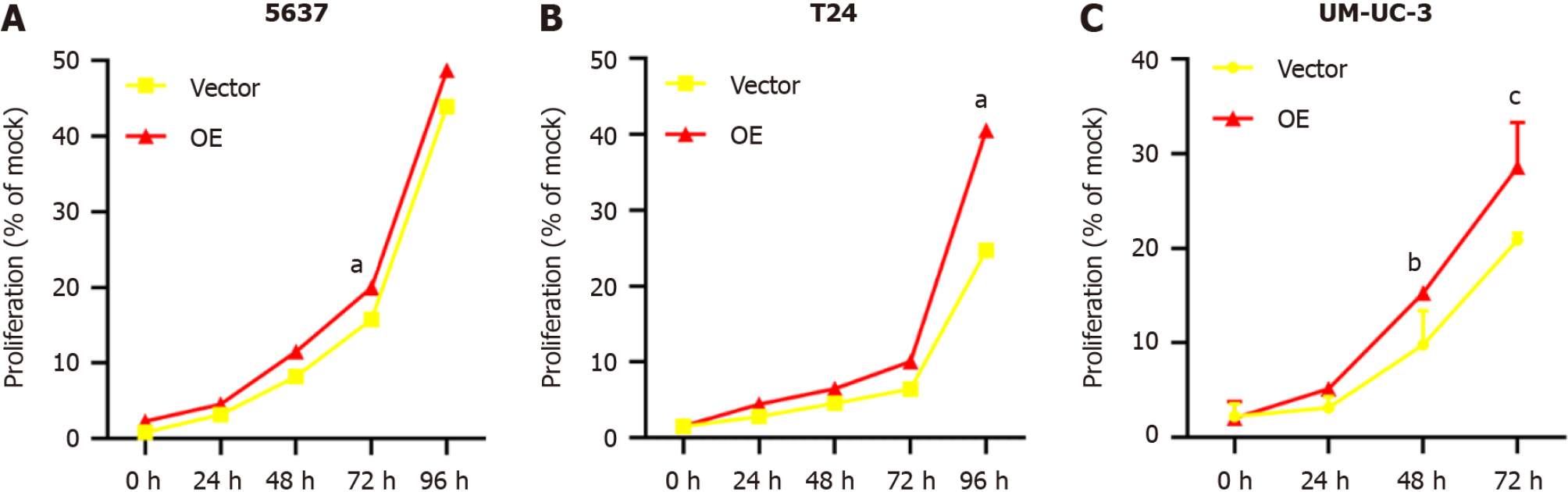
TM9SF1 overexpression promotes BC cell proliferation in vitro: The cell activity was tested by a CCK8 assay. The results showed that compared with the control group transfected with the empty vector, the proliferation rate of BC cells in the TM9SF1 overexpression group was significantly higher at 72 h in the 5637 cells, 96 h in the T24 cells, and 48 h and 72 h in the UM-UC-3 cells (P < 0.05) (Figure 2). This finding indicates that TM9SF1 overexpression plays a role in promoting BC cell proliferation in vitro.
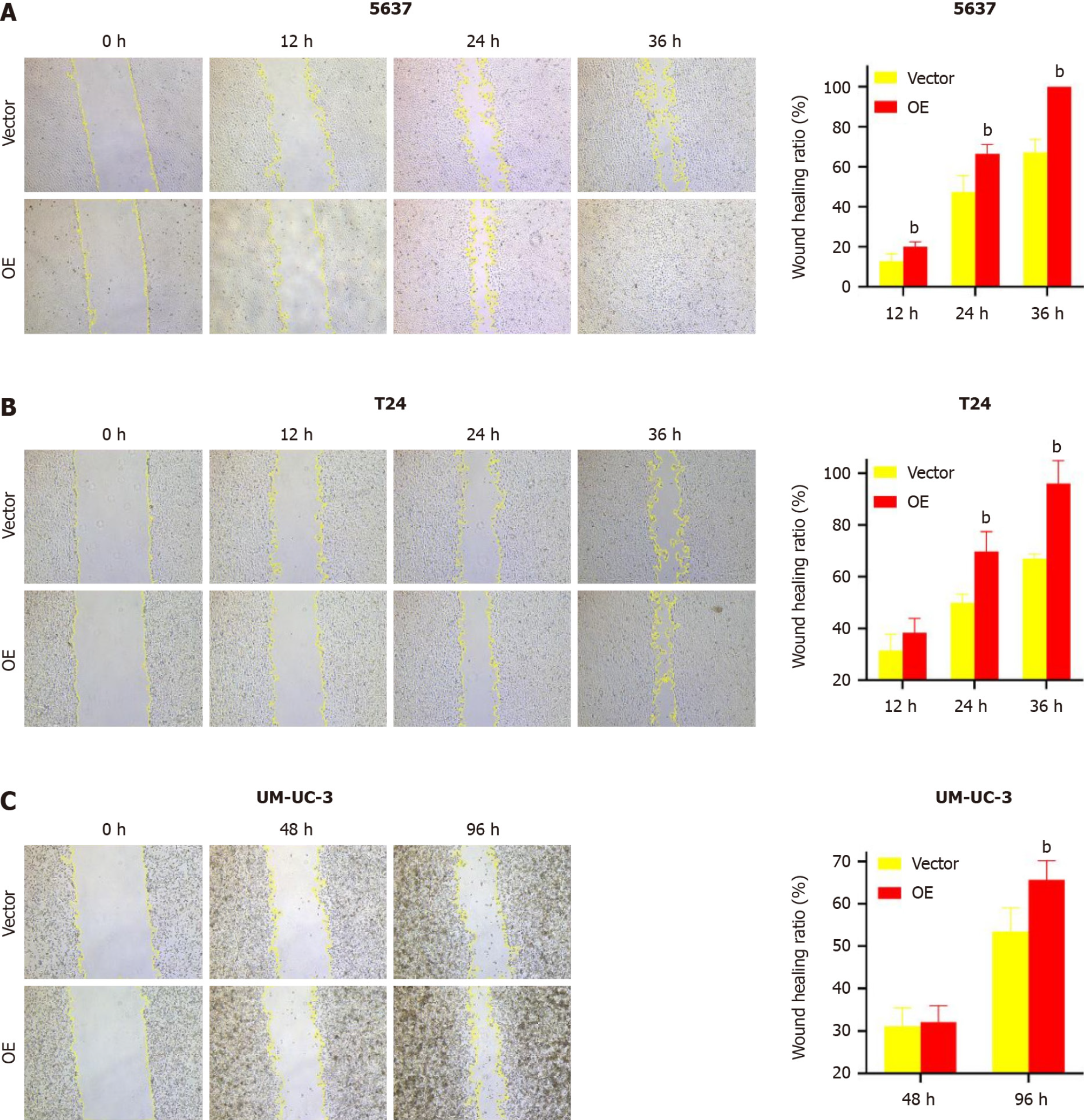
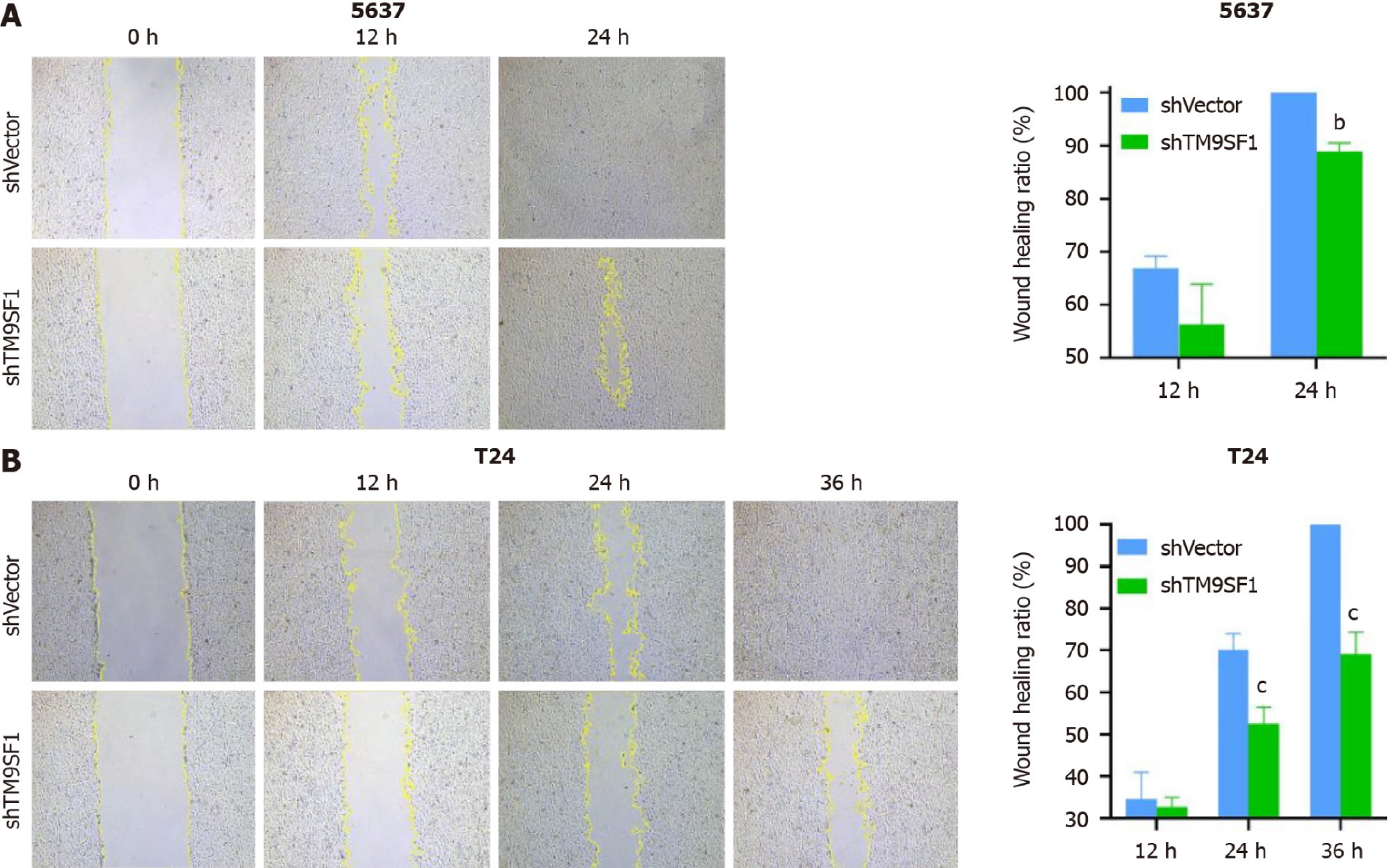
