Published online Jan 24, 2023. doi: 10.5306/wjco.v14.i1.13
Peer-review started: July 25, 2022
First decision: October 24, 2022
Revised: October 25, 2022
Accepted: December 6, 2022
Article in press: December 6, 2022
Published online: January 24, 2023
Processing time: 169 Days and 19.6 Hours
Esophageal squamous cell carcinoma (ESCC) is causing a high mortality rate due to the lack of efficient early prognosis markers and suitable therapeutic regimens. The prognostic role of genes responsible for the acquisition of radioresistance in ESCC has not been fully elucidated.
To establish a prognostic model by studying gene expression patterns pertinent to radioresistance in ESCC patients.
Datasets were obtained from the Gene Expression Omnibus and The Cancer Genome Atlas databases. The edgeR, a Bioconductor package, was used to analyze mRNA expression between different groups. We screened genes specifically responsible for radioresistance to estimate overall survival. Pearson correlation analysis was performed to confirm whether the expression of those genes correlated with each other. Genes contributing to radioresistance and overall survival were assessed by the multivariate Cox regression model through the calculation of βi and risk score using the following formula: 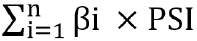 .
.
We identified three prognostic mRNAs (cathepsin S [CTSS], cluster of differentiation 180 [CD180], and SLP adapter and CSK-interacting membrane protein [SCIMP]) indicative of radioresistance. The expression of the three identified mRNAs was related to each other (r > 0.70 and P < 0.05). As to 1-year and 3-year overall survival prediction, the area under the time-dependent receiver operating characteristic curve of the signature consisting of the three mRNAs was 0.716 and 0.841, respectively. When stratifying patients based on the risk score derived from the signature, the high-risk group exhibited a higher death risk and shorter survival time than the low-risk group (P < 0.0001). Overall survival of the low-risk patients was significantly better than that of the high-risk patients (P = 0.018).
We have developed a novel three-gene prognostic signature consisting of CTSS, CD180, and SCIMO for ESCC, which may facilitate the prediction of early prognosis of this malignancy.
Core Tip: The current study identified a novel three-gene prognostic signature consisting of CTSS, CD180, and SCIMO for esophageal squamous cell carcinoma, which may facilitate the prediction of early prognosis of this malignancy.
- Citation: Wang XY, Beeraka NM, Xue NN, Yu HM, Yang Y, Liu MX, Nikolenko VN, Liu JQ, Zhao D. Identification of a three-gene prognostic signature for radioresistant esophageal squamous cell carcinoma. World J Clin Oncol 2023; 14(1): 13-26
- URL: https://www.wjgnet.com/2218-4333/full/v14/i1/13.htm
- DOI: https://dx.doi.org/10.5306/wjco.v14.i1.13
Esophageal cancer is one of the most commonly occurring gastrointestinal tumors and ranks 7th in incidence and 6th in death among all malignancies worldwide. The highest incidence rate was reported in China[1]. Esophageal cancer includes two main pathological types, namely, esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC), and 88% of ESCC cases originate in central and southern Asia[2]. Surgery is the conventional method of treatment for early-stage esophageal cancer patients. Neoadjuvant radiotherapy is also reported to be a crucial therapeutic modality for treating advanced stage ESCC patients[3]. However, the differences in sensitivity of each patient to radiation therapy result in variable prognoses among ESCC patients. ESCC is an aggressive malignancy with a poor overall survival[4]. The available staging system is not very satisfactory in predicting the treatment outcome in ESCC patients, and the application of cancer genomics to predict clinical outcomes may improve the treatment of ESCC[5,6].
Tumor radiotherapy can induce either direct damage to DNA by inducing DNA double-strand breaks, or indirectly modulate cell signaling cascades to foster tumor cell death[7]. However, the clinical outcomes of radiotherapy in most esophageal tumor patients predominantly depend on the inherent sensitivity of tumor cells to radioactive rays. Furthermore, tumor cell insensitivity can lead to the occurrence of radioresistance, which involves several cellular mechanisms including cell cycle checkpoint regulation[8], stemness acquisition[9,10], epithelial mesenchymal transformation (EMT)[11], and activation of multiple pro-survival and pro-proliferation signaling pathways[12,13]. Furthermore, radioresistance is also mediated by tumor-associated microenvironment factors, such as hypoxia-induced HIF-1 signaling factors[14,15], tumor-associated fibroblasts (CAFs)[16], and tumor-associated macrophages[17,18]. Hence, radioresistance is one of the significant reasons for the failure of radiotherapy in ESCC patients. High-throughput sequencing technology is a promising novel approach to identify genes that are related to tumor radioresistance in ESCC. Maher et al[19] identified a set of five genes including EPB41L3, RTKN, STAT5B, NMES1, and RNPC1 as biomarkers for response to neoadjuvant radiotherapy in esophageal cancer. Overexpression of PTK7 can activate NF-kB to enhance raidoresistance in radiosensitive ESCC cells[20]. Transcriptome analysis delineated that the MALAT1-ATG9B and DDIT4-MB-PLAT genes could regulate radioresistance in in vitro models of ESCC cells by modulation of autophagy and hypoxia pathways[21]. The prognostic role and underlying genomic pathways pertinent to the acquisition of radioresistance in ESCC patients have not yet been fully unraveled. Therefore, it is crucial to identify biomarkers and genes pertaining to radioresistance in ESCC for selecting novel therapeutic modalities to mitigate radioresistance in this malignancy.
The current study identified mRNAs as potential radioresistance markers in ESCC cells with the aid of merged mRNA data collected from the Gene Expression Omnibus (GEO) and Cancer Genome Atlas (TCGA) databases. The study identified a three-gene signature, including CTSS, CD180, and SCIMP, that may predict the development of radioresistance in ESCC cells. Furthermore, we constructed a prognostic model for radioresistant ESCC based on the risk scores derived from clinical features and the three-gene signature.
Primarily, the microarray profiles in GSE81812 dataset pertaining to ‘non-radiated KYSE-180 cells’ and “12 and 30 Gy radiated KYSE-180 cells” were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) to identify mRNAs contributing to radioresistance in ESCC cells. The edgeR package (www.bioconductor.org/packages/release/bioc/html/edgeR.html) was used to analyze the differential expression of mRNAs between different groups (‘0 Gy group vs 12 Gy group’ and ‘0 Gy group vs 30 Gy group’) to identify genes related to radioresistance. The cutoff parameters were false discovery rate < 0.05 and|Log2 fold change|>2.
Gene expression profile and clinical information of ESCC patients in the TCGA database were downloaded (https://gdc-portal.nci.nih.gov/). Overall survival rates were determined to ascertain the prognostic significance of the identified radioresistance promoting mRNAs in the TCGA database; the overall survival rates were analyzed by using survival package in R through Kaplan-Meier analysis and finally compared using the Log-rank test and Cox proportional hazards regression analysis. Then, radioresistance-promoting mRNAs associated with overall survival were screened.
The association of radioresistance-promoting mRNAs with overall survival was estimated using the multivariate Cox regression model, adjusted for age, gender, grade, and stage, to calculate βi. The forest plot was plotted to exhibit the hazards regression (HR) of the multivariate Cox regression model results. Later, risk score was estimated by using the following formula: 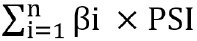 . By using the maximally selected rank statistics from the ‘survminer’ package in R, all samples were divided into a low-risk group and a high-risk group subsequently, and survival analysis was conducted to assess prognosis differences between the two groups.
. By using the maximally selected rank statistics from the ‘survminer’ package in R, all samples were divided into a low-risk group and a high-risk group subsequently, and survival analysis was conducted to assess prognosis differences between the two groups.
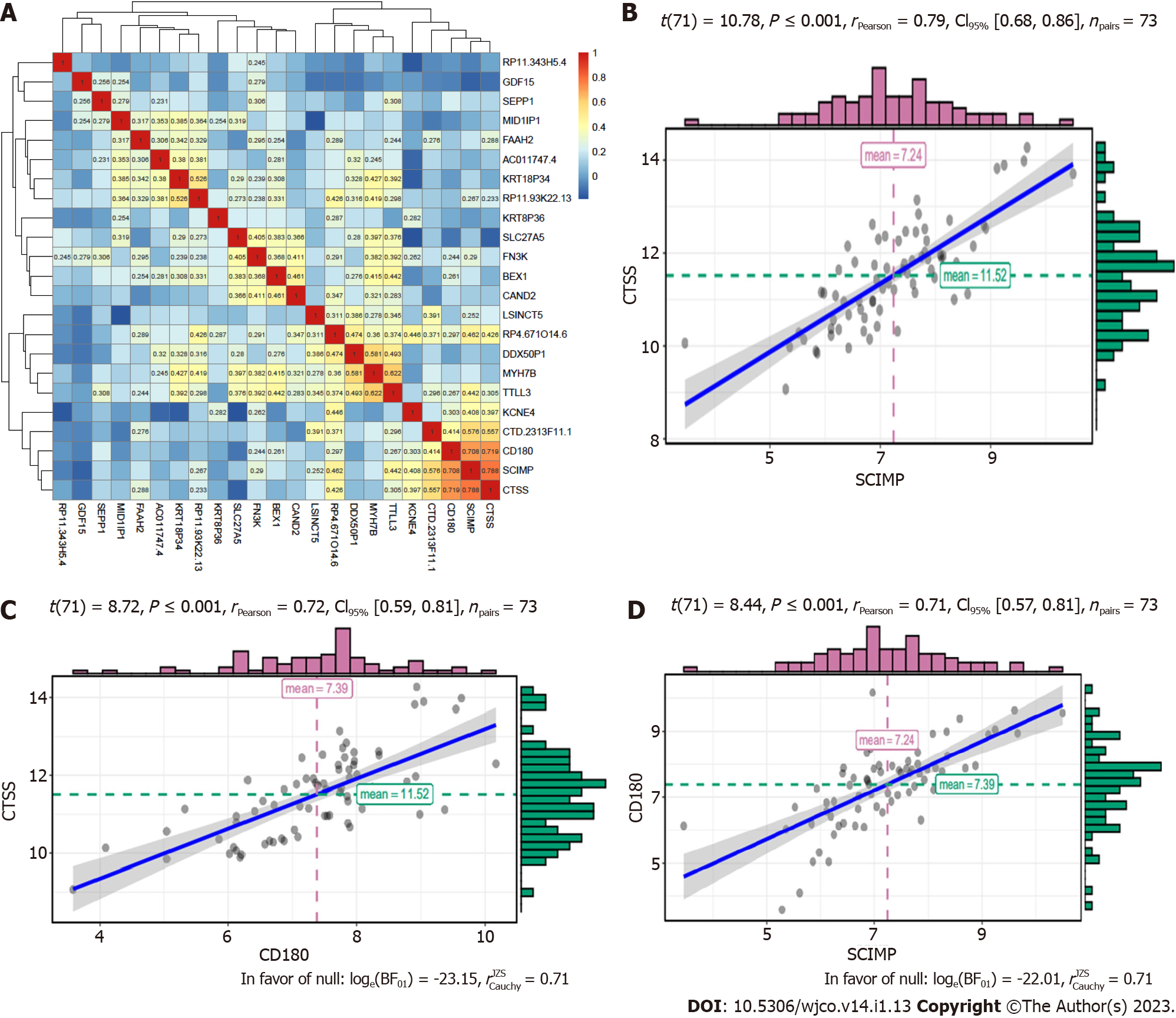
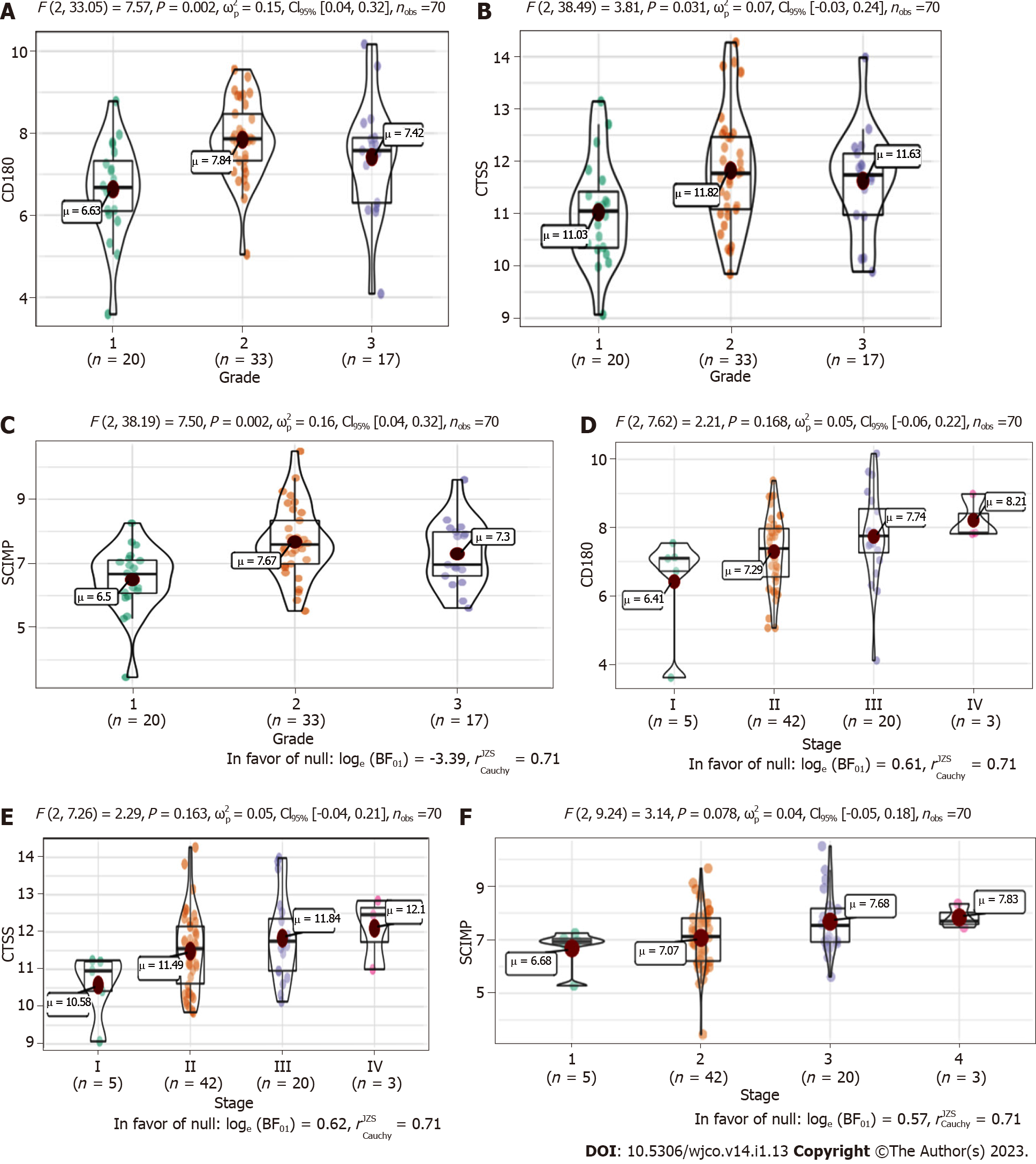
Pearson correlation coefficients (P < 0.05) were calculated using r.test () in R to confirm whether the identified radioresistance-associated mRNAs were typically related to the stage and grade of ESCC. The results are shown in violin plots.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of mRNAs associated with radioresistance in ESCC was performed using the ClusterProfile package (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) for a more comprehensive under
Statistical analyses were executed with the aid of SPSS 22.0 software (IBM, Chicago, IL, United States) and R version 3.6.0. Overall survival rate was estimated using the Kaplan-Meier method. Multivariate Cox proportional HR analysis was executed to identify prognostic factors (the three-gene signature, age, gender, tumor stage, and tumor grade). Differences between groups were compared using the Student’s t-test or paired samples t-test. P < 0.05 was considered to have statistical significance.
