Published online Nov 24, 2023. doi: 10.5306/wjco.v14.i11.479
Peer-review started: August 28, 2023
First decision: September 14, 2023
Revised: September 20, 2023
Accepted: October 23, 2023
Article in press: October 23, 2023
Published online: November 24, 2023
Processing time: 85 Days and 20.8 Hours
The COP9 signalosome subunit 6 (COPS6) has been implicated in cancer progression, while its precise role in most types of cancer remains elusive.
To investigate the functional and clinical relevance of COPS6 across various tumor types using publicly available databases.
We used R software and online analysis databases to analyze the differential expression, prognosis, mutation and related functions of COPS6 in pan-cancer.
Differential expression analysis and survival analysis demonstrated that COPS6 was highly expressed and associated with high-risk profiles in the majority of cancer types. Possible associations between COPS6 expression level and pro
In conclusion, this study systematically explored the significance of COPS6 across different tumor types, providing a solid foundation for considering COPS6 as a novel biomarker in cancer research.
Core Tip: The COP9 signalosome subunit 6 COPS6 has been implicated in several cancer types. However, its precise role in most cancer types remains poorly understood. Therefore, we aimed to investigate the function of COPS6 in various tumor types. Through our analysis, we discovered that COPS6 is highly expressed and associated with high-risk profiles in most cancers. Meanwhile, COPS6 expression was positively correlated with tumor mutation burden, microsatellite instability, and immune infiltration of the tumor microenvironment. Our findings suggest that COPS6 could be a potential biomarker for cancer research. Our study contributes to the understanding of the role of COPS6 in cancer progression and highlights the clinical applications.
- Citation: Wang SL, Zhuo GZ, Wang LP, Jiang XH, Liu GH, Pan YB, Li YR. Computational exploration of the significance of COPS6 in cancer: Functional and clinical relevance across tumor types. World J Clin Oncol 2023; 14(11): 479-503
- URL: https://www.wjgnet.com/2218-4333/full/v14/i11/479.htm
- DOI: https://dx.doi.org/10.5306/wjco.v14.i11.479
According to recent data from the American Cancer Society, it is projected that the United States will witness 1958310 new cancer cases and 609820 cancer-related deaths by 2023[1]. While there has been a 1.5% decrease in cancer mortality rates between 2019 and 2020, with an overall decline of 33% since 1991, the incidence of breast cancer, prostate cancer, and uterine cancer is on the rise, imposing potential challenges to the future progress. Furthermore, these types of cancer demonstrate significant disparities in mortality rates among different ethnic groups. Conversely, gastric cancer, esophageal cancer, and cervical cancer have shown downward mortality rates, while lung cancer, colorectal cancer (CRC), and female breast cancer continue to exhibit gradually upward mortality rates[2,3]. These trends underscore the persisting challenges caused by cancer. Hence, it is imperative to explore new targets for early diagnosis and personalized treatment, as emphasized by previous studies[4,5]. The identification and analysis of novel pan-cancer genes can provide valuable insights into the intricate process of tumorigenesis. Public databases and online analysis tools, such as The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) provide convenient access to comprehensive cancer-related functional genomics datasets across diverse cancer types, enabling in-depth pan-cancer analysis[6,7].
The COP9 signalosome (COPS) is a multiprotein complex involved in protein degradation, transcriptional activation, signal transduction, and tumor progression[8,9]. COPS6, together with its dimerization partner COPS5, plays a crucial role in the activation process of deneddylase activity by embedding into the core of the helical bundle[10]. While the literature has reported the mechanisms of COPS6 in human malignancies, such as cervical cancer, papillary thyroid carcinoma (THCA), CRC, breast cancer, lung adenocarcinoma (LUAD), and glioblastoma, the available information is incomplete and the underlying mechanisms remain to be fully elucidated[11].
The present study aimed to comprehensively elucidate the involvement and clinical implications of COPS6 in many diverse types of cancer. This involved an in-depth analysis of differential expression patterns, prognostic values, gene mutations, immune infiltration, correlation analysis, and functional enrichment assessment, utilizing publicly available databases.
GEPIA2 (http://gepia2.cancer-pku.cn/#degenes) was utilized for analysis of RNA-seq expression data collected from TCGA and Genotype-Tissue Expression (GTEx) projects, enabling differential analysis, correlation analysis, and survival analysis[12]. Genetic difference analysis was conducted using TIMER2 (http://timer.cistrome.org/)[13]. COPS6 expression level in tumor and normal samples was compared using R (ver. 4.0.3), TIMER2, and GEPIA2. Box plots were generated using the ggpubr R (ver. 4.0.3) package, and differential expression of COPS6 in TCGA samples was determined using the Wilcoxon test. For cancers lacking normal controls in the TCGA database, the TIMER2 website was employed for differential analysis of the COPS6 gene.
For protein analysis, UALCAN online portal (http://ualcan.path.uab.edu/analysis-prot.html) was utilized to examine gene, protein, methylation, and phosphorylation differences[14]. UALCAN facilitated the comparison of differential expression of COPS6 protein between tumor and normal tissues. Age-differential expression data was obtained using the limma and ggpubr R packages, while clinical stage differential expression of COPS6 was obtained from the GEPIA2 website.
To perform survival analysis for COPS6, the "Survival Map" feature of GEPIA2 was utilized. This facilitated plotting heatmaps representing overall survival (OS) and disease-free survival (DFS) using data from TCGA database. Forest plots, encompassing OS, progression-free interval (PFI), disease-specific survival (DSS), and disease-free interval (DFI), were generated using the survival and forestplot R packages in association with Cox analysis.
In March 2022, the pan-cancer data were downloaded from the TCGA database, including tumor stage, tumor grade, survival time, and mutation information. The raw data were preprocessed by the R programming language. The survminer R package was utilized to generate Kaplan-Meier survival curves for OS, PFI, DSS, and DFI, with a significance level set at P < 0.05.
For mutation analysis, cBioPortal (https://www.cbioportal.org/) was utilized[15]. In this study, the "Quick Search" feature of cBioPortal was employed to examine the mutation frequency, type, copy number alteration (CNA), and structural variants of TCGA tumors involving COPS6. Furthermore, information related to the specific mutation sites and three-dimensional (3D) structure of the COPS6 protein was collected. To assess the impact of COPS6 alterations on patient survival, "TCGA, PanCancer Atlas" and "Compassion/Survival" modes were utilized to plot OS, DSS, DFS, and progression-free survival curves for TCGA cases with and without COPS6 alterations, respectively, using the log-rank test.