TM9SF1 overexpression promotes BC cell migration in vitro: In this study, we used a wound-healing assay to detect the migration ability of cells. After calculating the change of scratch area, we found that in the 5637 cells, the wound healing rate of the TM9SF1 overexpression group was significantly higher than that of the control group at 12 h, 24 h, and 36 h (P < 0.01). The scratch healing rate of the cells in the T24 overexpression group was also significantly higher than that of the control group at 24 h and 36 h (P < 0.01). Although UM-UC-3 cells were less likely to migrate toward the scratches, the rate of scratch closure in the OE group was still slightly higher than that in the control group and was statistically significant at 96 h (P < 0.01) (Figure 3).
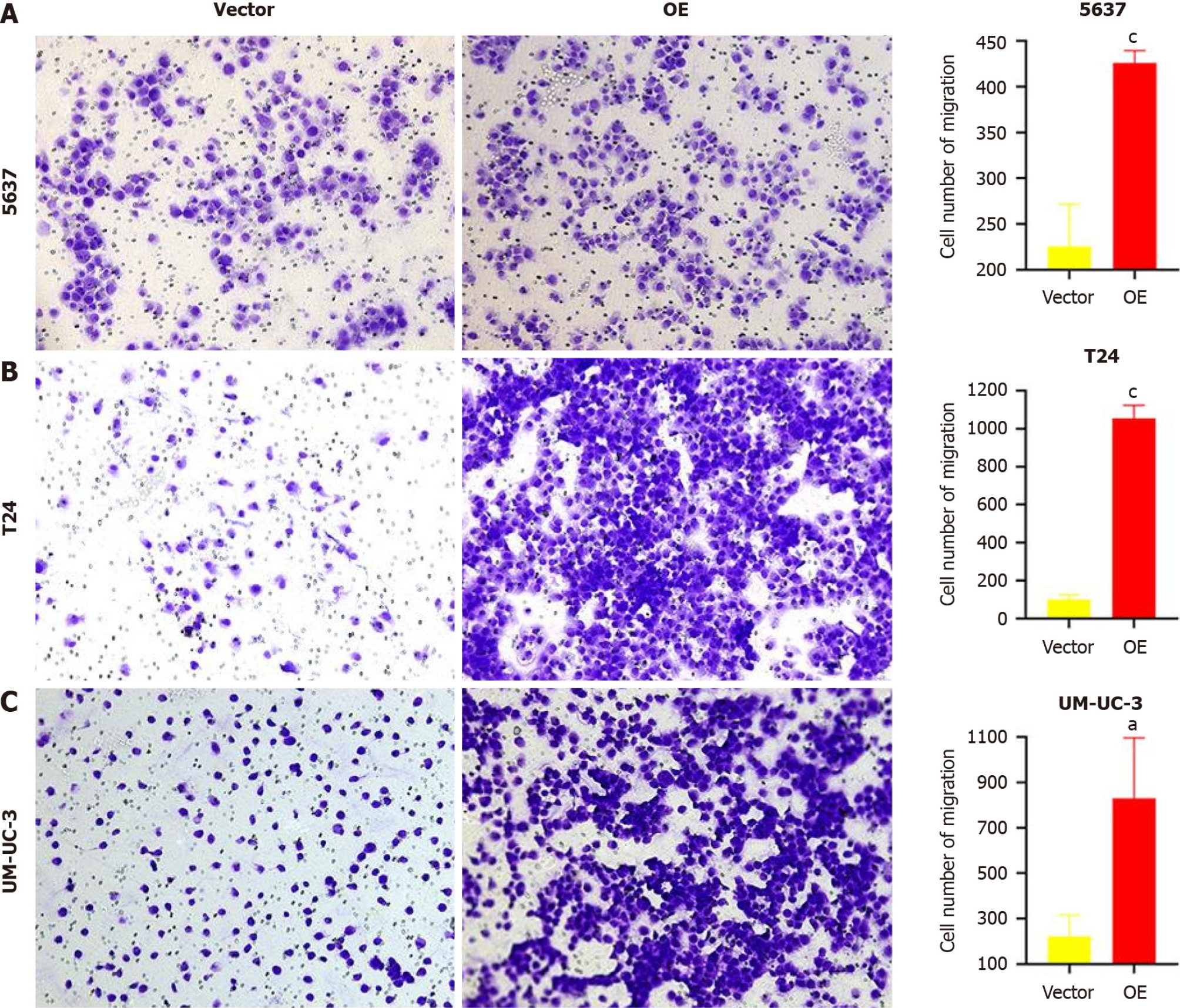