The gene count data of expression profiles of 22456 mRNAs in 41 samples of 0 Gy, 92 samples of 12 Gy, and 89 samples of 30 Gy were obtained from the GSE81812 dataset downloaded from GEO. We identified upregulation of 1168 mRNAs in the ‘0 Gy group vs 12 Gy group’ comparison and 497 mRNAs in the ‘0 Gy group vs 30 Gy group’ comparison by using the edgeR package. To distinguish the differentially expressed mRNAs at different X-ray levels, the top 50 mRNAs are shown in a heatmap and principal component analysis (PCA) was performed (Figure 1A-D). A total of 379 intersection mRNAs were identified from the 0-12 Gy and 0-30 Gy comparisons as radioresistance associated genes.
Log-rank test and Cox proportional hazards regression were adjusted for other confounding factors such as gender, age, stage, and grade. These statistical analyses were used to screen for prognostic genes, and a total of 5293 mRNAs were selected. Among them, 44 mRNAs were significantly associated with radioresistance. We selected 23 mRNAs that were negatively correlated with prognosis for further analysis. The intersection of radioresistant prognostic mRNAs is visualized in a Venn diagram (Figure 1E).
For the 23 mRNAs mentioned above, we primarily investigated whether their expression correlated with each other based on the data in the TCGA database. Although they were expressed at different levels in ESCC patients, the results showed strong correlations among three mRNAs, namely, CTSS, CD180, and SCIMP (r > 0.70 and P < 0.05). The correlations of 23 mRNAs are shown in a heatmap (Figure 2A). Hence, we selected these three mRNAs as radioresistance-promoting mRNAs of interest. Correlations of these three mRNAs are shown in a scattergram (Figure 2B-D).
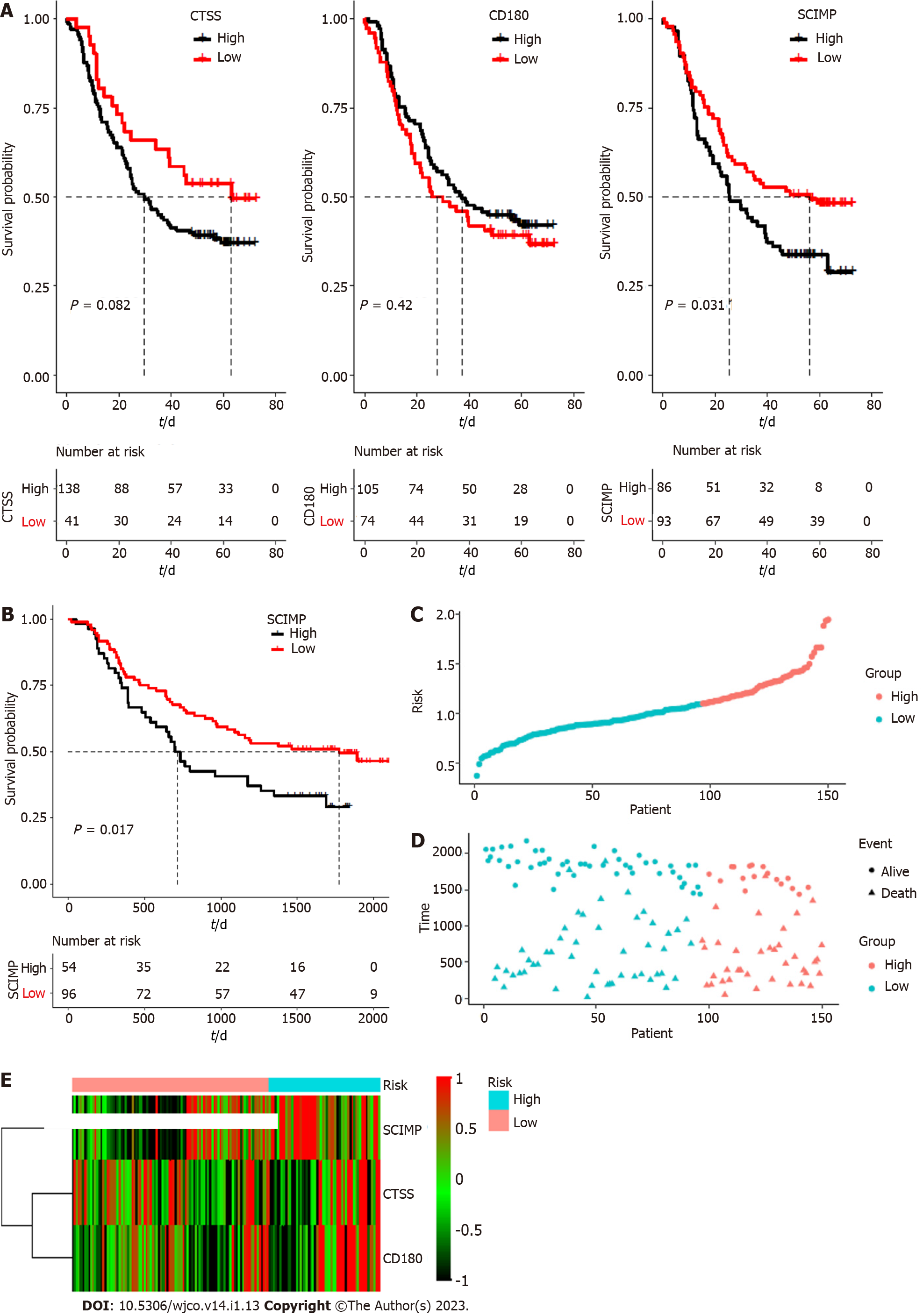
To explore the potential prognostic value of the above three mRNAs pertinent to radioresistance, we evaluated the overall survival rates of ESCC patients based on the expression patterns of these three mRNAs based on the data in the TCGA database by using Kaplan-Meier curves. As shown in Figure 3A, their low expression was associated with a good overall survival (TCGA database), and the median survival time was statistically significant (P < 0.05) for all the three mRNAs.