To investigate immune infiltration, TIMER2 web server was used. In the present study, the presence of CD8+ T cells, cancer-associated fibroblasts, natural killer (NK) cells, and macrophages was assessed using the "Immune" module. To explore the relationship between immune inflammatory cells and COPS6 expression, multiple algorithms were utilized, including XCELL, EPIC, TIMER, MCPCOUNTER, CIBERSORT-ABS, TIDE, CIBERSORT, and QUANTISEQ. P values and partial correlation values were obtained using the purity-adjusted Spearman's rank correlation test to quantify the strength and significance of the observed correlations.
The "Similar Gene Detection" module on the GEPIA2 website was utilized to identify the top 100 genes correlated with COPS6. Further analysis using the "correlation analysis" module on the same website narrowed down the selection to the top 5 genes with the highest correlation coefficients. Heatmap analysis of these genes was conducted using the "Gene_Corr" module available on the TIMER2 website. The purity-adjusted Spearman's rank correlation test was applied to obtain P-values and partial correlation values.
For protein-protein interaction (PPI) analysis, the STRING database (https://string-db.org/) was employed[16]. COPS6 was submitted to the database to generate a PPI network with Homo sapiens as the reference organism. The network settings included a full network type, evidence-based network edges, experiments as active interaction sources, a minimum required interaction score of high confidence (0.7), and a maximum number of interactors shown in the 1st and 2nd shells.
To identify overlapping genes between the COPS6-related genes obtained from the GEPIA2 and STRING, a Venn diagram was generated using GraphPad Prism 9.0.0 software (GraphPad Software Inc., San Diego, CA, United States). The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the DAVID database (https://david.ncifcrf.gov/home.jsp) with involvement of parameters, such as "OFFICIAL_GENE_SYMBOL", "Homo sapiens", and "functional annotation chart".
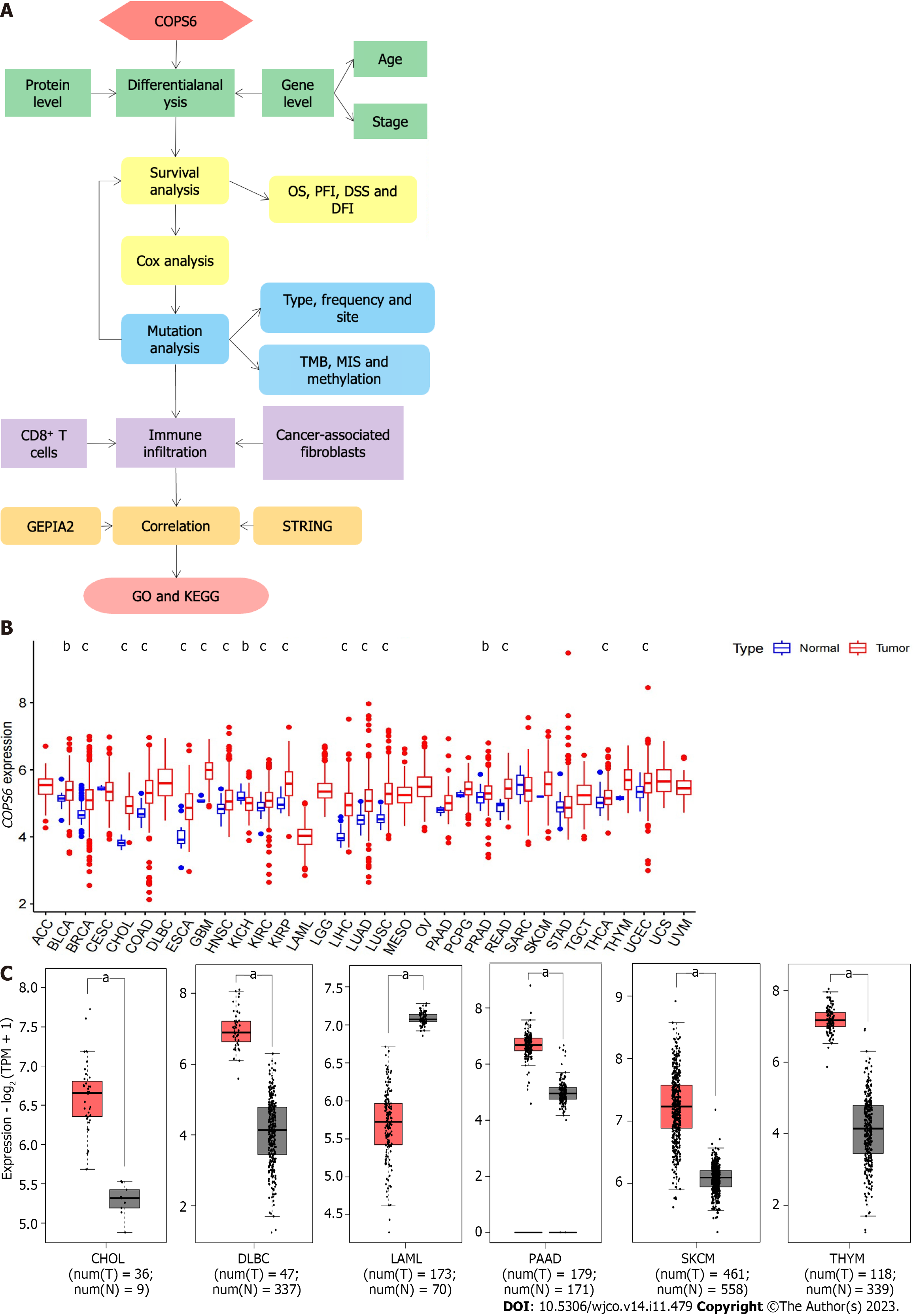
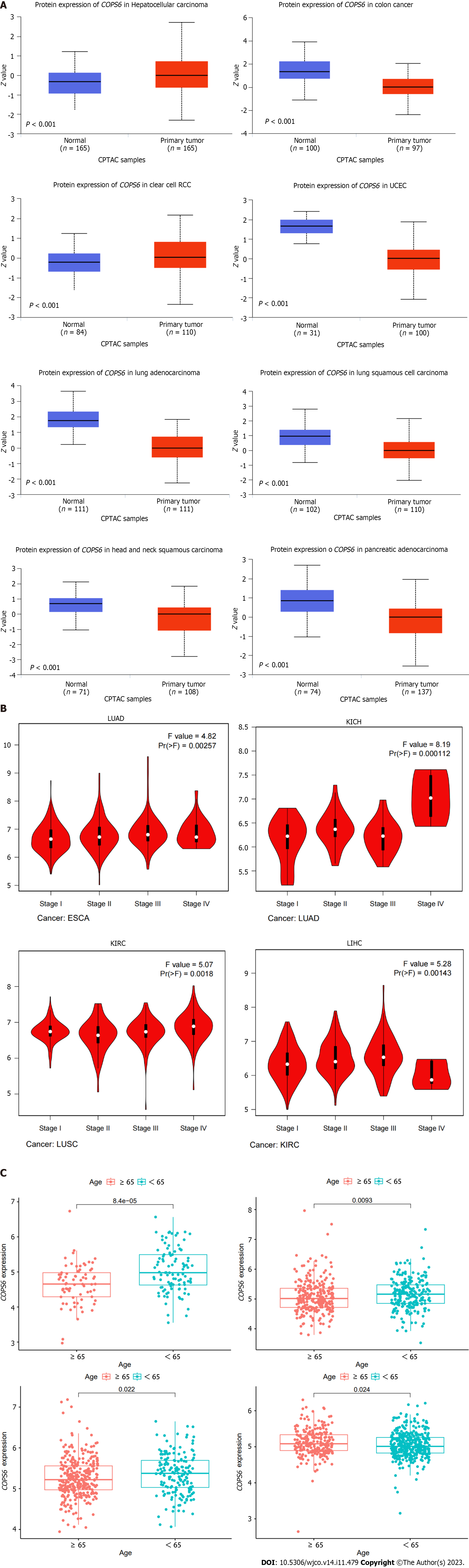
The overview of the pan-cancer analysis workflow is shown in Figure 1A. Differential expression analysis of COPS6 was conducted on TCGA data using R programming language. Significant differential expression (P < 0.05) of COPS6 was found between normal and tumor tissues in several cancer types, including bladder cancer (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), LUAD, lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), THCA, and uterine corpus endometrial carcinoma (UCEC) (Figure 1B). Differential expression analysis results of COPS6 in these tumors and normal samples were obtained from the GEPIA2 website. Furthermore, COPS6 expression level exhibited significant differences in skin cutaneous melanoma (SKCM), pancreatic adenocarcinoma (PAAD), thymoma (THYM), diffuse large B-cell lymphoma (DLBCL), acute myeloid leukemia (LAML), and CHOL (Figure 1C) compared with TCGA normal samples using GTEx data.
To compare protein expression level of COPS6 among multiple types of cancer, protein expression differences between tumor and normal tissues from the CPTAC database on the UALCAN website were compared. The results revealed the elevated expression level of COPS6 in hepatocellular carcinoma and clear cell renal cell carcinoma tissues (Figure 2A). Additionally, the relationship between COPS6 expression level and clinical parameters was investigated using the GEPIA2 website, indicating the presence of association between COPS6 expression level and clinical stages of LUAD, KICH, KIRP, and LIHC (P < 0.05) (Figure 2B). Furthermore, significantly upregulated COPS6 expression level in ESCA, LUAD, and LUSC in cases who aged < 65-years-old in TCGA database was found, whereas COPS6 expression level was reduced in KIRC (P < 0.05) (Figure 2C).
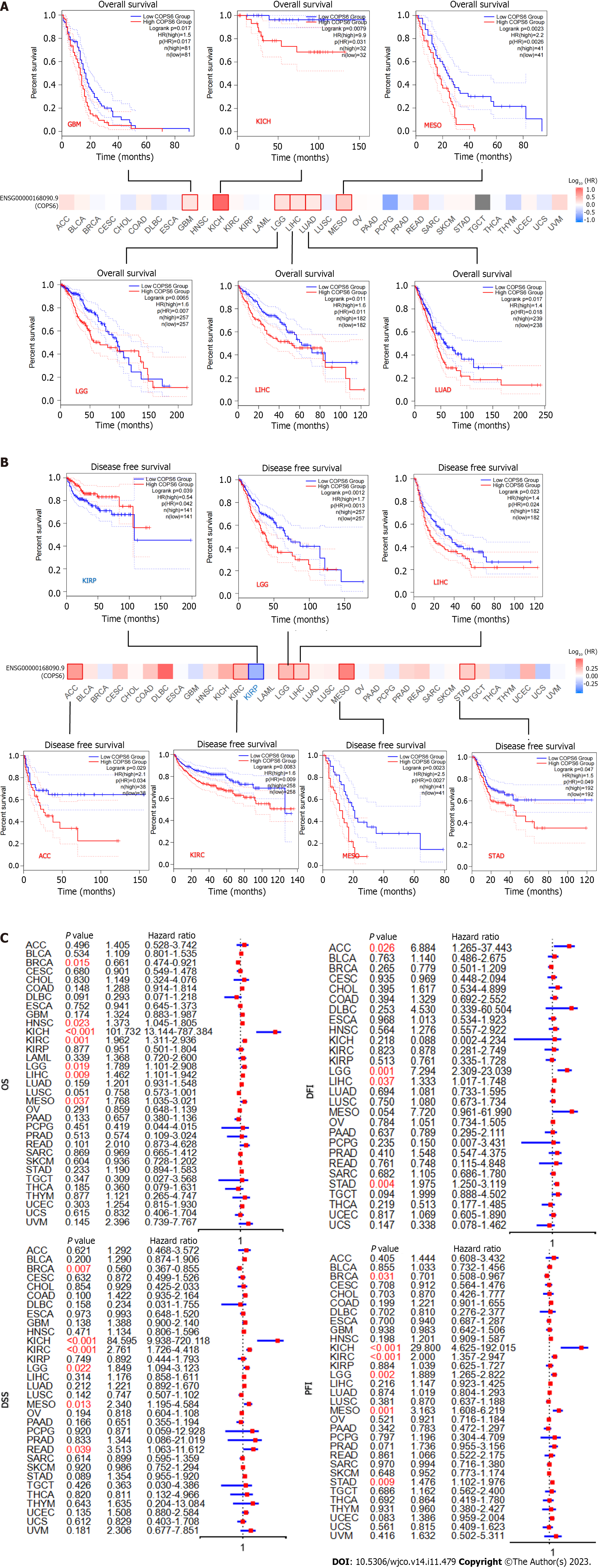
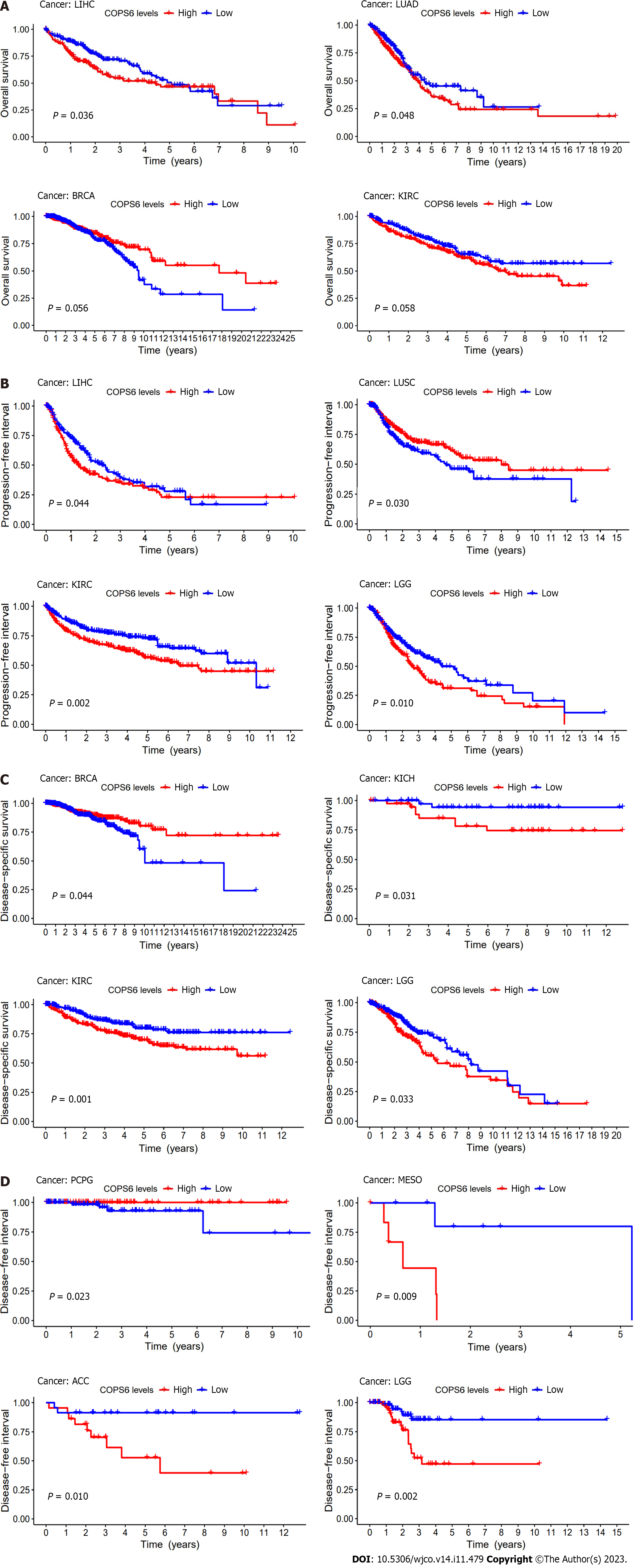
To assess the relationship between COPS6 expression level and patient prognosis across various tumors, patients were divided into high and low COPS6 expression groups based on the median COPS6 expression level. Utilizing the GEPIA2 website, it was revealed that high expression level of COPS6 was significantly associated with poor OS in GBM (P = 0.017), KICH (P = 0.031), mesothelioma (MESO) (P = 0.0026), lower grade glioma (LGG) (P = 0.007), LIHC (P = 0.011), and LUAD (P = 0.018) (Figure 3A). Conversely, low expression level of COPS6 was correlated with poor DFS in KIRP (P = 0.042), while high expression level of COPS6 was associated with poor DFS in LGG (P = 0.0013), LIHC (P = 0.024), adrenocortical carcinoma (ACC) (P = 0.034), KIRC (P = 0.009), MESO (P = 0.0027), and stomach adenocarcinoma (STAD) (P = 0.049) (Figure 3B). Cox regression analysis indicated that COPS6 was a high-risk gene for OS in HNSC, KICH, KIRC, LGG, LIHC, and MESO (P < 0.05), while it was appeared as a low-risk gene for OS in BRCA (P < 0.05). Additionally, COPS6 was identified as a high-risk gene for DSS in KICH, KIRC, LGG, MESO, and READ (P < 0.05), as well as a low-risk gene for DSS in BRCA (P < 0.05). Moreover, COPS6 was found as a high-risk gene for DFI in ACC, LGG, LIHC, and STAD (P < 0.001), as well as a high-risk gene for PFI in KICH, KIRC, LGG, MESO, and STAD (P < 0.05), while a low-risk gene for PFI in BRCA (P < 0.05). These findings were derived from TCGA database using the survival and forestplot R package (Figure 3C). The association between COPS6 expression level and OS (Figure 4A), PFI (Figure 4B), DSS (Figure 4C), and DFI (Figure 4D) was further confirmed through Kaplan-Meier survival analysis in pan-cancer patients from TCGA database.
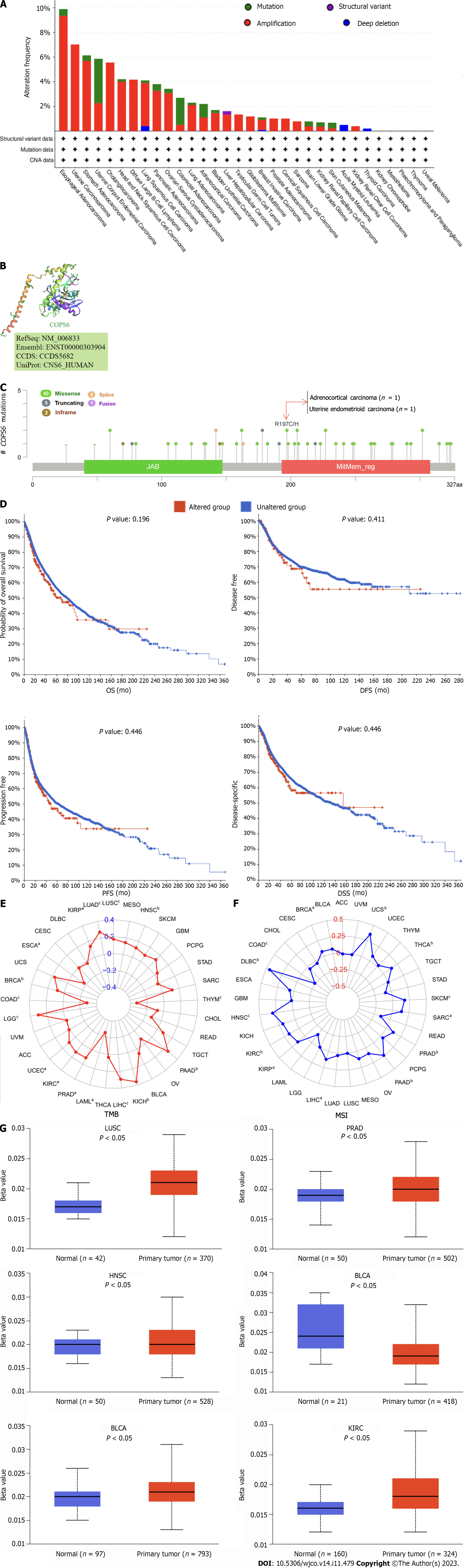
Using the cBioPortal website, comprehensive information was obtained regarding the mutation types, frequency, CNAs, and structural variants of COPS6 across all TCGA tumors. Missense mutations were identified as the predominant mutation type. Among all TCGA tumors, the highest frequency of variations was found in esophageal adenocarcinoma (9.89%), with amplification being the most frequent alteration (9.34%) (Figure 5A). A 3D representation of the COPS6 protein (Figure 5B) was constructed, revealing a notable mutation site, R197C/H, observed in one case each of adrenocortical carcinoma and endometrioid carcinoma (Figure 5C). Investigation of the relationship between COPS6 mutations and prognosis in TCGA cases revealed no significant impact of mutation status on the prognosis of all types of cancer (Figure 5D).
Furthermore, the correlations between COPS6 expression level and tumor mutational burden (TMB) and microsatellite instability (MSI) were analyzed. Positive correlations were identified between COPS6 expression level and TMB in LUAD, KIRP, LUSC, HNSC, PAAD, KICH, LIHC, KIRC, UCEC, LGG, BRCA, and PRAD (P < 0.05), while negative correlations were found in THYM, COAD, ESCA, and LAML (P < 0.05) (Figure 5E). Positive associations between COPS6 expression level and MSI were observed in BRCA, USC, THCA, SKCM, SARC, PRAD, PAAD, KIRP, KIRC, HNSC, DLBC, and LIHC (P < 0.05), with a positive association observed in COAD (P < 0.05) (Figure 5F). Additionally, comparison of the COPS6 promoter methylation level between normal and tumor samples revealed a higher methylation level in the tumor group in PRAD, LUSC, HNSC, BRCA, and KIRC, whereas a lower methylation level in BLCA (Figure 5G).
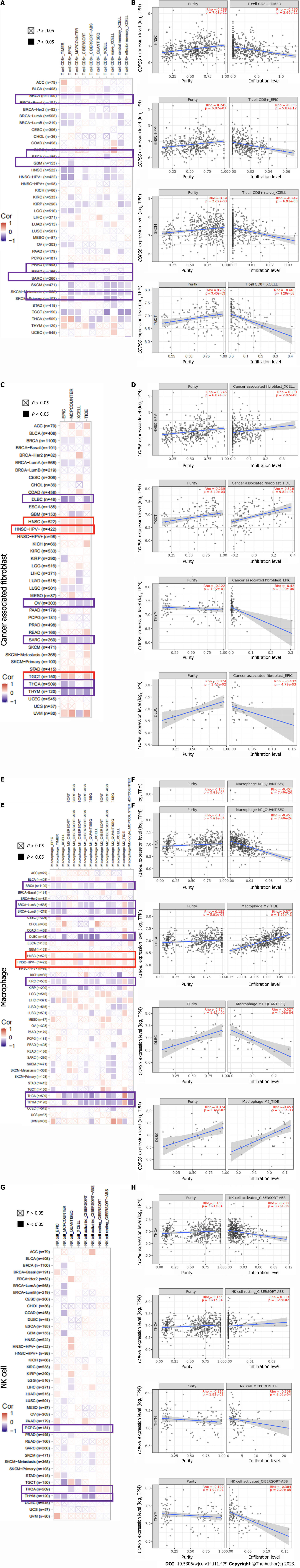
The relationship between COPS6 expression level and immune-infiltrating cells in TCGA tumors was examined using the TIMER2 website. Across multiple algorithms, a negative correlation was found between COPS6 expression level and CD8+ T cell infiltration in BRCA-LumA, HNSC, HNSC-HPV-, SKCM, SKCM-metastasis, and tenosynovial giant cell tumor (TGCT) (Figure 6A and B). Conversely, the correlation between cancer-associated fibroblast infiltration and COPS6 expression level exhibited heterogeneity. Negative correlations were identified in DLBCL, OV, SARC, THYM, and THCA, while positive correlations were found in HNSC, HNSC-HPV-, and TGCT (Figure 6C and D). The relationship between COPS6 expression level and macrophage infiltration varied depending on macrophage subtype. An inverse association was detected between COPS6 expression level and M1 macrophages, along with a positive association between COPS6 expression level and M2 macrophages in certain tumors (Figure 6E and F). For instance, in DLBCL, four algorithms demonstrated a negative association between COPS6 expression level and M1 macrophages, while TIDE algorithm revealed a positive association between COPS6 expression level and M2 macrophages. Moreover, in BRCA, KIRC, THCA, and THYM, the TIDE algorithm indicated a positive correlation between COPS6 expression level and M2 macrophages. In most of the tumors, NK cell infiltration exhibited a weak correlation with COPS6 expression level, and clear associations were found only in a few tumors (Figure 6G and H). For instance, in THCA and THYM, COPS6 expression level showed a negative correlation with NK cell infiltration. Further analysis of NK cell subtypes revealed a negative correlation between COPS6 expression level and activated NK cell infiltration, as well as a positive correlation between COPS6 expression level and resting NK cell infiltration.
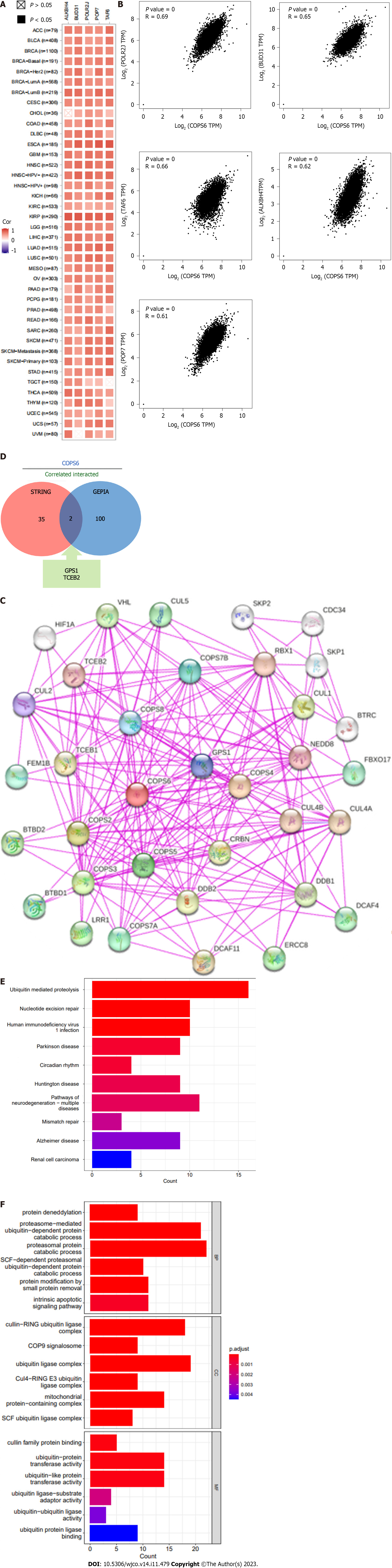
To gain deeper insights into the molecular mechanisms involving COPS6 in growth and progression of tumors, the GEPIA2 website was employed to screen the top 100 COPS6-associated genes. Subsequently, the top 5 genes with the highest correlation coefficients were identified and summarized as follows: POLR2J (r = 0.69, P < 0.001), BUD31 (r = 0.65, P < 0.001), TAF6 (r = 0.66, P < 0.001), ALKBH4 (r = 0.62, P < 0.001), and POP7 (r = 0.61, P < 0.001) (Figure 7A and B). The PPI network analysis was performed using the STRING website, resulting in the establishment of a network of 35 node genes (Figure 7C). The intersection of COPS6-associated genes obtained from the GEPIA2 and STRING led to the identification of GPS1 and TCEB2 (Figure 7D). Furthermore, the genes derived from both databases were merged, resulting in the detection of a total of 135 COPS6-related genes. Subsequently, GO and KEGG pathway enrichment analyses were conducted (Figure 7E and F). The KEGG pathway analysis revealed that COPS6-associated genes were enriched in pathways, such as ubiquitin-mediated proteolysis, nucleotide excision repair, human immunodeficiency virus 1 infection, Parkinson's disease, and circadian rhythm. The GO enrichment analysis indicated enrichment in the proteasomal protein catabolic process, proteasome-mediated ubiquitin-dependent protein catabolic process, protein modification by small protein removal, intrinsic apoptotic signaling pathway, protein deneddylation, COPS, Cullin-RING ubiquitin ligase (CRL) complex, SCF ubiquitin ligase complex, Cul4A-RING E3 ubiquitin ligase complex, Cullin family protein binding, ubiquitin-protein transferase activity, and ubiquitin-like protein transferase activity.
The COP9 signalosome (CSN) is a complex protein composed of eight subunits (CSN1-CSN8), participating in various physiological processes. The CSN1, 2, 3, 4, 7, and 8 subunits contain a percutaneous coronary intervention domain, which acts as a scaffold in CSN assembly, while the COPS6 and COPS5 subunits possess an Mpr1-Pad 1-N-terminal (MPN) domain[17]. COPS5 primarily exerts catalytic enzymatic activity, whereas COPS6, as an essential component of CSN, lacks the metal-binding site and isopeptidase activity associated with the COPS5 MPN domain. The precise function of COPS6 remains has still remained elusive[18]. COPS6 is involved in various processes, including the ubiquitin proteasome system, signal transduction, DNA damage response, and tumor progression. It exhibits a high expression level in diverse tumors, and studies have explored its role in cancer[19].
At present, there is a growing interest among researchers in investigating the role of COPS6 in tumors, as it has been shown to predominantly promote cancer. The CRLs are involved in the ubiquitination of Myc, and Fbxw7, a CRL component, which participates in Myc ubiquitination. In mouse experiments, Chen et al[20] demonstrated that COPS6 enhances Fbxw7 degradation through binding, thereby maintaining Myc stability and promoting tumor progression. Additionally, in a mouse model, Zhao et al[21] revealed that COPS6 attenuates p53-mediated tumor suppression, promotes tumor growth by stabilizing MDM2 protein, and participates in DNA damage-associated apoptosis. In human tumors, COPS6 also plays a significant role in tumor progression. Fang et al[22] demonstrated that COPS6 overexpression in CRC is associated with a worse prognosis. Mechanistic studies suggested that ERK2 directly binds to CSN6 Leu163/Val165 and phosphorylates COPS6 at Ser148, thereby regulating β-Trcp and stabilizing β-catenin expression, consequently blocking the ubiquitin-proteasome pathway and promoting CRC development.
Programmed death-1/programmed death-ligand 1 (PD-1/PD-L1) checkpoint blockade is an emerging immunotherapy modality in various tumors, while its regulatory mechanism remains uncertain. Su et al[23] demonstrated that COPS6 expression level could be regulated by the EGFR-ERK pathway, inhibiting PD-L1 degradation and maintaining PD-L1 stability in GBM. Additionally, several studies have reported the involvement of COPS6 in the epithelial-mesenchymal transition process in various tumors, promoting tumor invasion and metastasis. For instance, Zhang et al[24] revealed that the COPS6-UBR5-CDK9 axis could control melanoma proliferation and metastasis, while Mao et al[25] found that COPS6 could promote migration and invasion of cervical cancer cells by regulating the expression level of cathepsin L through the autophagy-lysosomal system. Furthermore, COPS6 was found to maintain the key transcription factor Snail1, promoting the invasion of breast cancer cells by inhibiting Snail1 ubiquitination[26].
While previous studies have highlighted the significant role of COPS6 in the progression of specific tumors, the heterogeneity of tumors suggests potential variations in its function across different cancer types. Therefore, a comprehensive analysis and screening are necessary to validate existing findings and provide direction for the future COPS6-related studies. In the present study, it was attempted to conduct comprehensive multilevel differential analysis and survival analysis of COPS6 in pan-cancer data collected from various public databases and online analysis tools, including TCGA, GEO, CPTAC, GEPIA2, TIMER2, and UALCAN. The findings demonstrated that the expression level of the COPS6 gene was significantly upregulated in the most types of cancer compared with normal tissues, except for KICH and LAML. Prognostic analysis revealed that the high expression level of COPS6 was typically associated with worse prognosis in the majority of tumors, while showing a favorable prognosis in KIRP, BRCA, LUSC, and PCPG. Mutational analysis indicated that missense mutations were the predominant mutation type found in COPS6. Additionally, TMB and MSI exhibited a positive correlation with COPS6 expression level in most of the tumors, and only few tumors showed a negative correlation. Further exploration of the impact of COPS6 mutations on patient outcomes revealed that these mutations did not significantly contribute to a worse prognosis in any specific tumor types. However, a comprehensive analysis across all tumors indicated a trend towards a shorter OS associated with COPS6 mutations. Therefore, it can be concluded that COPS6 mutations have a limited effect on patient prognosis.
Subsequently, the association between COPS6 expression level and the tumor immune microenvironment (TMIE) was investigated in various types of cancer. The TMIE plays a pivotal role in tumor progression, immune evasion, and therapeutic resistance, involving key components, such as CD8+ T cells, cancer-associated fibroblasts, macrophages, and NK cells[27]. The findings of the present study demonstrated a negative correlation between COPS6 expression level and CD8+ T cell infiltration in several tumors, such as BRCA, HNSC, and TGCT. This aligns with Du et al[28]’s results, demonstrating that COPS6 could inhibit CD8+ T cell infiltration within the tumor microenvironment (TME), thereby facilitating tumor immune evasion. Furthermore, a negative correlation was identified between COPS6 expression level and the M1 phenotype of tumor-associated macrophages (TAMs), while a positive correlation was found with the M2 phenotype. TAMs, which are macrophages that infiltrate tumor tissue and differentiate from monocytes, predominantly adopt the immunosuppressive M2 phenotype in the TMIE[29]. The present study revealed a positive correlation between COPS6 expression level and the M2 phenotype in the TIDE algorithm for DLBCL, BRCA, KIRC, THCA, THYM, and other tumors, while other algorithms exhibited a negative correlation with the M1 phenotype. However, it is noteworthy that in some tumors, only the TIDE algorithm yielded consistent results, while other algorithms suggested a negative or no correlation between COPS6 expression level and the M2 phenotype. This discrepancy could be attributed to variations in the statistical methods employed by each algorithm, necessitating further experimental validation of these findings.
There is a scarcity of research regarding the interaction between COPS6 Level and TME, highlighting the urgent need to explore the role of COPS6 in the TME. Furthermore, in the present study, correlation and enrichment analyses of COPS6 were conducted, and GPS1 and TCEB2 were identified as the two genes, exhibiting the strongest correlation. This investigation sheds light on the potential function and significance of COPS6 as a novel biomarker in cancer, setting the stage for further research on its molecular mechanisms and the development of targeted therapies. Moreover, the findings emphasize the importance of studying the COPS6-related TIME. However, it should be noted that the current study of COPS6 is preliminary, and the specific mechanisms of its action in different types of cancer remain elusive. Therefore, additional resources and efforts are warranted to delve deeper into the role of COPS6 in cancer.
The present study revealed a potential association of COPS6 with survival outcomes in various tumors. Notably, GPS1 and TCEB2 were identified as the two genes exhibiting the strongest correlation with COPS6 at both the gene and protein levels, making them promising targets for future investigations. Additionally, a significant association was found between COPS6 expression level and immune infiltration in diverse types of cancer, such as BRCA, HNSC, and TGCT, where research on the TIME remains limited.
This study is the first to explore the role of COPS6 in pan-cancer, taking full use of the existing public database to investigate COPS6 from the aspects of gene expression level, mutation, TIME, and prognosis. However, there are also some deficiencies in this study. For instance, only a multifaceted analysis of COPS6 was conducted through bioinformatics, while no experiment was carried out to verify the results, hindering the generalization of the findings.
In conclusion, the present study provided early evidence that COPS6 could be associated with clinicopathological characteristics in various tumors and could play a role in several cancer hallmarks. Additional research is needed to further elucidate the role of COPS6 in cancer progression.
The COP9 signaling body subunit 6 (COPS6) has been implicated in cancer progression, but its precise role in most types of cancer is unknown.
This study aimed to investigate the functional and clinical relevance of COPS6 in different tumor types, using publicly available databases.
This study hopes to provide a basis for COPS6 as a novel biomarker for cancer research by exploring the role of COPS6 in different cancer types.
We used R software and online analysis databases to analyze the differential expression, prognosis, mutation and related functions of COPS6 in pan-cancer.
Differential expression analysis and survival analysis demonstrated that COPS6 was highly expressed and associated with high-risk profiles in the majority of cancer types. Missense mutations are the main type of COPS6 mutations, and in most types of cancer, the levels of COPS6 expression are positively correlated with tumor mutation burden and microsatellite instability. Immune infiltration analysis found COPS6 to play different roles in different cancers. Gene co-expression and enrichment analysis highlighted COPS6-related genes were predominantly involved in processes, such as ubiquitin-mediated proteolysis and human immunodeficiency virus 1 infection.
This study provides early evidence that COPS6 may be associated with the clinicopathological features of various tumors and may play a role in several cancer features, providing a basis for subsequent studies related to COPS6.
Since this study mainly focused on data analysis, subsequent studies required experimental validation of relevant results.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Martinou E, United Kingdom S-Editor: Lin C L-Editor: Filipodia P-Editor: Zhang XD
| 1. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9945] [Article Influence: 4972.5] [Reference Citation Analysis (2)] |
| 2. | Feng R, Su Q, Huang X, Basnet T, Xu X, Ye W. Cancer situation in China: what does the China cancer map indicate from the first national death survey to the latest cancer registration? Cancer Commun (Lond). 2023;43:75-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 73] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 3. | Wang Y, Yan Q, Fan C, Mo Y, Wang Y, Li X, Liao Q, Guo C, Li G, Zeng Z, Xiong W, Huang H. Overview and countermeasures of cancer burden in China. Sci China Life Sci. 2023;1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 72] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 4. | Gupta S, Harper A, Ruan Y, Barr R, Frazier AL, Ferlay J, Steliarova-Foucher E, Fidler-Benaoudia MM. International Trends in the Incidence of Cancer Among Adolescents and Young Adults. J Natl Cancer Inst. 2020;112:1105-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 5. | Bhakta N, Force LM, Allemani C, Atun R, Bray F, Coleman MP, Steliarova-Foucher E, Frazier AL, Robison LL, Rodriguez-Galindo C, Fitzmaurice C. Childhood cancer burden: a review of global estimates. Lancet Oncol. 2019;20:e42-e53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 251] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 6. | Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016;1418:93-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 1445] [Article Influence: 160.6] [Reference Citation Analysis (0)] |
| 7. | Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-Analyzed Tumors. Cell. 2018;173:530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 454] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 8. | Ma XL, Xu M, Jiang T. Crystal structure of the human CSN6 MPN domain. Biochem Biophys Res Commun. 2014;453:25-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thomä NH. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 10. | Faull SV, Lau AMC, Martens C, Ahdash Z, Hansen K, Yebenes H, Schmidt C, Beuron F, Cronin NB, Morris EP, Politis A. Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat Commun. 2019;10:3814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 11. | Hou J, Cui H. CSN6: a promising target for cancer prevention and therapy. Histol Histopathol. 2020;35:645-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3076] [Article Influence: 512.7] [Reference Citation Analysis (0)] |
| 13. | Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2451] [Cited by in RCA: 3367] [Article Influence: 673.4] [Reference Citation Analysis (0)] |
| 14. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4227] [Article Influence: 528.4] [Reference Citation Analysis (0)] |
| 15. | Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, Schultz N, Fenyö D, Gao J. Integration and Analysis of CPTAC Proteomics Data in the Context of Cancer Genomics in the cBioPortal. Mol Cell Proteomics. 2019;18:1893-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 16. | Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605-D612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1211] [Cited by in RCA: 4825] [Article Influence: 1206.3] [Reference Citation Analysis (0)] |
| 17. | Birol M, Enchev RI, Padilla A, Stengel F, Aebersold R, Betzi S, Yang Y, Hoh F, Peter M, Dumas C, Echalier A. Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS One. 2014;9:e105688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Xu M, Zhen L, Lin L, Wu K, Wang Y, Cai X. Overexpression of CSN6 promotes the epithelial-mesenchymal transition and predicts poor prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2020;44:340-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Choi HH, Lee MH. CSN6-COP1 axis in cancer. Aging (Albany NY). 2015;7:461-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, Post SM, Ho Choi H, Chen JS, Wang E, Zhou Z, Tseng C, Gully C, Velazquez-Torres G, Fuentes-Mattei E, Yeung G, Qiao Y, Chou PC, Su CH, Hsieh YC, Hsu SL, Ohshiro K, Shaikenov T, Yeung SC, Lee MH. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun. 2014;5:5384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 21. | Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, Qu C, Zhang F, Chen YT, Lin YL, Lee DF, Jin F, Zhu R, Shaikenov T, Sarbassov D, Sahin A, Wang H, Lai CC, Tsai FJ, Lozano G, Lee MH. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Fang L, Lu W, Choi HH, Yeung SC, Tung JY, Hsiao CD, Fuentes-Mattei E, Menter D, Chen C, Wang L, Wang J, Lee MH. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell. 2015;28:183-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Su L, Guo W, Lou L, Nie S, Zhang Q, Liu Y, Chang Y, Zhang X, Li Y, Shen H. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol Carcinog. 2020;59:520-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 24. | Zhang Y, Hou J, Shi S, Du J, Liu Y, Huang P, Li Q, Liu L, Hu H, Ji Y, Guo L, Shi Y, Cui H. CSN6 promotes melanoma proliferation and metastasis by controlling the UBR5-mediated ubiquitination and degradation of CDK9. Cell Death Dis. 2021;12:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Mao Z, Sang MM, Chen C, Zhu WT, Gong YS, Pei DS. CSN6 Promotes the Migration and Invasion of Cervical Cancer Cells by Inhibiting Autophagic Degradation of Cathepsin L. Int J Biol Sci. 2019;15:1310-1324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Mou J, Wei L, Liang J, Du W, Pei D. CSN6 promotes the cell migration of breast cancer cells by positively regulating Snail1 stability. Int J Med Sci. 2020;17:2809-2818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 27. | Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer. 2023;23:173-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 160] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 28. | Du WQ, Zhu ZM, Jiang X, Kang MJ, Pei DS. COPS6 promotes tumor progression and reduces CD8(+) T cell infiltration by repressing IL-6 production to facilitate tumor immune evasion in breast cancer. Acta Pharmacol Sin. 2023;44:1890-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Wang S, Liu G, Li Y, Pan Y. Metabolic Reprogramming Induces Macrophage Polarization in the Tumor Microenvironment. Front Immunol. 2022;13:840029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |