Published online Jun 24, 2022. doi: 10.5306/wjco.v13.i6.496
Peer-review started: August 8, 2021
First decision: September 2, 2021
Revised: September 16, 2021
Accepted: May 28, 2022
Article in press: May 28, 2022
Published online: June 24, 2022
Processing time: 317 Days and 10 Hours
It is known that p53 suppression is an important marker of poor prognosis of cancers, especially in solid tumors of the breast, lung, stomach, and esophagus; liposarcomas, glioblastomas, and leukemias. Because p53 has mouse double minute 2 (MDM2) as its primary negative regulator, this molecular docking study seeks to answer the following hypotheses: Is the interaction between DS-3032B and MDM2 stable enough for this drug to be considered as a promising neoplastic inhibitor?
To analyze, in silico, the chemical bonds between the antagonist DS-3032B and its binding site in MDM2.
For molecular docking simulations, the file containing structures of MDM2 (receptor) and the drug DS-3032B (ligand) were selected. The three-dimensional structure of MDM2 was obtained from Protein Data Bank, and the one for DS-3032B was obtained from PubChem database. The location and dimensions of the Grid box was determined using AutoDock Tools software. In this case, the dimensions of the Grid encompassed the entire receptor. The ligand DS-3032B interacts with the MDM2 receptor in a physiological environment with pH 7.4; thus, to simulate more reliably, its interaction was made with the calculation for the prediction of its protonation state using the MarvinSketch® software. Both ligands, with and without the protonation, were prepared for molecular docking using the AutoDock Tools software. This software detects the torsion points of the drug and calculates the angle of the torsions. Molecular docking simulations were performed using the tools of the AutoDock platform connected to the Vina software. The analyses of the amino acid residues involved in the interactions between the receptor and the ligand as well as the twists of the ligand, atoms involved in the interactions, and type, strength, and length of the interactions were performed using the PyMol software (pymol.org/2) and Discovery Studio from BIOVIA®.
The global alignment indicated crystal structure 5SWK was more suitable for docking simulations by presenting the p53 binding site. The three-dimensional structure 5SWK for MDM2 was selected from Protein Data Bank and the three-dimensional structure of DS-3032B was selected from PubChem (Compound CID: 73297272; Milademetan). After molecular docking simulations, the most stable conformer was selected for both protonated and non-protonated DS-3032B. The interaction between MDM2 and DS-3032B occurs with high affinity; no significant difference was observed in the affinity energies between the MDM2/pronated DS-3032B (-9.9 kcal/mol) and MDM2/non-protonated DS-3032B conformers (-10.0 kcal/mol). Sixteen amino acid residues of MDM2 are involved in chemical bonds with the protonated DS-3032B; these 16 residues of MDM2 belong to the p53 biding site region and provide high affinity to interaction and stability to drug-protein complex.
Molecular docking indicated that DS-3032B antagonist binds to the same region of the p53 binding site in the MDM2 with high affinity and stability, and this suggests therapeutic efficiency.
Core Tip: The knowledge, at the molecular level, of the complexes formed by therapeutic drugs and their target in the body are relevant to understand the efficiency of the drug. These data can be provided, with high reliability, by bioinformatics tools, which saves time in relation to in vitro and in vivo analyses. The drug DS-3032B has been a potential candidate for oncogenic treatment in preclinical trials, but clinical studies are scarce. This work shows data on chemical interactions between this drug and its target, mouse double minute 2, that corroborate the preclinical data and demonstrate the stability of the therapeutic complex.
- Citation: da Mota VHS, Freire de Melo F, de Brito BB, Silva FAFD, Teixeira KN. Molecular docking of DS-3032B, a mouse double minute 2 enzyme antagonist with potential for oncology treatment development. World J Clin Oncol 2022; 13(6): 496-504
- URL: https://www.wjgnet.com/2218-4333/full/v13/i6/496.htm
- DOI: https://dx.doi.org/10.5306/wjco.v13.i6.496
Cancer is a genetic disease whose evolution leads to numerous changes in DNA. According to a survey conducted by the International Agency for Research on Cancer in 2018, cancer was considered the second leading cause of death in the world, affecting 18.1 million people and causing the death of 9.6 million people around the world[1]. For Brazil, the estimate for the triennium 2020-2022 predicts that 625000 new cases of cancer will occur[2].
While proto-oncogenes are genes responsible for the positive regulation of cell proliferation, tumor suppressor genes are genes responsible for negative regulation; in other words, it inhibits cell multiplication. An example of this class is the p53 gene, which is found mutated in about half of human cancers[3]. p53 is a transcription factor activated by signs of stress, such as DNA damage, oncogenes activation, and nutritional deprivation[4]; it also has an essential function in DNA damage repair and antioxidant response regulation[5].
Therefore, overactivation of p53 is considered as an option for selective therapies against cancer, providing a targeting of neoplastic cells and sparing unaffected normal tissue[6]. One proposed treatment aims to inhibit tumor growth by activating the tumor suppressor protein p53 through inhibition of the mouse double minute 2 (MDM2)[7].
MDM2 is an E3 ubiquitin ligase enzyme that has a negative regulatory role of the tumor suppressor p53. This enzyme controls transcriptional activity and stability of p53. MDM2 expression is regulated in several tumors, resulting in loss of p53-dependent activities, such as apoptosis and cell cycle arrest[8].
p53 is targeted for degradation by the proteasome by MDM2. Through E3 ubiquitin ligase activity, MDM2 promotes ubiquitination of p53, leading to increased p53 degradation. In some human tumors, MDM2 has been shown to be abnormally upregulated leading to enhanced degradation and reduction of p53 activity[9].
The molecule DS-3032B (Milademetan or RAIN-32) is an MDM2 antagonist that prevents its interaction with p53. Clinical trials with DS-3032B have been conducted by the National Institute of Health (phase I) in patients with leukemia and lymphoma and showed clinical efficacy[10]. A phase I trial evaluated the safety, tolerability, efficacy, and pharmacokinetics of DS-3032B in Japanese patients with solid tumors who relapsed after or refractory to standard therapy, and dose-limiting toxicities, safety, tolerability, maximum tolerated dose, pharmacokinetics, and recommended dose for phase II clinical trial were determined[11].
However, the trials are empirical, as the mechanism and biochemistry of interaction of DS-3032B with MDM2 are not known. Although there is a lot of preclinical evidence of the action of MDM2 inhibitors as monotherapy or in combination, clinical experience with these agents is limited. Thus, information, at the molecular level, about the complex formed between the inhibitor and its target will help to clarify the nature of the interaction and its stability[9].
Such information is important to understand the functioning of the compound and even increase its efficiency through structural alterations. Since obtaining these data by crystallization and X-ray diffraction is laborious and time consuming, a plausible alternative has been facilitated by computational methods, such as molecular docking that has proven useful and reliable for predicting the possible interactions and affinity of ligands with macromolecules. In silico methods have been gaining increasing prominence since the experimental determination of complex three-dimensional structures is quite complex and costly[12]. Thus, the aim of this study was to analyze the interaction of the DS-3032B to its binding site in MDM2, the chemical bonds between drug and protein, and the affinity of the formed complex in order to clarify the stability of the interaction and thus help in elucidating the molecular mechanism of therapeutic action of the antagonist.
For molecular docking simulations, the structures of MDM2 (receptor) and the drug DS-3032B (ligand) were selected. The three-dimensional structure of MDM2 was selected from Protein Data Bank (PDB) (http://www.rcsb.org/pdb/home/home.do) after a previous global alignment of all available primary sequences using CLUSTAL X 2.0 software. The 3D structure file was obtained in the extension ".pdb" (input file). The selected receptor’s three-dimensional structure was prepared for molecular docking simulations using AutoDock Tools software; the water molecules were deleted, since they do not belong to the molecule and can interfere in the docking process and hydrogen atoms were also added. Then it was determined through the AutoDock Tools software the location and dimensions of the Grid (virtual box that delimits the region where the ligand will perform possible interactions with the receptor). In this case, the dimensions of the Grid encompassed the entire receptor. The Grid data and coordinates were used in molecular docking.
The three-dimensional structure of the drug DS-3032B was solved experimentally and deposited in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/); the file was obtained in the extension ".sdf" and converted to “.pdb” (input file). The ligand DS-3032B interacts with the MDM2 receptor in a physiological environment with pH 7.4; thus, to simulate more reliably, its interaction was made with the calculation for the prediction of its protonation state using the MarvinSketch 5.7® software from ChemAxon®. Both ligands, with and without the protonation, were prepared for molecular docking using the AutoDock Tools 1.5.6 software. This software detects the torsion points of the drug and calculates the angle of the torsions.
Molecular docking procedures for a rigid protein and a flexible ligand were used. A grid of points in x, y, and z directions was built with a grid spacing of 1.0 Å using the AutoGrid component of the software. Molecular docking simulations were performed using the tools of the AutoDock platform (http://autodock.scripps.edu/) connected to the Vina 1.1.2 software (http://vina.scripps.edu/). The software used associates two components: A search algorithm and a score function. First, the algorithm was responsible for the search of possible combinations in the bonds, exploring the rotational, translational, and conformational degrees of freedom of the ligand as well as of the proteins. Then, the score function was used to choose the best binding modes. These functions were obtained according to the force fields of molecular mechanics and empirical parameters from free energy calculations.
The analyses of the amino acid residues involved in the interactions between the receptor and the ligand, as well as the twists of the ligand, atoms involved in the interactions, and type, strength, and length of the interactions were performed using the PyMol 2.5 software (pymol.org/2) and Discovery Studio from BIOVIA®.
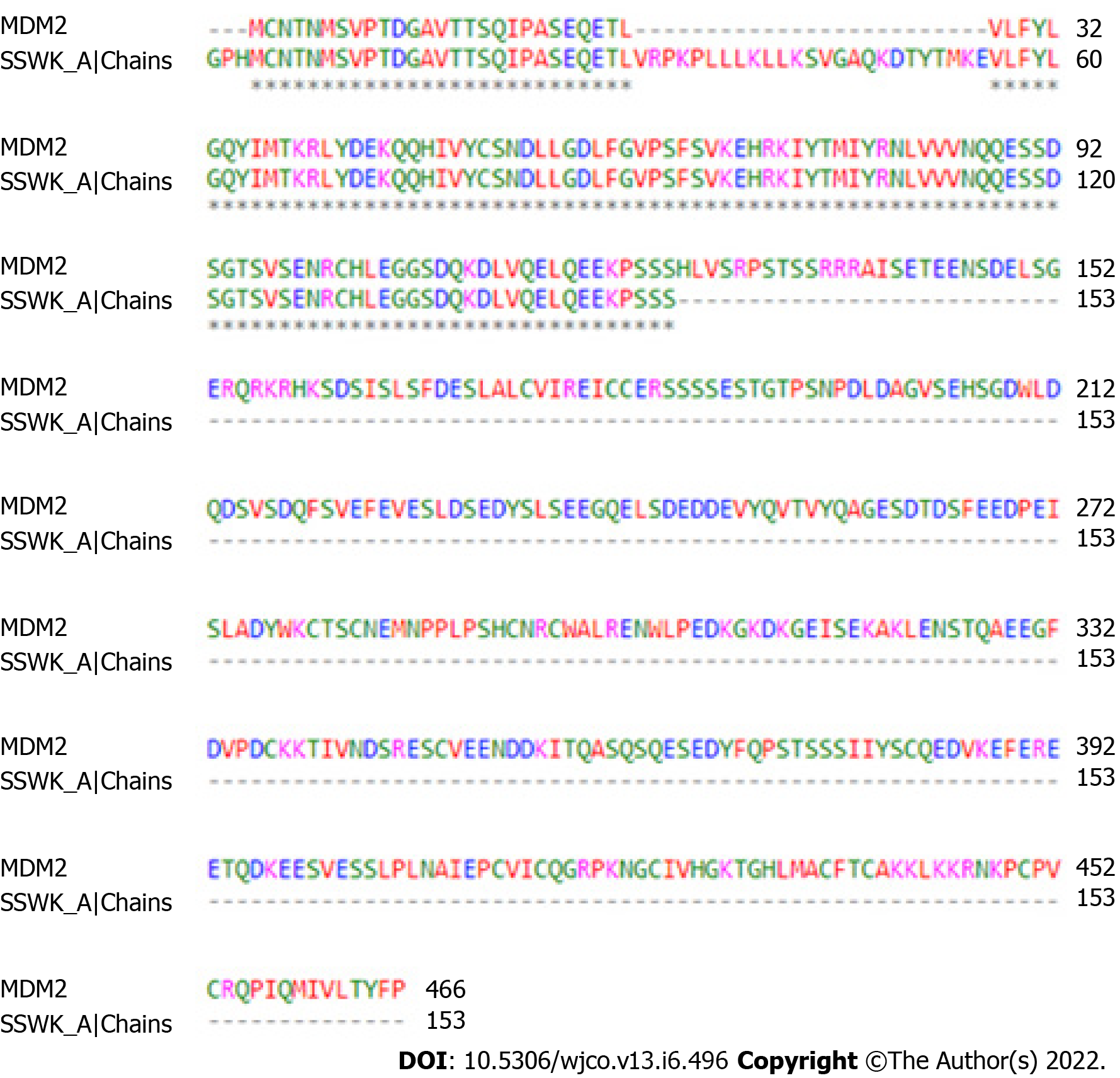
During the search for the three-dimensional structure of the receptor, the alignment of primary sequences of three-dimensional structures available in the PDB corresponding to MDM2 was performed. The 5SWK, from Homo sapiens, was indicated as the best structure by having structure resolved by X-ray crystallography with high resolution (1.92 Å) and greater coverage of the site responsible for the antagonism of the protein. The antagonist binding site is located in chain A of MDM2, so chain A was isolated to perform molecular docking simulations. Figure 1 shows the primary sequence alignment between 5SWK chain A (153 residues) from PDB and whole MDM2 (466 residues). The consensus region presents the binding site for the MDM2 receptor antagonist. The three-dimensional structure of the antagonist DS-3032B was obtained from PubChem database with the CID (compound identification number) 73297272. MarvinSketch® software showed that at pH 7.4, 97.37% of DS-3021B was distributed in its protonated form.
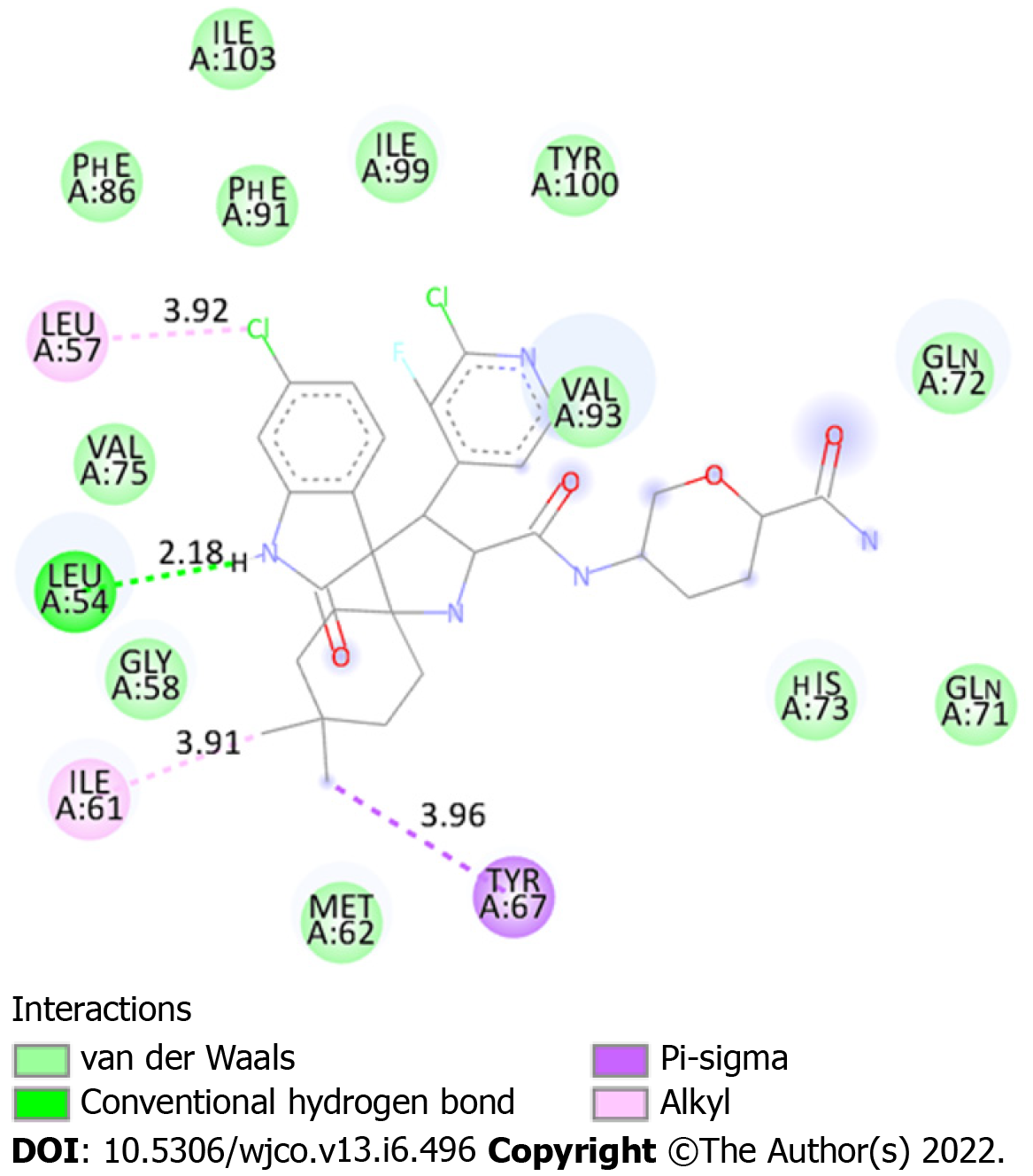
A ranking of nine conformations presenting different affinity energies was obtained in molecular docking for each ligand (protonated and non-protonated), and the conformation with the lowest energy (Figure 2) was selected for subsequent analysis of drug-protein interactions. In addition, two root mean square deviation (RMSD) metric variants are also available: RMSD/L.b. (lower limit of RMSD) and RMSD/u.b. (upper limit RMSD), which differ by the way the atoms are matched in the distance calculation. While in RMSD/u.b. each atom is matched in one conformation to itself in the other conformation, ignoring any symmetry, in RMSD/L.b., each atom is matched in one conformation to the nearest atom of the same element type in the other conformation. No significant difference was observed between the affinity energies between both MDM2/protonated DS-3032B and MDM2/non-protonated DS-3032B conformers (Table 1); therefore, the protonated form, which prevails under physiological conditions, was better analyzed in this study. Sixteen amino acid residues of MDM2 are involved in chemical bonds with the protonated DS-3032B. Polar bond, hydrophobic interactions (pi-sigma and alkyl), and Van der Waals were observed (Table 2). These 16 residues of MDM2 chain A belong to the p53 biding site region. Four out of 16 interactions are more relevant, and they range from 2.18 to 3.96 Å; the shortest bond is a hydrogen bond between an oxygen atom of leucine residue 54 (LEU 54) of MDM2 and a nitrogen atom of one of the rings of the DS-3032B antagonist (Figure 3).
| Conformer | Affinity energy (kcal/mol) | Dist. From | |
| RMSD l.b. | RMSD u.b. | ||
| MDM2/protonated DS-3032B | -10.0 | 0.000 | 0.000 |
| MDM2/non-protonated DS-3032B | -9.9 | 0.000 | 0.000 |
| Residues of MDM2 | Interaction | Bond size (Å) |
| Leu 54 | Hydrogen bond | 2.18 |
| Leu 57 | Alkyl | 3.92 |
| Ile 61 | Alkyl | 3.91 |
| Tyr 67 | Pi-Sigma | 3.96 |
| Ile 103 | Van der Waals | - |
| Phe 86 | Van der Waals | - |
| Phe 91 | Van der Waals | - |
| Ile 99 | Van der Waals | - |
| Tyr 100 | Van der Waals | - |
| Val 75 | Van der Waals | - |
| Val 93 | Van der Waals | - |
| Gln 71 | Van der Waals | - |
| Gln 72 | Van der Waals | - |
| Gly 58 | Van der Waals | - |
| His 73 | Van der Waals | - |
| Met 62 | Van der Waals | - |
The MDM2, also called E3 ubiquitin ligase enzyme, is commonly overexpressed in various cancers[13], inactivating directly p53 by interacting with its transcriptional activation domain and inducing its degradation through ubiquitination[4]. DS-3032B is a compound derived from dispiropyrrolidine, also called milademetan. It impairs the binding of MDM2 to the transcriptional activation domain of p53[14].
Pharmacological inhibition of the p53-MDM2 interaction has been evaluated as a therapeutic approach to exert p53-mediated antitumor effects. Because MDM2 antagonists can produce nongenotoxic activation of wild-type p53, leading to anticancer activity, these agents are candidates to improve the therapeutic index of current chemotherapy regimens while minimizing the risk of resistance to single-agent MDM2 inhibition[15]. MDM2 inhibitors have demonstrated in preclinical and clinical studies their antineoplastic effects arising from p53 activation caused by negative regulation of MDM2 in solid and hematological tumors.
Identifying the interactions between drugs and their targets is critical in the discovery of new drugs. This helps to understand better the mechanism of the disease and to identify unexpected therapeutic activity or adverse side effects of the drugs. Therefore, the prediction of interaction between drugs and targets becomes important in the context of pharmacology and drug redefinition[16]. The precise and efficient identification of interactions between drugs and their targets in the body can reveal hidden functions of these drugs and target proteins as well as speed up the drug development process[17]. Drug development is a time-consuming process, the experimental identification of interactions between drugs and their targets is very costly, and modern technologies have mitigated this problem. The computational prediction of drug target interactions has been shown to be fundamental for the study of drugs, since it reduces the time and costs of the process[18].
In fact, through in silico approach, it was possible to observe that DS-3032B is able to connect to the p53 binding site of MDM2 chain A with a significant affinity. The interaction between the antagonist and MDM2 involves 16 amino acid residues by polar and nonpolar bonds throughout the entire structure of the drug. The arrangement of the connections contributes to the low bind energy value and consequently to the stability of the complex. Complex stability and high affinity indicate promising therapeutic potential for DS-3032B.
There are three clinical studies of MDM2 inhibitors in various cancers, including the MDM2 antagonist, DS-3032B, still in early phase (solid tumors and lymphoma: NCT01877382; myeloma: NCT02579824; leukemia: NCT02319369; acute myeloid leukemia: NCT03671564; acute myeloid leukemia, being associated with Quizartinib: NCT03552029; refractory leukemia, being associated with Cicarabine: NCT03634228)[19]. Clinical responses in these trials have been limited overall, but some patients have clearly achieved clinical benefit through monotherapy with MDM2 inhibitors[20]. Although monotherapy with MDM2 inhibitors has benefits, clinical responses are usually modest; association with other inhibitors has shown synergism and more efficient clinical responses[9]. However, the association of more medications can increase the presence of unwanted effects and even decrease patients' adherence to therapy. The action of drugs is related to their interaction with their therapeutic target; drug-protein affinity predicts complex stability, longer interaction time, and greater pharmacological effectiveness.
Thus, knowledge about the molecular interactions between the test drug and its therapeutic target becomes interesting to predict the behavior of the drug in the body, predict its efficiency and stability, and can help predict how this drug can affect the physiology of the individual. Since in silico studies can provide this data satisfactorily and reliably and much faster compared to in vitro and in vivo studies, this methodology has been used to support scientific studies. The analyses performed in this study, with the DS-3032B, indicate that the complex formed between this drug and its target is quite stable, indicating high therapeutic efficiency. This efficiency, measured indirectly, through affinity energy may be responsible for the good preclinical results of ds-3032B, and it may be more effective as monotherapy than current inhibitors.
Regarding the in silico analyses performed, the ligand position is selected based on calculations that are ranked according to docking score that represents the binding affinity between the ligand and the receptor and is expressed in kcal/mol[21]. Molecular docking is an established in silico structure-based method widely used in drug discovery. Docking enables the identification of novel compounds of therapeutic interest, predicting ligand-target interactions at a molecular level[22]. In this study it was obtained as a result of molecular docking, the ranking of the nine conformations with the highest affinities for the receptor, in parallel with two metric variants of RMSD: RMSD/L.b. and RMSD/u.b., which differ by the way the atoms are matched in the distance calculation. While in RMSD/u.b. each atom is matched in one conformation to itself in the other conformation, ignoring any symmetry, in RMSD/L.b. each atom is matched in one conformation to the nearest atom of the same element type in the other conformation. Whereas that for RMSD, the tolerance value respected is at most 2.0Å[23]. In addition, the affinity of the conformation presents the binding energy between the receptor and the ligand, being considered significant values less than -6.0 kcal/mol[13]. The compound that requires lower energy for the interaction to occur forms a more stable complex, in other words, has greater biological activity[24].
It is important to highlight that the DS-3032B, in addition to presenting satisfactory preclinical data regarding its anti-tumor potential, which are supported by computational findings, this drug is administered orally[11]. Thus, the DS-3032B becomes an attractive therapy compared to invasive and uncomfortable administrations for the patient.
In this study, higher biological activity means greater antagonism of MDM2 and consequent restoration of the tumor suppressor, p53. The confirmation was provided by the results obtained during molecular docking calculations, given that the conformer with the highest affinity showed -9.9 kcal/mol and therefore can be considered a promising candidate to inhibit the MDM2 protein. These dyspyrolidine-derived compounds may represent a starting point for the development of new drugs to treat cancers with overexpression of the MDM2 protein. The identified results reinforce that bioinformatics offers great direction in the search and validation of treatment targets, because it presents itself as a starting point for improving the knowledge involving drug- protein interactions. In addition, it also promotes cost and time reduction when compared to traditional research methods and directs the treatment so that the new drugs have their side effects minimized. However, in silico processes are complementary and do not rule out the need for in vitro and in vivo tests.
Mouse double minute 2 (MDM2) is the main negative regulator of tumor suppressor p53; in this context, the effective inhibition of MDM2 is an alternative for cancer treatments.
DS-3032B is an MDM2 antagonist, and its activity is known only empirically, so bioinformatics analyses can point to molecular characteristics of complex interaction.
To analyze, in silico, the interactions between the antagonist DS-3032B and MDM2 and infer the antineoplastic potential of the drug.
The analysis of chemical bonds, interaction of the drug-protein complex, and its stability were done by molecular docking.
Molecular docking simulations between MDM2 chain A (PDB: 5SWK) and DS-3032B (CID: 73297272) in its protonabed form indicated a complex with significant affinity energy, -10.0 kcal/mol. The results indicate a stable complex, maintained by hydrophilic and hydrophobic bonds involving 16 amino acid residues of MDM2.
DS-3032B is able to bind to MDM2 with high affinity and stability, suggesting therapeutic efficiency.
Analyze the DS-3032B/MDM2 complex using molecular dynamics and verify the possibility of structural changes of the drug to increase its efficiency.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Odhar HA, Iraq A-Editor: Kołat D, Poland S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Gong ZM
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55839] [Article Influence: 7977.0] [Reference Citation Analysis (132)] |
| 2. | Santos MO. Estimativa 2018: Incidência de Câncer no Brasil. Revista Brasileira de Cancerologia. 2018;64:119-120. [RCA] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Bieging KT, Attardi LD. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 4. | Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci. 2020;133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 210] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 5. | Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med. 2016;6:a026104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 810] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 6. | Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 808] [Cited by in RCA: 802] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 7. | Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254-271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 250] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 8. | Oliner JD, Saiki AY, Caenepeel S. The Role of MDM2 Amplification and Overexpression in Tumorigenesis. Cold Spring Harb Perspect Med. 2016;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 9. | Konopleva M, Martinelli G, Daver N, Papayannidis C, Wei A, Higgins B, Ott M, Mascarenhas J, Andreeff M. MDM2 inhibition: an important step forward in cancer therapy. Leukemia. 2020;34:2858-2874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 251] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 10. | DiNardo CD, Rosenthal J, Andreeff M, Zernovak O, Kumar P, Gajee R, Chen S, Rosen M, Song S, Kochan J, Limsakun T, Olin R. Phase 1 Dose Escalation Study of MDM2 Inhibitor DS-3032b in Patients with Hematological Malignancies - Preliminary Results. Blood. 2016;128:593. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Takahashi S, Fujiwara Y, Nakano K, Shimizu T, Tomomatsu J, Koyama T, Ogura M, Tachibana M, Kakurai Y, Yamashita T, Sakajiri S, Yamamoto N. Safety and pharmacokinetics of milademetan, a MDM2 inhibitor, in Japanese patients with solid tumors: A phase I study. Cancer Sci. 2021;112:2361-2370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 12. | Hobani Y, Jerah A, Bidwai A. A comparative molecular docking study of curcumin and methotrexate to dihydrofolate reductase. Bioinformation. 2017;13:63-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 13. | Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 961] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 14. | Arnhold V, Schmelz K, Proba J, Winkler A, Wünschel J, Toedling J, Deubzer HE, Künkele A, Eggert A, Schulte JH, Hundsdoerfer P. Reactivating TP53 signaling by the novel MDM2 inhibitor DS-3032b as a therapeutic option for high-risk neuroblastoma. Oncotarget. 2018;9:2304-2319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Chen L, Rousseau RF, Middleton SA, Nichols GL, Newell DR, Lunec J, Tweddle DA. Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget. 2015;6:10207-10221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Anusuya S, Kesherwani M, Priya KV, Vimala A, Shanmugam G, Velmurugan D, Gromiha MM. Drug-Target Interactions: Prediction Methods and Applications. Curr Protein Pept Sci. 2018;19:537-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Núñez S, Venhorst J, Kruse CG. Target-drug interactions: first principles and their application to drug discovery. Drug Discov Today. 2012;17:10-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 18. | Pliakos K, Vens C. Drug-target interaction prediction with tree-ensemble learning and output space reconstruction. BMC Bioinformatics. 2020;21:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 19. | Tisato V, Voltan R, Gonelli A, Secchiero P, Zauli G. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J Hematol Oncol. 2017;10: 133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 202] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 20. | Ishizawa J, Nakamaru K, Seki T, Tazaki K, Kojima K, Chachad D, Zhao R, Heese L, Ma W, Ma MCJ, DiNardo C, Pierce S, Patel KP, Tse A, Davis RE, Rao A, Andreeff M. Predictive Gene Signatures Determine Tumor Sensitivity to MDM2 Inhibition. Cancer Res. 2018;78:2721-2731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Lee HS, Jo S, Lim HS, Im W. Application of binding free energy calculations to prediction of binding modes and affinities of MDM2 and MDMX inhibitors. J Chem Inf Model. 2012;52:1821-1832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 325] [Cited by in RCA: 1120] [Article Influence: 186.7] [Reference Citation Analysis (0)] |
| 23. | Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26714] [Cited by in RCA: 14574] [Article Influence: 971.6] [Reference Citation Analysis (0)] |
| 24. | Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384-13421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1277] [Cited by in RCA: 1177] [Article Influence: 117.7] [Reference Citation Analysis (0)] |