Subsequently, the connection between the three-gene signature and overall survival was explored through multivariate Cox regression model adjusted for patient age, gender, tumor grade, and tumor stage, for which, the HR with 95% confidence interval was depicted through the forest plot (Figure 3B). ROC analysis for the model is shown in Figure 3C (area under the curve: 0.716 and 0.841 for 1- and 3-year survival, respectively). Accordingly, the risk score of each patient was calculated, and all the patients were divided into either a high risk group or a low risk group based on the risk score.
The patients of the high-risk group exhibited a ‘higher death risk and shorter survival time’ than the patients in the low-risk group; the heatmap of the three genes (CTSS, CD180, and SCIMP) showed that the high-risk patients typically had higher expression of these genes than the low-risk patients (Figure 3D-F). The Kaplan-Meier curves revealed that the low-risk patients typically with low expression of these three genes exhibited a good overall survival (Figure 3G).
To further validate the prognostic value of the three mRNAs, GSE53625 dataset was downloaded from the GEO database. As shown in Figure 4A, downregulation of SCIMP expression was associated with a good survival outcome. When patients were divided into two groups based on CTSS expression, there was no statistically significant difference in their survival. CD180 expression also showed no significant correlation with survival. In the same manner, the risk score of GEO samples was calculated, and the overall survival of patient samples in the low-risk group was also higher than that of patient samples in the high-risk group (Figure 4B). The risk curve, scatter plot, and heatmap results were also similar to those obtained based on TCGA dataset (Figure 4C-E).
We next explored the association between the three radioresistance-promoting mRNAs and pathological grade (Figure 5A-C) and tumor-node-metastasis (TNM) stage (Figure 5D-F). CTSS, CD180, and SCIMP exhibited significantly higher expression in advanced pathological grades (2-3 vs 1) and tumor stages (II-IV vs I).
To further explore the underlying biological features of the three mRNAs in ESCC, we performed Pearson correlation between the three mRNAs, namely, CTSS, CD180, and SCIMP, and the other mRNAs to identify co-expressed mRNAs. A total of 539 mRNAs were selected for KEGG pathway enrichment analysis (P < 0.01, r > 0.4). Our results showed that the co-expressed mRNAs were mainly enriched in 50 pathways, including NF-kB, JAK-STAT, cell adhesion molecules signaling, and PD-L1 expression & PD-1 checkpoint pathways (Figure 6).
Prolonged and fractionated irradiation during radiotherapy in ESCC patients could confer radioresistance and result in distant metastasis, which may lead to treatment failure[22,23]. CAFs can foster radioresistance in ESCC tumor cells through the long noncoding RNA DNM3OS by modulating the PDGFβ/PDGFRβ/FOXO1 signaling pathway, suggesting that CAFs-promoted DNM3OS could be a crucial target to reverse radioresistance in ESCC tumor cells. A study by Zhao et al[24] in 2020, showed that three genes (FOXL2, TCF4, and NR2F2) exhibited a significant correlation with the prognosis of endometrial carcinoma; biological pathways associated with the low expression of these three genes were significantly enriched in cell cycle and fatty acid metabolism of cancer cells. However, there is limited evidence to validate the gene signatures involved in conferring radioresistance in ESCC patients to delineate accurate and efficient disease prognosis[25]. Ma et al[26] demonstrated that HMGB1 promotes radioresistance through the activation of autophagy. Furthermore, differentially expressed genes (DEGs) including ‘CFLAR, LAMA5, ITGA6, ITGB4, and SDC4’ in five signaling cascades (PI3K-AKT pathway, CYCS gene-based apoptosis pathway, S100AX–AKT3-related pathway, SDC4 and HSPG2 pathway, and mTOR signaling pathway) were reported to be associated with radioresistance in in vitro ESCC models, and tissue biopsies of ESCC patients[27]. In the present study, we, for the first time, constructed a risk score model based on three radioresistance-associated mRNAs (CTSS, CD180, and SCIMP) and clinical features of ESCC patients; this model could facilitate oncologists to predict overall survival of ESCC patients with acquired radioresistance in radiotherapy.
A research study showed that the insulin-like growth factor 2 mRNA-binding protein 3 can contribute to the development of radioresistance in ESCC[28]. miR-205 promotes radioresistance in ESCC typically through enhancing DNA repair, impairing apoptosis, and stimulating EMT[29]. Another factor i.e., eEF2K, could foster the progression of radioresistance in ESCC[30]. In our study, the involvement of three mRNAs (CTSS, CD180, and SCIMP) in radioresistance was analyzed through the transcriptome profiling of ESCC samples between non-radiated KYSE-180 cells and 12 or 30 Gy far infrared radiation-treated KYSE-180 cells and by constructing a risk score model. However, the overall survival information in GSE81812 dataset is unavailable, so we conducted univariate and multivariate Cox regression analysis based on the TCGA database, and identified 49 radioresistance-associated mRNAs associated with survival, of which 23 were inversely correlated with survival. After comprehensive correlation analysis, we selected three radioresistance-associated mRNAs (CTSS, CD180, and SCIMP) that were strongly correlated with each other based on the data in the TCGA database. Subramanian et al[31] deciphered that the well-developed genomic signatures are significantly beneficial for improving clinical outcomes in ESCC patients. Results of the overall survival of patients in this study suggested that patients with a higher risk score exhibited a poorer prognosis. Moreover, we downloaded the GSE53625 dataset as independent validation data to validate the prognostic role of the three-mRNA signature. Our result confirmed that the risk score model could also predict the survival outcome based on the external validation datasets.
Among the three mRNAs investigated, CTSS encodes a cysteine protease. Seo et al[32] showed radiation-induced CTSS overexpression, which can consequently promote radioresistance, and knockdown of CTSS could induce impairment of radioresistance by modulating the ROS-IFN-γ pathway[32]. Additionally, a plethora of research studies have found that CTSS is particularly involved in modulating autophagy pathways[33], PI3K/Akt and Ras/Raf/MAPK signaling pathways[34], and EGFR-ERK signaling pathway[35] as these signaling cascades are more or less involved in conferring radioresistance. However, there are no reports available in the literature to delineate that CD180 and SCIMP are involved in causing radioresistance in ESCC patients. CD180 belongs to the family of Toll-like receptors. Its expression has been reported to be associated with acute or chronic leukemia[36]. SCIMP encodes a transmembrane adaptor protein that shapes host defense and inflammation via direct modulation of TLR4[37].
A report by Yang et al[27] described the activation of the PI3K-Akt signaling pathway (KEGG ID: hsa05200) with upregulation of DEGs such as LAMA5, LAMB2, LAMB3, ITGA6, and ITGB4 at 12-Gy and 30-Gy fractionated irradiation. Thus, PI3K-Akt is reported to be involved in protecting KYSE-180 cells from undergoing apoptosis after irradiation. CYCS gene-based apoptosis pathway (KEGG ID: hsa04210) is impaired after 12-Gy irradiation due to the induction of CYCS downregulation. KEGG pathway analysis of S100AX–AKT3 signaling depicted that the activation of this pathway could enhance the migration and metastasis of HSCC KYSE-180-12 Gy and KYE-180-30 Gy cells[27,38]. SDC4 and HSPG2 [KEGG ID: hsa05205] are two proteoglycans that were reported to be upregulated during the irradiation of KYSE-180 cells at doses of 12 Gy and 30 Gy. These genes are responsible for tumor cell invasion and metastasis[27]. In the present study, KEGG pathway analysis was performed to clarify the underlying mechanisms of the three mRNAs contributing to the radioresistance of ESCC cells. Our results showed that these mRNAs were mainly enriched in pathways that are related to radioresistance, such as the JAK-STAT signaling pathway[39] and NF-kB signaling pathway[40]. Our results also demonstrated the radioresistance-promoting ability of these three mRNAs. Besides, these mRNAs were enriched in immune-related pathways, such as antigen processing and presentation, cytokine-cytokine receptor interaction, and Th17 cell differentiation. Hence, these three radioresistance-associated mRNAs might be involved in the regulation of immune pathways contributing to ESCC cell radioresistance.
In summary, our study proved that CTSS, CD180 and SCIMP can promote the development of radioresistance in ESCC patients. The novel three-gene signature developed based on the three genes can be used as a prognostic model to predict the prognosis of patients with radioresistant ESCC.
Esophageal squamous cell carcinoma (ESCC) is causing a high mortality rate due to the lack of efficient early prognosis markers and suitable therapeutic regimens.
The prognostic role of genes responsible for the acquisition of radioresistance in ESCC has not been fully elucidated.
To establish a prognostic model by studying gene expression patterns pertinent to radioresistance in ESCC patients.
Datasets were obtained from the Gene Expression Omnibus and The Cancer Genome Atlas databases. The edgeR, a Bioconductor package, was used to analyze mRNA expression between different groups. We screened genes specifically responsible for radioresistance to estimate overall survival. Pearson correlation analysis was performed to confirm whether the expression of those genes correlated with each other. Genes contributing to radioresistance and overall survival were assessed by the multivariate Cox regression model through the calculation of βi and risk score using the following formula:  .
.
We identified three prognostic mRNAs (cathepsin S [CTSS], cluster of differentiation 180 [CD180], and SLP adapter and CSK-interacting membrane protein [SCIMP]) indicative of radioresistance. The expression of the three identified mRNAs was related to each other (r > 0.70 and P < 0.05). As to 1-year and 3-year overall survival prediction, the area under the time-dependent receiver operating characteristic curve of the signature consisting of the three mRNAs was 0.716 and 0.841, respectively. When stratifying patients based on the risk score derived from the signature, the high-risk group exhibited a higher death risk and shorter survival time than the low-risk group (P < 0.0001). Overall survival of the low-risk patients was significantly better than that of the high-risk patients (P = 0.018).
We have developed a novel three-gene prognostic signature consisting of CTSS, CD180, and SCIMO for ESCC.
The three-gene signature developed in this study may facilitate the prediction of early prognosis of this malignancy.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bagheri-Mohammadi S, Iran; Kao JT, Taiwan S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55699] [Article Influence: 7957.0] [Reference Citation Analysis (132)] |
| 2. | Wang Y, Lyu Z, Qin Y, Wang X, Sun L, Zhang Y, Gong L, Wu S, Han S, Tang Y, Jia Y, Kwong DL, Kam N, Guan XY. FOXO1 promotes tumor progression by increased M2 macrophage infiltration in esophageal squamous cell carcinoma. Theranostics. 2020;10:11535-11548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 3. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Corvera C, Das P, Denlinger CS, Enzinger PC, Fanta P, Farjah F, Gerdes H, Gibson M, Glasgow RE, Hayman JA, Hochwald S, Hofstetter WL, Ilson DH, Jaroszewski D, Johung KL, Keswani RN, Kleinberg LR, Leong S, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Paluri RK, Park H, Perry KA, Pimiento J, Poultsides GA, Roses R, Strong VE, Wiesner G, Willett CG, Wright CD, McMillian NR, Pluchino LA. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl ComprCancNetw. 2019;17:855-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 678] [Article Influence: 113.0] [Reference Citation Analysis (0)] |
| 4. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21354] [Article Influence: 2135.4] [Reference Citation Analysis (3)] |
| 5. | Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2015;149:1700-1715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 412] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 6. | Zhan XH, Jiao JW, Zhang HF, Li CQ, Zhao JM, Liao LD, Wu JY, Wu BL, Wu ZY, Wang SH, Du ZP, Shen JH, Zou HY, Neufeld G, Xu LY, Li EM. A three-gene signature from protein-protein interaction network of LOXL2- and actin-related proteins for esophageal squamous cell carcinoma prognosis. Cancer Med. 2017;6:1707-1719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Bayo J, Tran TA, Wang L, Peña-Llopis S, Das AK, Martinez ED. Jumonji Inhibitors Overcome Radioresistance in Cancer through Changes in H3K4 Methylation at Double-Strand Breaks. Cell Rep. 2018;25:1040-1050.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 8. | Zhou Y, Chu L, Wang Q, Dai W, Zhang X, Chen J, Li L, Ding P, Zhang L, Gu H, Lv X, Zhang W, Zhou D, Zhang P, Cai G, Zhao K, Hu W. CD59 is a potential biomarker of esophageal squamous cell carcinoma radioresistance by affecting DNA repair. Cell Death Dis. 2018;9:887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Qian D, Zhang B, Zeng XL, Le Blanc JM, Guo YH, Xue C, Jiang C, Wang HH, Zhao TS, Meng MB, Zhao LJ, Hao JH, Wang P, Xie D, Lu B, Yuan ZY. Inhibition of human positive cofactor 4 radiosensitizes human esophageal squmaous cell carcinoma cells by suppressing XLF-mediated nonhomologous end joining. Cell Death Dis. 2014;5:e1461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Croagh D, Frede J, Jones PH, Kaur P, Partensky C, Phillips WA. Esophageal stem cells and genetics/epigenetics in esophageal cancer. Ann N Y AcadSci. 2014;1325:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | He E, Pan F, Li G, Li J. Fractionated Ionizing Radiation Promotes Epithelial-Mesenchymal Transition in Human Esophageal Cancer Cells through PTEN Deficiency-Mediated Akt Activation. PLoS One. 2015;10:e0126149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Hein AL, Ouellette MM, Yan Y. Radiation-induced signaling pathways that promote cancer cell survival (review). Int J Oncol. 2014;45:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 13. | Su H, Jin X, Zhang X, Zhao L, Lin B, Li L, Fei Z, Shen L, Fang Y, Pan H, Xie C. FH535 increases the radiosensitivity and reverses epithelial-to-mesenchymal transition of radioresistant esophageal cancer cell line KYSE-150R. J Transl Med. 2015;13:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Zhu H, Yang X, Ding Y, Liu J, Lu J, Zhan L, Qin Q, Zhang H, Chen X, Yang Y, Liu Z, Yang M, Zhou X, Cheng H, Sun X. Recombinant human endostatin enhances the radioresponse in esophageal squamous cell carcinoma by normalizing tumor vasculature and reducing hypoxia. Sci Rep. 2015;5:14503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Matsuo M, Matsumoto S, Mitchell JB, Krishna MC, Camphausen K. Magnetic resonance imaging of the tumor microenvironment in radiotherapy: perfusion, hypoxia, and metabolism. SeminRadiatOncol. 2014;24:210-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Underwood TJ, Hayden AL, Derouet M, Garcia E, Noble F, White MJ, Thirdborough S, Mead A, Clemons N, Mellone M, Uzoho C, Primrose JN, Blaydes JP, Thomas GJ. Cancer-associated fibroblasts predict poor outcome and promote periostin-dependent invasion in oesophageal adenocarcinoma. J Pathol. 2015;235:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 17. | Shigeoka M, Urakawa N, Nakamura T, Nishio M, Watajima T, Kuroda D, Komori T, Kakeji Y, Semba S, Yokozaki H. Tumor associated macrophage expressing CD204 is associated with tumor aggressiveness of esophageal squamous cell carcinoma. Cancer Sci. 2013;104:1112-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 18. | Izawa S, Mimura K, Watanabe M, Maruyama T, Kawaguchi Y, Fujii H, Kono K. Increased prevalence of tumor-infiltrating regulatory T cells is closely related to their lower sensitivity to H2O2-induced apoptosis in gastric and esophageal cancer. Cancer ImmunolImmunother. 2013;62:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Maher SG, Gillham CM, Duggan SP, Smyth PC, Miller N, Muldoon C, O'Byrne KJ, Sheils OM, Hollywood D, Reynolds JV. Gene expression analysis of diagnostic biopsies predicts pathological response to neoadjuvant chemoradiotherapy of esophageal cancer. Ann Surg. 2009;250:729-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. PTK7 regulates radioresistance through nuclear factor-kappa B in esophageal squamous cell carcinoma. TumourBiol. 2016;37:14217-14224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Wu H, Yu J, Kong D, Xu Y, Zhang Z, Shui J, Li Z, Luo H, Wang K. Population and singlecell transcriptome analyses reveal diverse transcriptional changes associated with radioresistance in esophageal squamous cell carcinoma. Int J Oncol. 2019;55:1237-1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, Gebski V; Australasian Gastro-Intestinal Trials Group. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectableoesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12:681-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 1263] [Article Influence: 90.2] [Reference Citation Analysis (0)] |
| 23. | Ding M, Zhang E, He R, Wang X. Newly developed strategies for improving sensitivity to radiation by targeting signal pathways in cancer therapy. Cancer Sci. 2013;104:1401-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Zhao H, Jiang A, Yu M, Bao H. Identification of biomarkers correlated with diagnosis and prognosis of endometrial cancer using bioinformatics analysis. J Cell Biochem. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X, Zhang X, Yang L, Zhou R, Wu S. Cancer-associated Fibroblast-promoted LncRNADNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25:1989-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 26. | Ma H, Zheng S, Zhang X, Gong T, Lv X, Fu S, Zhang S, Yin X, Hao J, Shan C, Huang S. High mobility group box 1 promotes radioresistance in esophageal squamous cell carcinoma cell lines by modulating autophagy. Cell Death Dis. 2019;10:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Yang L, Zhang X, Hou Q, Huang M, Zhang H, Jiang Z, Yue J, Wu S. Single-cell RNA-seq of esophageal squamous cell carcinoma cell line with fractionated irradiation reveals radioresistant gene expression patterns. BMC Genomics. 2019;20:611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Yoshino K, Motoyama S, Koyota S, Shibuya K, Sato Y, Sasaki T, Wakita A, Saito H, Minamiya Y, Sugiyama T, Ogawa J. Identification of insulin-like growth factor 2 mRNA-binding protein 3 as a radioresistance factor in squamous esophageal cancer cells. Dis Esophagus. 2014;27:479-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Pan F, Mao H, Bu F, Tong X, Li J, Zhang S, Liu X, Wang L, Wu L, Chen R, Wei H, Li B, Li C, Yang Y, Steer CJ, Zhao J, Guo Y. Sp1-mediated transcriptional activation of miR-205 promotes radioresistance in esophageal squamous cell carcinoma. Oncotarget. 2017;8:5735-5752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 30. | Zhu H, Song H, Chen G, Yang X, Liu J, Ge Y, Lu J, Qin Q, Zhang C, Xu L, Di X, Cai J, Ma J, Zhang S, Sun X. eEF2K promotes progression and radioresistance of esophageal squamous cell carcinoma. RadiotherOncol. 2017;124:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Subramanian J, Simon R. What should physicians look for in evaluating prognostic gene-expression signatures? Nat Rev ClinOncol. 2010;7:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Seo HR, Bae S, Lee YS. Radiation-induced cathepsin S is involved in radioresistance. Int J Cancer. 2009;124:1794-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Hsin MC, Hsieh YH, Wang PH, Ko JL, Hsin IL, Yang SF. Hispolon suppresses metastasis via autophagic degradation of cathepsin S in cervical cancer cells. Cell Death Dis. 2017;8:e3089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 34. | Gautam J, Bae YK, Kim JA. Up-regulation of cathepsin S expression by HSP90 and 5-HT7 receptor-dependent serotonin signaling correlates with triple negativity of human breast cancer. Breast Cancer Res Treat. 2017;161:29-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 35. | Chen KL, Chang WS, Cheung CH, Lin CC, Huang CC, Yang YN, Kuo CP, Kuo CC, Chang YH, Liu KJ, Wu CM, Chang JY. Targeting cathepsin S induces tumor cell autophagy via the EGFR-ERK signaling pathway. Cancer Lett. 2012;317:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 36. | Chaplin JW, Kasahara S, Clark EA, Ledbetter JA. Anti-CD180 (RP105) activates B cells to rapidly produce polyclonal Ig via a T cell and MyD88-independent pathway. J Immunol. 2011;187:4199-4209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 37. | Luo L, Bokil NJ, Wall AA, Kapetanovic R, Lansdaal NM, Marceline F, Burgess BJ, Tong SJ, Guo Z, Alexandrov K, Ross IL, Hibbs ML, Stow JL, Sweet MJ. SCIMP is a transmembrane non-TIR TLR adaptor that promotes proinflammatory cytokine production from macrophages. Nat Commun. 2017;8:14133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 38. | Grottke A, Ewald F, Lange T, Nörz D, Herzberger C, Bach J, Grabinski N, Gräser L, Höppner F, Nashan B, Schumacher U, Jücker M. Downregulation of AKT3 Increases Migration and Metastasis in Triple Negative Breast Cancer Cells by Upregulating S100A4. PLoS One. 2016;11:e0146370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 39. | Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, Nam JS. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J ExpClin Cancer Res. 2019;38:399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 40. | Hou Y, Liang H, Rao E, Zheng W, Huang X, Deng L, Zhang Y, Yu X, Xu M, Mauceri H, Arina A, Weichselbaum RR, Fu YX. Non-canonical NF-κB Antagonizes STING Sensor-Mediated DNA Sensing in Radiotherapy. Immunity. 2018;49:490-503.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 198] [Article Influence: 28.3] [Reference Citation Analysis (0)] |