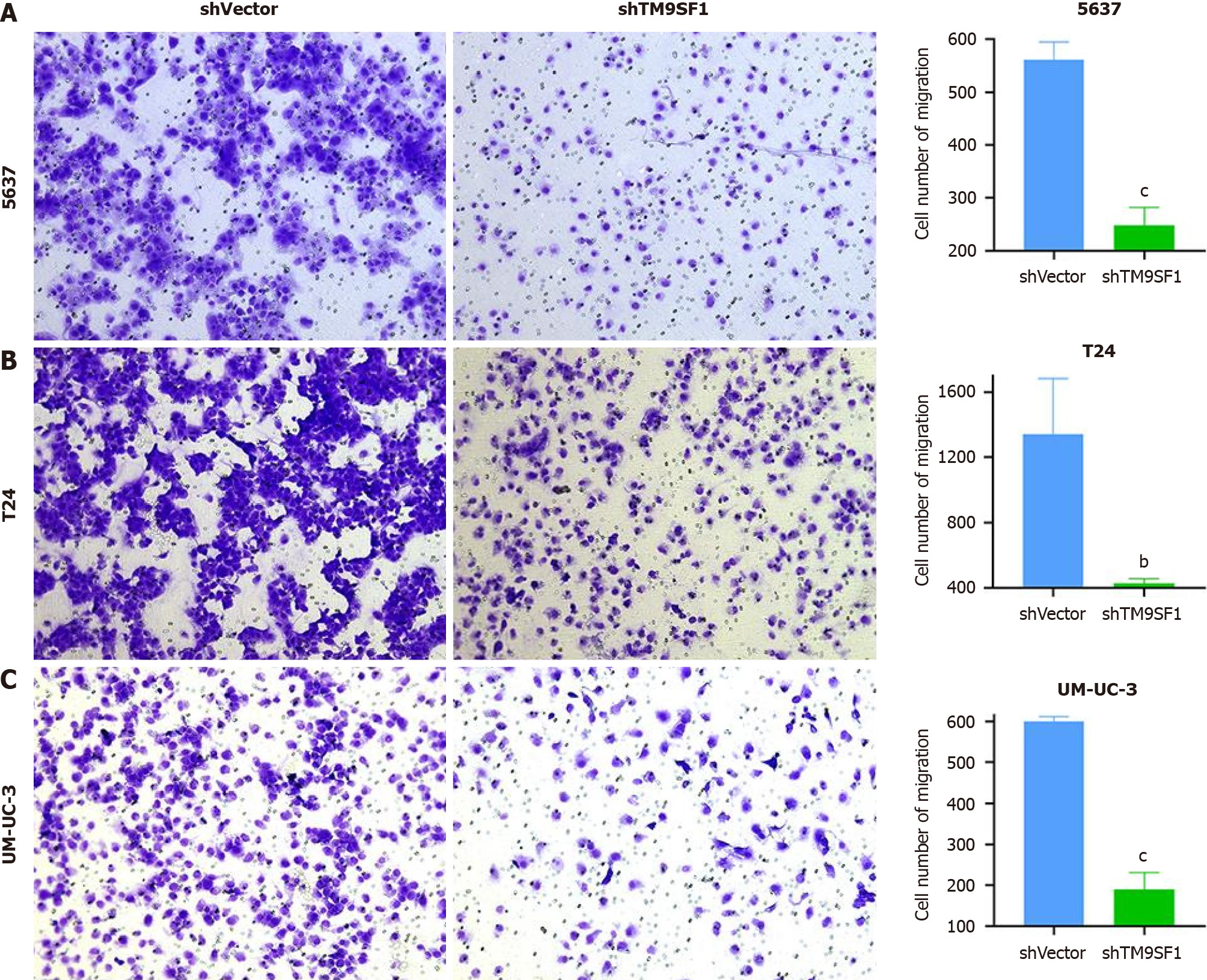
A transwell migration assay was performed to verify the changes in cell migration ability. After 24 h of incubation, the number of cells passing through the transwell membrane was counted. As shown in Figure 4, the number of cells passing through the membrane was significantly higher in the three BC cell lines with TM9SF1 overexpression than in the control group (P < 0.05).
By combining the above two migration experiments, we speculate that the overexpression of TM9SF1 can significantly improves the migration ability of BC cells.
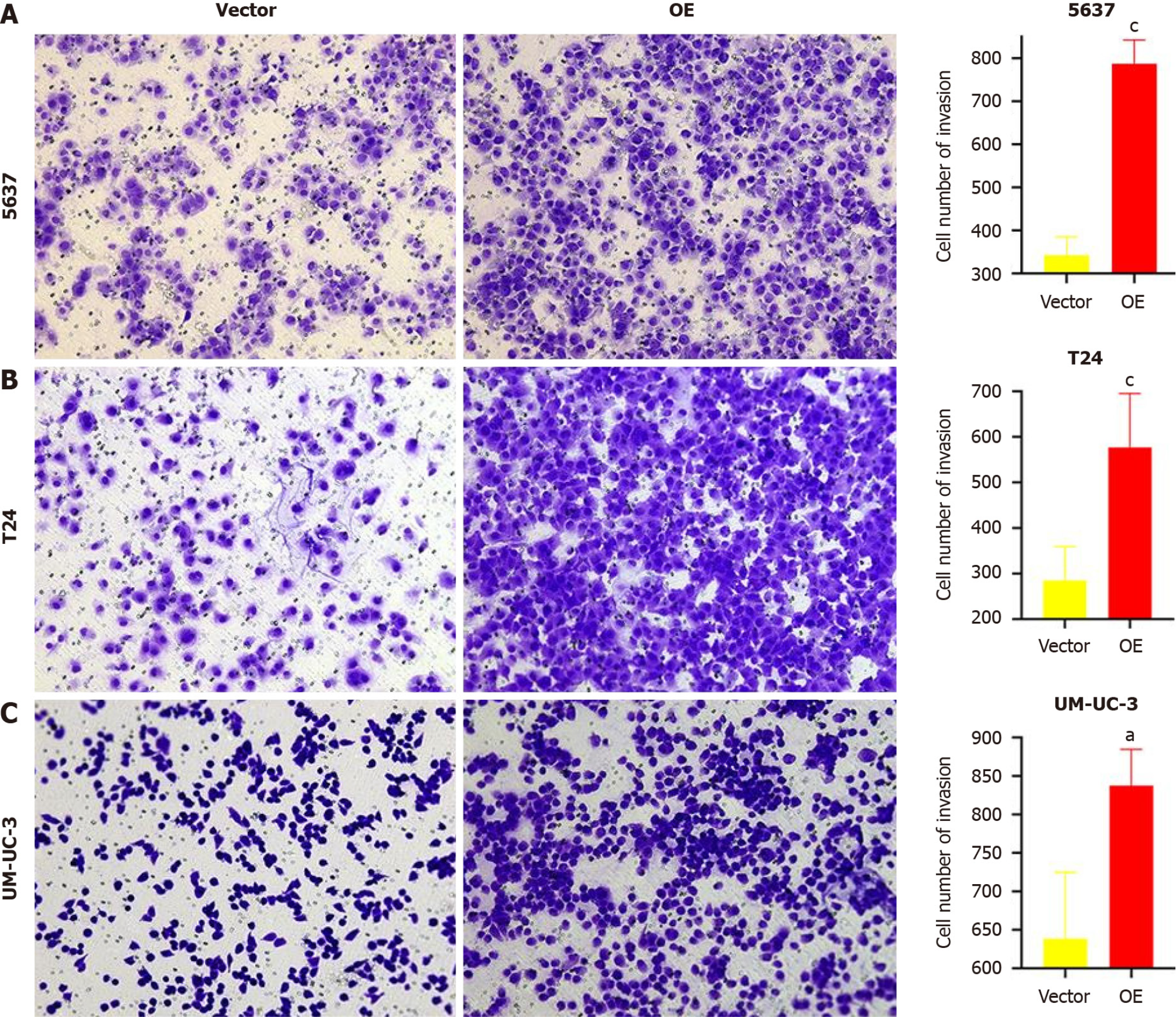
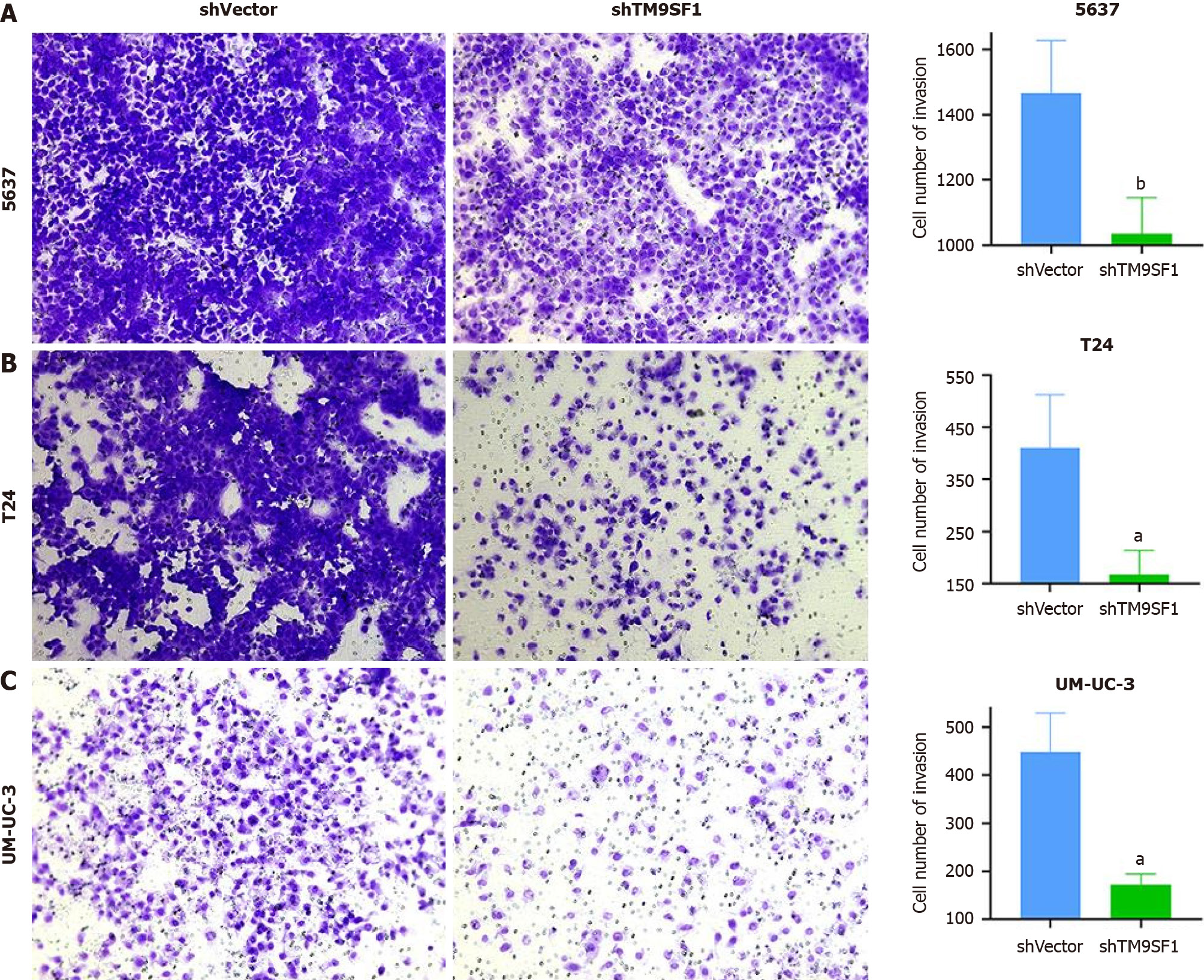
TM9SF1 overexpression promotes BC cell invasion in vitro: Matrigel matrix gel was used to mimic the extracellular matrix. The high-nutrient culture medium in the lower chamber of the transwell was separated from the low-nutrient culture medium in the upper chamber, which caused the BC cells to secrete hydrolases and move to pass through the filter membrane lined with matrix gel. This can be used to assess the invasive ability of the BC cells. After analyzing the fixed stained cells, we found that the number of cells in the OE group of the three BC cell lines was 1.3 times more than that in the empty vector group (P < 0.05), indicating that the overexpression of TM9SF1 has a significant effect in promoting the invasive ability of BC cells (Figure 5).
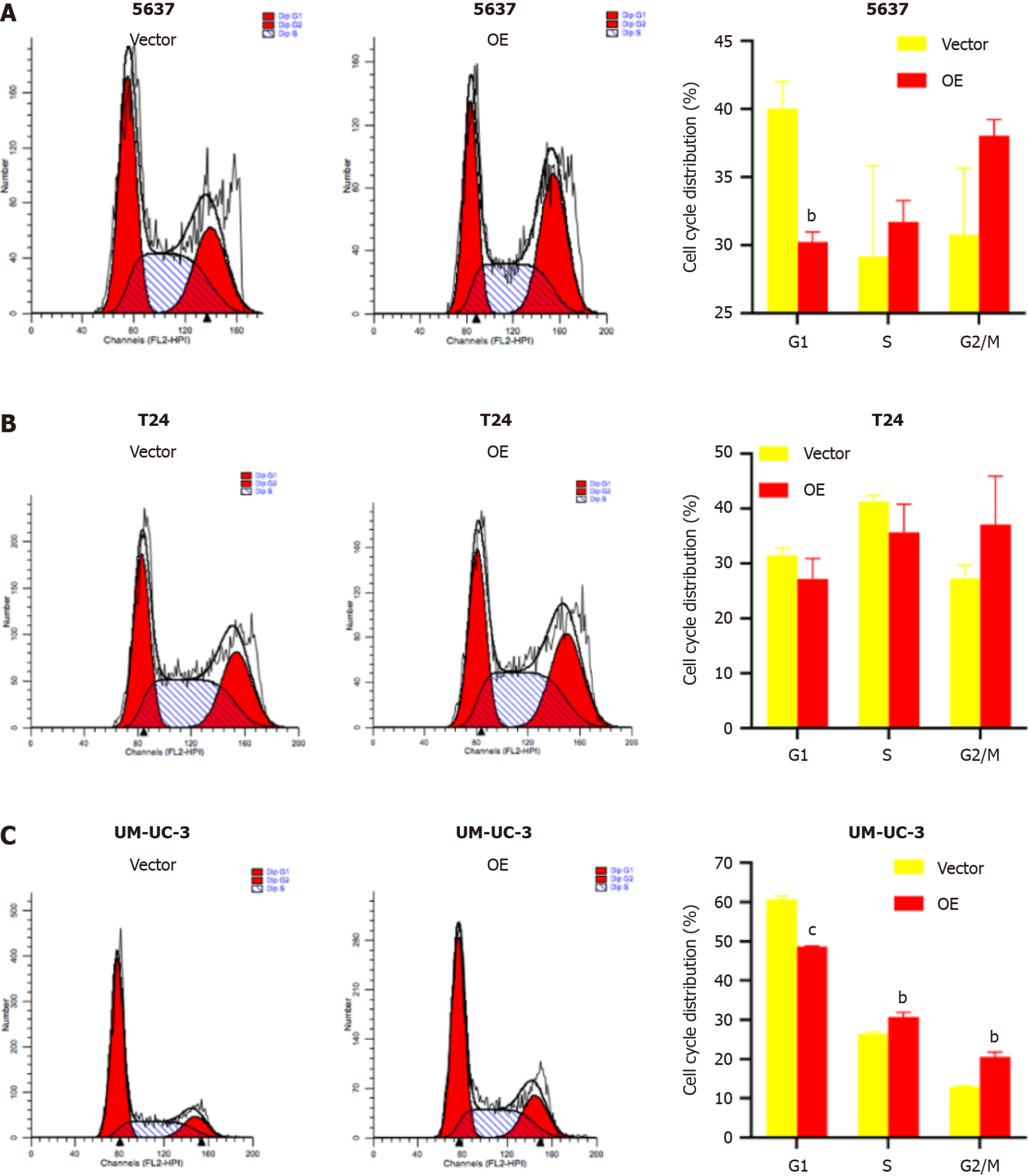
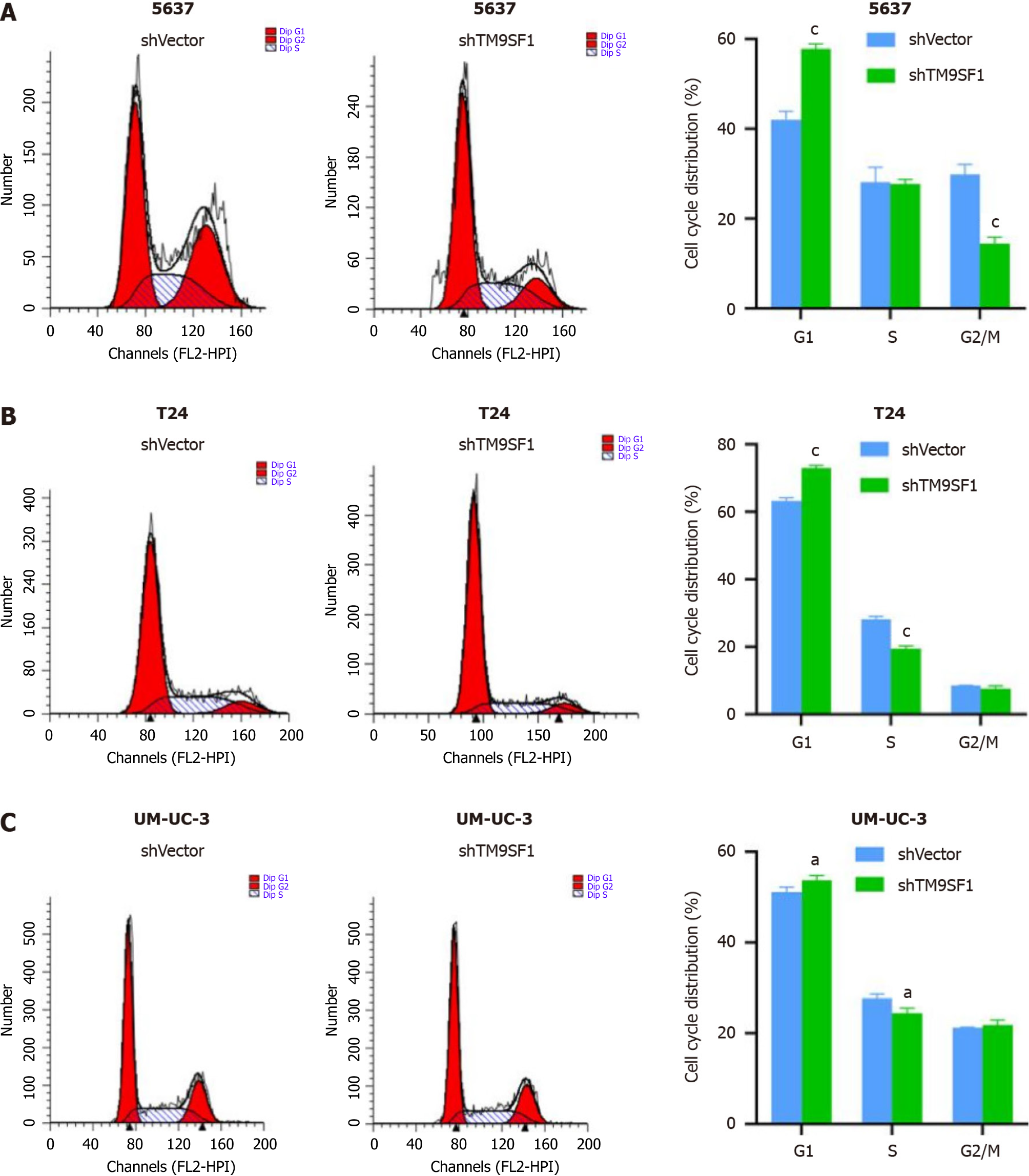
TM9SF1 overexpression promotes cell entry into the G2/M phase: Flow cytometry was used to investigate the effect of TM9SF1 overexpression by detecting the percentage of cells distributed in different phases of the cell cycle. The results showed that the proportion of cells distributed in the G1 phase in all three BC cell lines was smaller in the OE group than in the empty vector group, while the proportion of cells in the G2/M phase was higher in the OE group than in the empty vector group. However, for the 5637 and UM-UC-3 cell lines, the OE group had more cells in the S phase than in the empty vector group, while the opposite was true regarding the T24 cell line. Although the statistical results of 5637 cells in the S, G2/N, and G1 phases as well as the T24 cells in the G1, S, and G2/M phases were not significant, the changing trend combined with the statistical results of the UM-UC-3 cells in the G1, S, and G2/M phases suggests that TM9SF1 overexpression has a certain effect on BC cell cycle progression, reducing the proportion of cells in the G1 phase and promoting the entry of cells into the G2/M phase (Figure 6).
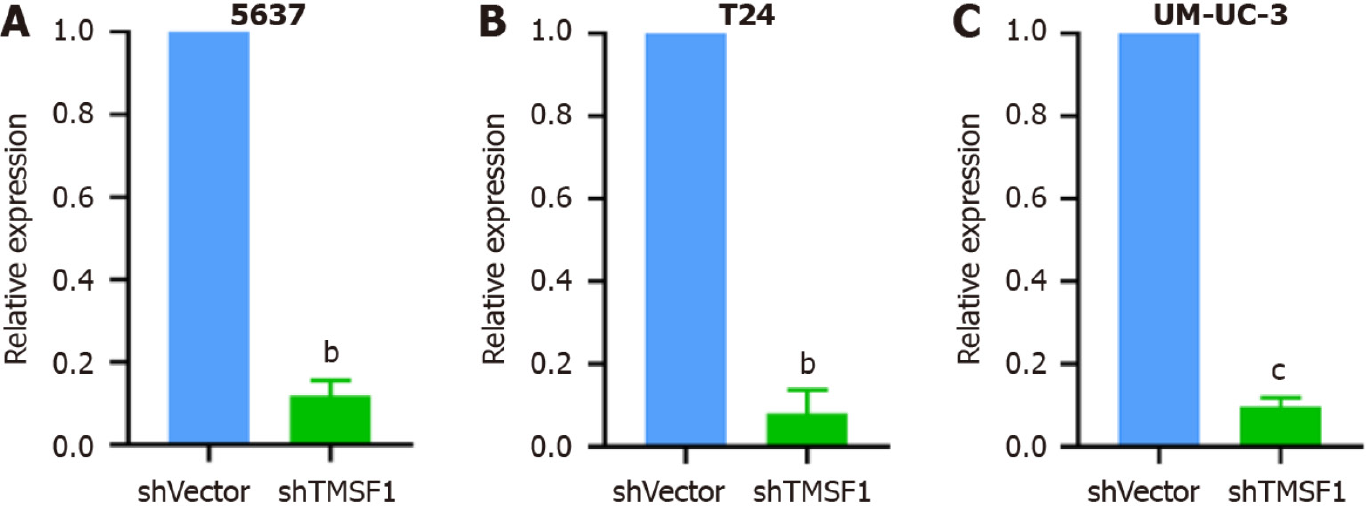
Stable transfectant construction for TM9SF1 silencing: shRNA was used to construct TM9SF1-silenced stable transfectants in three cell lines: 5637, T24, and UM-UC-3. The TM9SF1-silenced stable transfectants and the cells transfected with the empty lentiviral vector as a control group [short hairpin vector (shVector) control group] were used. qRT-PCR was used to detect the TM9SF1 silencing efficiency in BC cell transfectants with TM9SF1 silencing (shTM9SF1 group). The quantitative results showed that the silencing efficiency was 88.09%, 90.39%, and 92.04% in the 5637, UM-UC-3, and T24 cells, respectively. All these values were statistically significant (P < 0.01), suggesting that the TM9SF1-silencing stable transfectants were successfully constructed in all three BC cell lines (Figure 7).
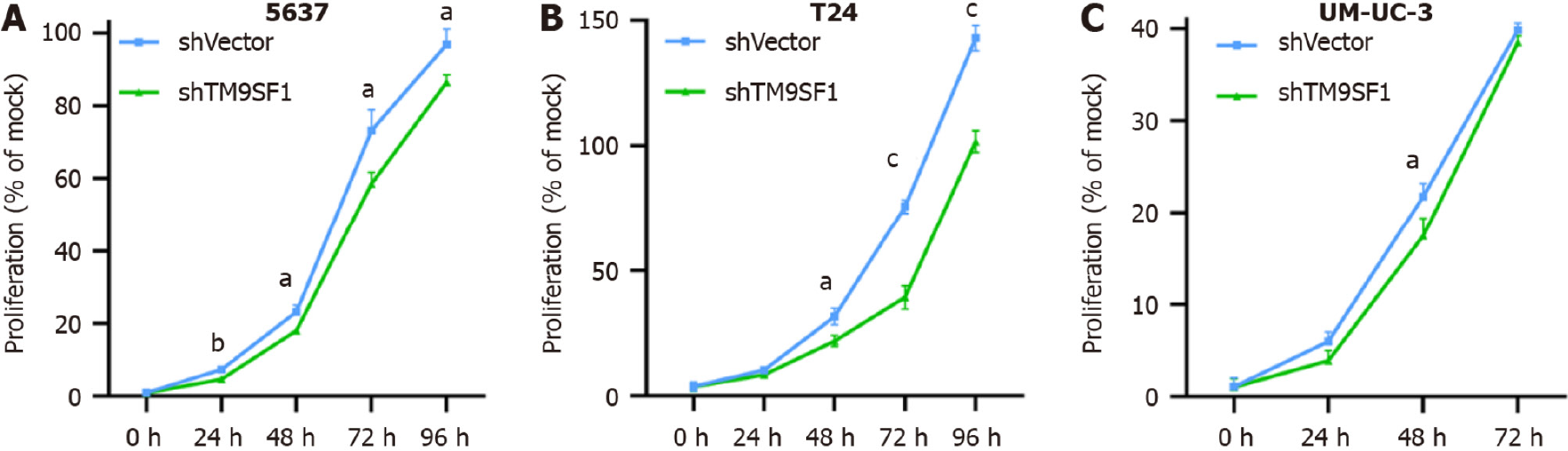
TM9SF1 silencing inhibits BC cell proliferation in vitro: To test the effect of silencing TM9SF1 on cell proliferation in BC cells, we examined cell viability using a CCK8 assay. Compared with the shVector control group transfected with the empty vector, the proliferation rate of BC cells in the shTM9SF1 group was significantly reduced in the 5637 cells at 24 h, 48 h, 72 h, and 96 h; in the T24 cells at 48 h, 72 h, and 96 h; and in the UM-UC-3 cells at 48 h (P < 0.05). This finding indicates that silencing TM9SF1 has an inhibitory effect on BC cell proliferation in vitro (Figure 8).
TM9SF1 silencing inhibits BC cell migration in vitro: Cell scratch and transwell migration assays were used to detect the migration ability of the cells. Of the three BC cell lines, the 5637 and T24 cells were selected for the cell scratch assay (Figure 9). The closure rate of the TM9SF1-silenced 5637 cells was higher than that of the control group at 12 h and 24 h but was statistically significant only at 24 h (P < 0.01). The closure rate of the TM9SF1-silenced T24 cells compared to the empty control group was statistically significant at 12 h, 24 h, and 36 h. Moreover, the T24 cells’ closure rate significantly decreased at 24 h and 36 h (P < 0.001).
All three BC cell lines were subjected to transwell migration assays to validate the inhibitory effect of TM9SF1 silencing on the migratory ability of BC cells. After spreading the cells in the transwell chambers, they were incubated for 24 h. The chambers were then removed and washed with phosphate buffered solution (PBS), and the cells that passed through the filter membrane were fixed with methanol. The cells under the filter membrane were then stained purple with crystal violet (the excess dye was removed with PBS). Next, the chambers were air dried at room temperature. Once dried, images of the cells were captured via a microscope, and the cells were counted using ImageJ software. This information was statistically analyzed using SPSS and graphed using GraphPad Prism 8.0. As can be seen in Figure 10, the number of cells that passed through the filter membrane was significantly lower in all three BC cell strains than in the shVector group (P < 0.01). By comprehensively analyzing both assays, we can conclude that silencing the expression of TM9SF1 in BC cells can significantly reduce their migration ability.
TM9SF1 silencing inhibits BC cell invasion in vitro: Invasion occurs when malignant tumor cells enter adjacent host tissue by secreting proteins that digest the tissue cells’ extracellular matrix. Detecting a tumor’s invasion ability can help determine the rate of metastasis, as cell invasion is the first step: Tumor cells break through the basement membrane in situ, infiltrate blood and lymphatic vessels, colonize other tissues, and proliferate. The transwell invasion assay mimics the in vivo environment of the human body and can therefore determine the invasive ability of tumor cells.
Therefore, in this study, BC cells were induced to secrete hydrolytic enzymes to pass through a layer of Matrigel matrix gel (which mimics the extracellular matrix in vivo) on a transwell filter membrane. After that, the BC cells that had passed through the matrix gel and reached the bottom of the membrane were washed, fixed, stained, dried, photographed, and counted to determine their invasive ability.
The number of 5637 cells in the shTM9SF1 group with TM9SF1 silencing was 1/1.42 of that in the shVector group. The number of T24 cells in the TM9SF1-silenced group was 1/2.46 of that in the control group. The number of UM-UC-3 cells in the shTM9SF1 group was 1/2.6 of that in the shVector group. In all groups, P value was less than 0.05. The results suggest that the invasion ability of BC cells is significantly reduced after TM9SF1 is silenced (Figure 11).
TM9SF1 silencing blocks cells in the G1 phase: To investigate the effect of silenced TM9SF1 on the BC cell cycle, the shTM9SF1 cells collected from the 5637, T24, and UM-UC-3 cell lines were stained and detected by flow cytometry. The number of cells distributed in each phase of the cell cycle was analyzed and compared. In all three BC lines, the shTM9SF1 group had a higher proportion of BC cells in the G1 phase than did the shVector group (P < 0.05). The number of 5637 cells in the S phase was lower in the TM9SF1-silenced group than in the shVector group, but the difference was not significant. However, the number of 5637 cells in the G2/M phase was significantly lower in the shTM9SF1 group than in the shVector group. The number of T24 cells in the S phase in the shTM9SF1 group was significantly lower than that in the shVector group (P < 0.001). However, silencing TM9SF1 did not have much effect on T24 cells in the G2/M phase. The number of the shTM9SF1 group’s UM-UC-3 cells in the S phase was significantly less than the same cells in the shVector group (P < 0.05), but there was no significant difference found in cells in the G2/M phase. These combined results suggest that silencing TM9SF1 expression inhibits BC cell proliferation by arresting BC cells in the G1 phase and prolonging the time between mitosis completion and DNA replication (Figure 12).
In this study, we confirmed that TM9SF1 is a pro-carcinogenic gene in BC by overexpressing and silencing it in the 5637, T24, and UM-UC-3 cell lines using cellular function assays. TM9SF1 is expressed not only in BC but also in esophageal squamous cell carcinoma and cervical cancer, and because of its pro-cancer role, it can aggravate poor prognoses in patients[18-20]. The experimental results of the present study revealed that overexpression of TM9SF1 reduces the number of cells in the G1 phase and prompts them to enter the G2/M phase and start mitosis, thereby promoting BC cell proliferation. However, the silencing of TM9SF1 blocks cells in the G1 phase and prevents them from entering the DNA replication phase, thereby inhibiting BC cell proliferation. It is hypothesized that TM9SF1 may promote BC cell proliferation by mainly affecting the G1 phase of BC cells, which in turn promotes the development of BC. Although TM9SF1 was experimentally demonstrated to be an oncogene of BC in this study, Wei et al found TM9SF1 to be a gastric cancer suppressor gene via N6,2'-O-dimethyladenosine (m6Am) sequencing. TM9SF1 is a target of phosphorylated C-terminal domain-interactingfactor 1 (PCIF1); PCIF1 can reduce TM9SF1 translation by m6Am modification, while TM9SF1 can reverse the effect of PCIF1 on the invasiveness of gastric cancer cells[21]. This indicates that TM9SF1 has different functions and effects in different tumor types.
In other studies, TM9SF1 has been shown to synergistically interact with the tumor marker gene estrogen receptor-binding fragment-associated antigen 9 (EBAG9) to regulate the epithelial-mesenchymal transition in cancer cells via an attenuated anti-tumor immune response, leading to immune escape, which results in tumor growth[22]. Therefore, in addition to affecting the biological phenotype of BC and promoting BC development, TM9SF1 can also affect an anti-tumor immune response to BC, the mechanism of which must be studied in future research.
Furthermore, it has been shown through a combination of suppression subtractive hybridization and transmembrane trapping techniques that Myc-tagged TM9SF1 is localized on the surface of transfected COS-7L cells[23]. If the same technology can be used for BC, researching targeted therapy may go further. TM9SF1 also plays a crucial role in autophagy. One study showed that its high expression can worsen the prognosis of patients with cervical cancer[24]. From this perspective, TM9SF1 may affect the occurrence and development of BC by regulating autophagy. However, this study was based on the results obtained from in vitro cell experiments, and expression detection in BC tissues still needs to be improved.
This study was the first to attempt to construct a stable BC cell line to investigate the overexpression and silencing of TM9SF1 using in vitro experiments for the purpose of exploring the pro-cancer effect of TM9SF1 in BC. We verified that overexpressed TM9SF1 enhances the growth, migration, and invasion of BC cells and promotes their entry into the G2/M phase of the cell cycle. This information not only provides a new target in developing treatments for BC but is also a source of hope for BC patients.
TM9SF1 is suspected to function as an oncogene in BC, as it has been shown to enhance the growth, migration, and invasion of BC cells while also promoting their entry into the G2/M phase of the cell cycle. Moreover, beyond its influence on the biological phenotype of BC and its role in advancing BC development, there is a need for further investigation into the mechanisms underlying these effects, specifically focusing on TM9SF1’s impact on autophagy and anti-tumor immune responses. Consequently, delving into genetic factor analysis and targeted therapies in the realm of cancer treatment holds promising prospects.
Bladder cancer (BC) is the most common urological tumor. It has a high recurrence rate, displays tutor heterogeneity, and resists chemotherapy. Furthermore, the long-term survival rate of BC patients has remained unchanged for decades, which seriously affects the quality of patient survival. To improve the survival rate and prognosis of BC patients, it is necessary to explore the molecular mechanisms of BC development and progression and identify targets for treatment and intervention. Transmembrane 9 superfamily member 1 (TM9SF1), also known as MP70 and HMP70, is a member of a family of nine transmembrane superfamily proteins that was first identified in 1997. TM9SF1 can be expressed in BC, but its biological function and mechanism in BC are not clear.
Previous studies have identified TM9SF1 expression in BC, and through genome-wide microarray analyses of tissue sections, TM9SF1 has been found to be a commonly differentially expressed gene in BC. However, the biological function of TM9SF1 in BC cells remains unclear. To discover better treatment options for this disease, it is crucial to conduct a more in-depth exploration of its biological behavior.
This study explored the effect of TM9SF1 on BC cells’ biological phenotype by constructing stable cell transfectants with overexpression or silencing of TM9SF1. The biological relationship between TM9SF1 and BC in vitro was validated, with an aim to provide a new way of thinking about targeted BC therapy.
Cells at 60%-80% confluence were transfected with lentiviral vectors to achieve stable overexpression or silencing of TM9SF1 in three BC cell lines (5637, T24, and UM-UC-3). The effect of TM9SF1 on the biological behavior of BC cells was investigated through CCK8, wound-healing assay, transwell assay, and flow cytometry.
In this study, we confirmed that TM9SF1 is a pro-carcinogenic gene in BC by overexpressing and silencing it in the 5637, T24, and UM-UC-3 cell lines using cellular function assays. The experimental results of the present study revealed that overexpression of TM9SF1 reduces the number of cells in the G1 phase and prompts them to enter the G2/M phase and start mitosis, thereby promoting BC cell proliferation. Yet, the silencing of TM9SF1 blocks cells in the G1 phase and prevents them from entering the DNA replication phase, thereby inhibiting BC cell proliferation. It is hypothesized that TM9SF1 may promote BC cell proliferation mainly by affecting the cell cycle progression of BC cells, which in turn promotes BC development.
This study was the first to attempt to construct a stable BC cell line to investigate the overexpression and silencing of TM9SF1 using in vitro experiments for the purpose of exploring the pro-cancer effect of TM9SF1 in BC. We verified that overexpression of TM9SF1 enhances the growth, migration, and invasion of BC cells and promotes their entry into the G2/M phase of the cell cycle. TM9SF1 may be an oncogene in BC.
In addition to influencing the biological phenotype of BC and promoting its development, TM9SF1 may also impact the anti-tumor immune response in BC. Further research is needed to elucidate the mechanisms underlying this effect. Furthermore, genetic factor analysis and targeted therapy aimed at inhibiting carcinogenesis offer promising prospects.
Thanks to Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis for providing technical support. Thanks to China Undergraduate lnnovation and Entrepreneurship Training Program (202310598044) and Future Academic Star of Guangxi Medical University (WLXSZX23112) for the experimental platforms.
Provenance and peer review: Unsolicited article; externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pavlidis TE, Greece S-Editor: Lin C L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | Matuszczak M, Kiljańczyk A, Salagierski M. A Liquid Biopsy in Bladder Cancer-The Current Landscape in Urinary Biomarkers. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 2. | Chang CH, Lin BJ, Chen CH, Nguyen NL, Hsieh TH, Su JH, Chen MC. Stellettin B Induces Cell Death in Bladder Cancer Via Activating the Autophagy/DAPK2/Apoptosis Signaling Cascade. Mar Drugs. 2023;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 3. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 63990] [Article Influence: 15997.5] [Reference Citation Analysis (174)] |
| 4. | Afonso J, Santos LL, Longatto-Filho A, Baltazar F. Competitive glucose metabolism as a target to boost bladder cancer immunotherapy. Nat Rev Urol. 2020;17:77-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 5. | Wong MCS, Fung FDH, Leung C, Cheung WWL, Goggins WB, Ng CF. The global epidemiology of bladder cancer: a joinpoint regression analysis of its incidence and mortality trends and projection. Sci Rep. 2018;8:1129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 6. | Lobo N, Afferi L, Moschini M, Mostafid H, Porten S, Psutka SP, Gupta S, Smith AB, Williams SB, Lotan Y. Epidemiology, Screening, and Prevention of Bladder Cancer. Eur Urol Oncol. 2022;5:628-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 157] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 7. | Chen X, Wang M, Yu K, Xu S, Qiu P, Lyu Z, Zhang X, Xu Y. Chronic stress-induced immune dysregulation in breast cancer: Implications of psychosocial factors. J Transl Int Med. 2023;11:226-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 8. | Teoh JY, Huang J, Ko WY, Lok V, Choi P, Ng CF, Sengupta S, Mostafid H, Kamat AM, Black PC, Shariat S, Babjuk M, Wong MC. Global Trends of Bladder Cancer Incidence and Mortality, and Their Associations with Tobacco Use and Gross Domestic Product Per Capita. Eur Urol. 2020;78:893-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 156] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 9. | Charlton ME, Adamo MP, Sun L, Deorah S. Bladder cancer collaborative stage variables and their data quality, usage, and clinical implications: a review of SEER data, 2004-2010. Cancer. 2014;120 Suppl 23:3815-3825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 417] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 11. | Funt SA, Rosenberg JE. Systemic, perioperative management of muscle-invasive bladder cancer and future horizons. Nat Rev Clin Oncol. 2017;14:221-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 12. | Berdik C. Unlocking bladder cancer. Nature. 2017;551:S34-S35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 13. | Zhu S, Yu W, Yang X, Wu C, Cheng F. Traditional Classification and Novel Subtyping Systems for Bladder Cancer. Front Oncol. 2020;10:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796-2810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 1030] [Article Influence: 114.4] [Reference Citation Analysis (0)] |
| 15. | Dobruch J, Oszczudłowski M. Bladder Cancer: Current Challenges and Future Directions. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 208] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 16. | Wang L, Yu C, Lu Y, He P, Guo J, Zhang C, Song Q, Ma D, Shi T, Chen Y. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007;12:1489-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Chluba-de Tapia J, de Tapia M, Jäggin V, Eberle AN. Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene. 1997;197:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Zaravinos A, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Identification of common differentially expressed genes in urinary bladder cancer. PLoS One. 2011;6:e18135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Pan S, Sun S, Liu B, Hou Y. Pan-cancer Landscape of the RUNX Protein Family Reveals their Potential as Carcinogenic Biomarkers and the Mechanisms Underlying their Action. J Transl Int Med. 2022;10:156-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 20. | Hu YX, Zheng MJ, Zhang WC, Li X, Gou R, Nie X, Liu Q, Hao YY, Liu JJ, Lin B. Systematic profiling of alternative splicing signature reveals prognostic predictor for cervical cancer. J Transl Med. 2019;17:379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Zhuo W, Sun M, Wang K, Zhang L, Li K, Yi D, Li M, Sun Q, Ma X, Liu W, Teng L, Yi C, Zhou T. m(6)Am methyltransferase PCIF1 is essential for aggressiveness of gastric cancer cells by inhibiting TM9SF1 mRNA translation. Cell Discov. 2022;8:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Miyazaki T, Ikeda K, Sato W, Horie-Inoue K, Inoue S. Extracellular vesicle-mediated EBAG9 transfer from cancer cells to tumor microenvironment promotes immune escape and tumor progression. Oncogenesis. 2018;7:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 23. | Di Cristina M, Minenkova O, Pavoni E, Beghetto E, Spadoni A, Felici F, Gargano N. A novel approach for identification of tumor-associated antigens expressed on the surface of tumor cells. Int J Cancer. 2007;120:1293-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Chen H, Deng Q, Wang W, Tao H, Gao Y. Identification of an autophagy-related gene signature for survival prediction in patients with cervical cancer. J Ovarian Res. 2020;13:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |