INTRODUCTION
Approximately 85% of breast cancer cases are associated with a multifactorial beginning. Breast cancer has a high incidence and mortality among women. The majority of mammary tumors (> 80%) are invasive and grow mainly in the mammary ducts (invasive ductal carcinoma), and a minor fraction of them can also grow in the mammary lobules. The establishment of new biomarkers has been difficult because the molecular characteristics of mammary tumors are highly heterogeneous, defining breast cancer as a collection of mammary neoplasms[1-3].
More than 70% of all breast cancer cases are estrogen receptor alpha positive (ERα+), whereas approximately 30% are ERα-[4]. In addition to ERα, other biomarkers for breast cancer include the progesterone receptor (PR), androgen receptor, human epidermal growth factor receptor 2 (HER2), and Ki67 (a cellular marker for proliferation)[5,6]. Consequently, the classification of breast cancers is based on the detection of these biomarkers, resulting in four subtypes: (1) Luminal A-like (the most common): ERα+, PR ≥ 20%, HER2-, Ki67 < 20%; (2) Luminal B-like: ERα+, PR < 20% and/or HER2+ and/or Ki67 ≥ 20%; (3) HER2-overexpression: ERα-, PR-, HER2+; and (4) Basal-like: ERα-, PR-, HER2- (triple-negative). Luminal A is more responsive to endocrine therapy than luminal B. HER2-overexpressing subtypes are responsive to anti-HER2-targeted therapy; however, basal-like breast cancer is not responsive to these therapies[7,8].
Endocrine therapy includes selective estrogen receptor downregulators (SERD), selective estrogen receptor modulators (SERM), and aromatase inhibitors (AI)[9,10]. SERD and SERM negatively regulate the abundance and activity of ERα, respectively, whereas AI blocks estradiol production. These therapies are important because ERα is activated mainly by 17 β-estradiol to act as a transcription factor or as a mediator of signaling pathways. In breast cancer, the 17 beta-estradiol/ERα pathways have been associated with a pro-tumor activity. Despite the existence of endocrine therapies, a common problem in breast cancer management is de novo resistance or acquired resistance to endocrine therapy through yet unidentified mechanisms[11]. The panorama of breast cancer leads to the search for new biomarkers and molecular targets for therapies to control its progression. We reviewed TRIM25 as a potential biomarker and therapeutic target for breast cancer. TRIM25 is a protein affecting the proteome through its E3 ligase activity for interferon-stimulated gene 15 (ISG15) and ubiquitin, as described below.
ISGYLATION AND UBIQUITINATION
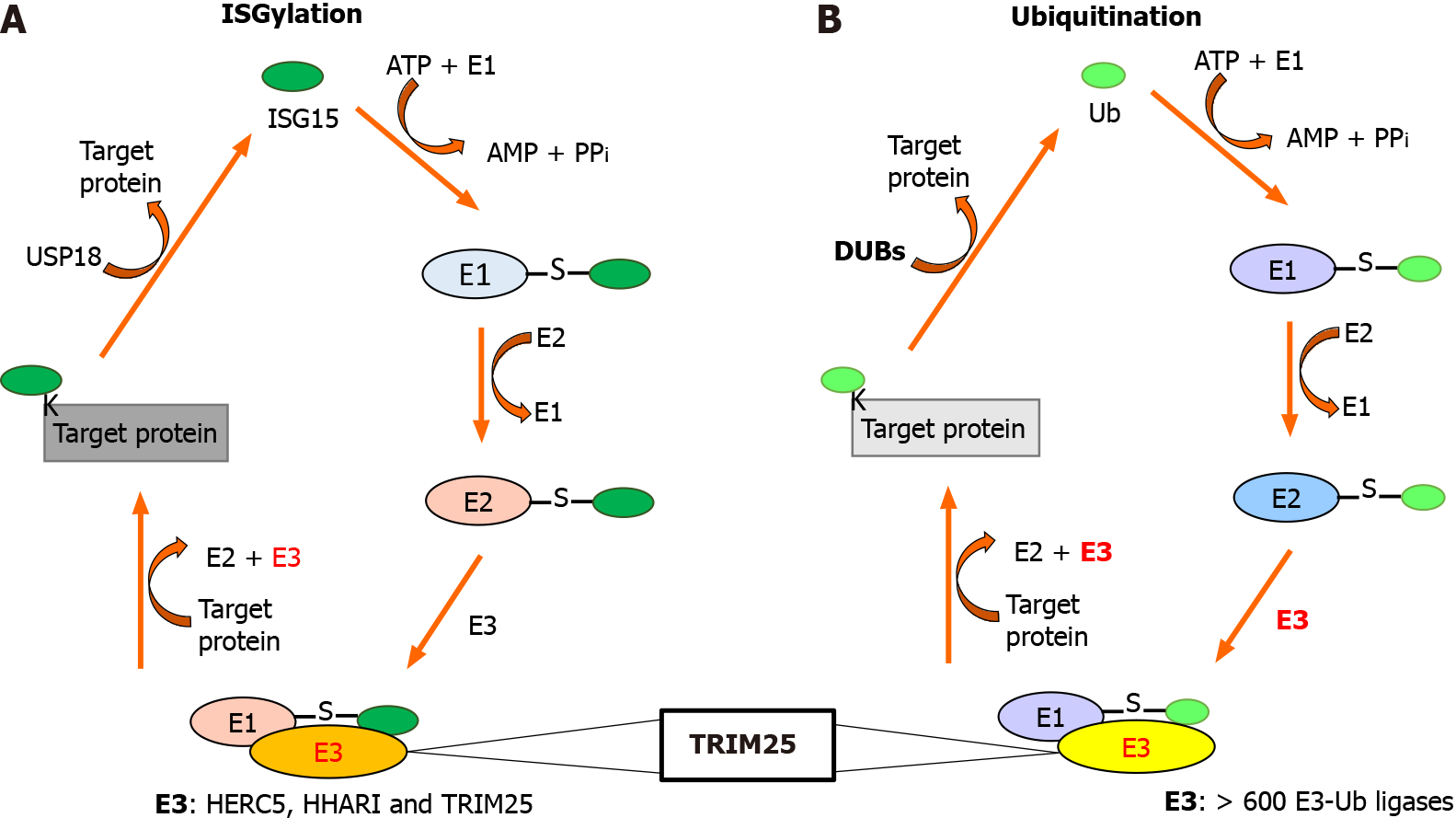
Ubiquitination and ISGylation are two processes that introduce posttranslational modifications to proteins by covalently adding ubiquitin (Ub) and ISG15 to target proteins, respectively (Figure 1). ISG15 is structurally related to the Ub, but its actions and mechanisms of regulation display differences[12,13].
Figure 1 TRIM25 is an E3-ligase common for interferon-stimulated gene 15 and ubiquitin proteins.
A: The ISGylation system; B: The Ubiquitination system. The ISGylation system and the ubiquitination system are similar, however, the ubiquitination system has more than 600 E3 ligases and the ISGyaltion system has three E3 ligases. Interestingly, both systems have in common the E3 ligase tripartite motif containing 25. ISG15: Interferon-stimulated gene 15; DUBs: Ubiquitin-proteasome system; Ub: Ubiquitin; USP18: Ubiquitin-specific protease 18.
Ub is an 8 kDa protein [76 amino acid (aa)] conserved from yeast to humans, which contains the LRLRGG motif in its C-terminal domain that mediates covalent binding with its target proteins during ubiquitination, which is executed by three enzymes: (1) E1-Ub activating enzymes; (2) E2-Ub conjugating enzymes; and (3) E3-Ub ligase enzymes. The E3-Ub ligases are responsible for providing substrate specificity, with more than 600 members in the ubiquitination system, one of them being TRIM25. Ubiquitination can be reverted by de-Ubiquitinases, which are approximately 100 different in humans. Some proteins can be monoubiquitinated in one or several lysine residues[13]. In addition, some proteins can be polyubiquitinated since the Ub lysine residues 6, 11, 27, 29, 33, 48, and 63 can be used for the sequential attachment of these proteins, forming homotypic Ub chains or branched Ub chains (by binding of several lysine residues of Ub) on their target proteins[14,15]. In general, monoubiquitination regulates DNA repair, gene expression, endocytosis, and protein stability, and polyubiquitination is a marker for degradation by the proteasome [ubiquitin-proteasome system (UPS)] and some non-proteolytic activities[16].
In contrast to Ub, ISG15 is produced from a precursor protein of 165 aa, which is processed by removing eight aa at the C–terminal as well as the N-terminal methionine. Mature ISG15 (15kDa) has two Ub-like domains connected by a “hinge” sequence, and contains the motive LRLRGG in its C-terminal domain as Ub[17]. Similar to ubiquitination, ISG15 requires the E1 activating enzyme, the E2 conjugating enzyme, and the E3 ligase enzyme to covalently bind to lysine residues on its target proteins, denominating this process as ISGylation[18]. In contrast to the ubiquitination system, only three E3 ligase enzymes are known to be involved in ISGylation systems: HERC5, HHARI, and estrogen-responsive finger protein (EFP)/TRIM25. The majority of ISGylated proteins have been reported as targets of HERC5, while one, 4EHP, is the target of HHARI. TRIM25 has been reported to have more targets[18,19]. Protein ISGylation can be reversed by USP18 (UBP43), an enzyme that mediates the de-ISGylation, generating the free form of ISG15 known as “free ISG15”[20]. ISGylation alters protein–protein interactions, modifications, and activities.
UBIQUITINATION AND ISG15/ISGYLATION IN BREAST CANCER
The study of protein posttranslational modifications is critical for understanding the molecular basis of disease progression, such as carcinogenesis[21], and given the importance of ubiquitination in protein function, the number of reports in this field has increased rapidly in recent years. Ubiquitination is an important cellular process to study during cancer progression, as it is related not only to degradation by the proteasome, but also to the abundance, localization, activity, and interaction of proteins[22]. Several analyses have shown different roles for ubiquitination related to topology; for example, Lys-48-linked monoubiquitination is a signal for endocytosis and nuclear trafficking, but polyubiquitination at that site is a signal for proteasome-dependent degradation.
The UPS regulates many cellular processes, such as cell cycle control in breast cancer, where expression of cyclins (D1, E1) and cyclin-dependent kinases is altered, and their cellular role is directly correlated with the clinical aspects of breast cancer. Among the best-known cancer clinical markers with E3-Ub ligase activity are BRCA1 (breast cancer type 1 susceptibility protein) and TRIM25; BRCA1 is an important player in breast cancer, and one of its diverse functions is to mediate the formation of Lys-6-linked polyubiquitination as well as DNA damage repair[23].
TRIM25, the subject of this review, has been reported to be an important regulator in breast cancer. Regarding the role of TRIM25 in ubiquitination, several studies have shown its interaction with a number of proteins that are modified by Ub and degraded, as described in the next section. Ub ligases regulate a number of cellular processes, including metabolism, homeostasis, cell cycle, stability of substrates and tumor suppressor and oncogenic activities. A high number of Ub ligases is deregulated in cancer as a consequence of altered posttranslational mechanisms, and this deregulation affects tumor induction, progression, and therapy resistance[21].
ISG15 and ISGylation are being studied in the context of breast cancer. It has been reported that the decrease in ISG15 or UBCH8 (an E2-conjugating enzyme for ISG15) by interfering RNA reduces proliferation, migration, and epithelial-mesenchymal transition in MDA-MB-231 breast cancer cells[24]. ISG15 or UBCH8 expression enhances the migration of breast cells, having a pro-tumor role. One mechanism proposed for ISGylation is interference with ubiquitination in breast cancer cell lines[24,25]. In contrast, ISG15 displays antitumor activity in vivo since xenotransplantation of athymic mice with ISG15 knockdown ZR-75-1 or MDA-MB-231 breast cancer cells, resulted in an increase in tumor development and a minor recruitment of NK cells in comparison to controls. Furthermore, subcutaneous injection of recombinant purified free ISG15 near the xenotransplant of MDA-MB-231 cells in these mice, led to a decrease in tumor growth, increased NK cell infiltration in tumors, and enhancement of major histocompatibility complex surface expression[26]. Despite the need for further studies, the results suggest that protein ISGylation may be coupled to carcinogenic events and that extracellular free ISG15 is an “antitumor” potential candidate factor.
The proteins targeted by TRIM25 and modulated by ubiquitination or ISGylation as well as the specific effects of these processes on breast cancer progression are discussed below.
TRIM25: STRUCTURE AND FUNCTION
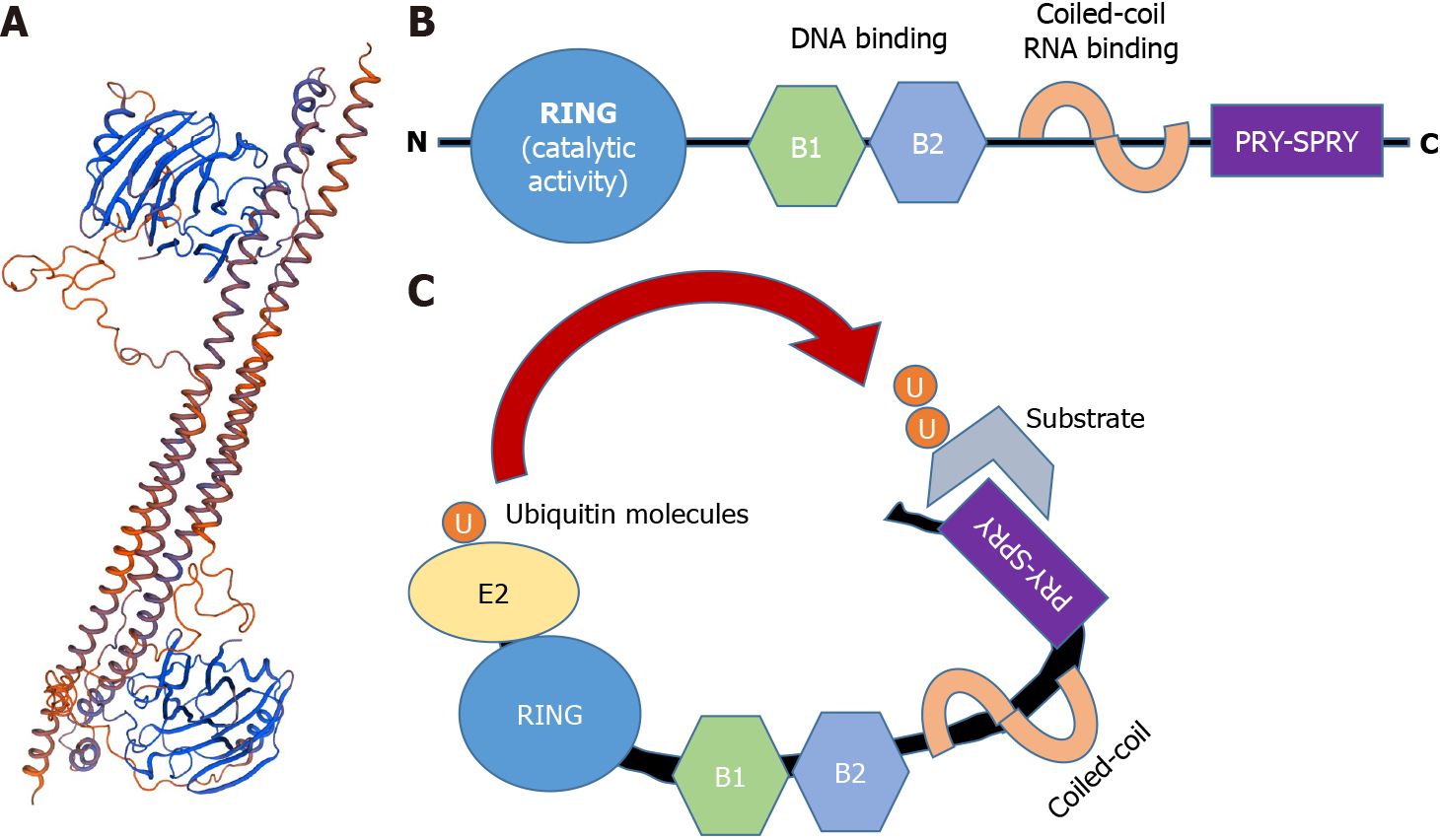
TRIM25 is a 17 beta-estradiol and type I IFN-inducible E3 Ligase, encoded by the TRIM25 gene, which is located on the human chromosome 17q22. TRIM25 belongs to the TRIM family of proteins that is characterized by the presence of three conserved N-terminal domains: a RING domain, one or two B-Boxes (B1/B2), and a coiled-coil (CC) domain[27]. The RING domain has a catalytic activity, as it promotes ubiquitin conjugation by binding to a ubiquitin-conjugating enzyme (E2)[27,28]. B-box domains regulate TRIM oligomerization, and the CC domain is responsible for the dimerization of TRIM ligases. The CC domain is also responsible for and binding to RNA. Additionally, TRIM proteins possess a variable C-terminal region that mediates the interaction with substrates; specifically. TRIM25 has a PRY-SPRY or B30.2 domain, which is involved in protein-protein interactions and RNA binding[29] (Figure 2).
Figure 2 Structure of TRIM25
. A: Crystal structure of the human TRIM25 coiled-coil and PRYSPRY domains. (DNA sequence retrieved from NCBI. Gene, https://www.ncbi.nlm.nih.gov/gene/7706#gene-expression 21/01/21, and 3D model created with Swiss Model: https://swissmodel.expasy.org/); B: Schematic representation of TRIM25, including the conserved RING, B boxes (B1 and B2), the coiled-coil domain and the C-terminal variable domain (CTD) PRY-SPRY; C: RING and CTD domains bind to the ubiquitin-loaded E2 and the substrate, respectively. Consequently, both molecules get closer, therefore favoring substrate ubiquitination.
TRIM25 is involved in many cellular processes, such as development, innate antiviral immunity, and tumor progression[30]. Regarding its role in development, it has been reported that TRIM25 is highly expressed in embryonic stem cells (ESCs)[31], and is an RNA-specific cofactor of Lin28a/TuT4-mediated uridylation[32] thus, it may be important for the maintenance of stemness[29]. Moreover, it is known that TRIM25 regulates estrogen-induced cell proliferation and uterine swelling, as mice that lack TRIM25 have underdeveloped uterus and diminished estrogen responsiveness[33].
Another important function of TRIM25 is related to innate antiviral immunity. In that sense, it has been shown that TRIM25 polyubiquitinates the K63 residue of the cytosolic receptor RIG-1, promotes the interaction with mitochondrial antiviral signaling protein, and leads to the activation of intracellular antiviral signaling and subsequent type I interferon production[27].
Finally, for the purpose of this review, it is important to highlight the contribution of TRIM25 in tumor progression. It has been reported that TRIM25 is highly expressed in breast, colorectal, urothelial, and endometrial tumors, promoting proliferation, migration, and invasion of tumor cells[29,34]. These functions are regulated by different molecular mechanisms. First, it has been reported that TRIM25 is related to tumor growth, as it increases p53 Levels by inhibiting its ubiquitination and degradation by the 26S proteasomes[35]. Moreover, Takayama et al[36] have suggested that TRIM25 modulates p53 signaling by interacting with the androgen-induced GTPase-activating protein-binding protein 2 protein. Additionally, it has been reported that TRIM25 promotes cell survival and growth by directly targeting Keap1 for ubiquitination and degradation, leading to Nrf2 activation, which consequently activates antioxidant defense and ameliorates oxidative stress[37]. TRIM25 has also been shown to act as a transcriptional regulator of breast cancer metastasis networks[30]. For instance, TRIM25 regulates some critical stemness signatures, such as POU5F1, SOX2, and NANOG, which enhance cellular colonization.
The findings presented in the previous paragraphs demonstrate the importance of TRIM25 in various cellular processes, and how some of them are involved in the development and progression of cancer. Hence, TRIM25 has been proposed as a regulator for breast cancer, and therefore, a molecular target for the diagnosis, prognosis, and treatment of this disease[29,38,39].
REGULATION OF TRIM25 EXPRESSION IN BREAST CANCER
The expression of TRIM25 gene can be induced by IFN–type I a and b[13]. In addition, it has been reported that TRIM25 has p53-responsive elements, inducing its expression in a p53–dependent manner upon DNA damage[40]. Among the most important stimuli that induce TRIM25 expression in breast cancer cells are estrogens. The mechanisms related to the regulation of TRIM25 expression are not fully understood. However, some of them have been investigated and the results indicate their complexity and possible association with several signaling pathways in several cellular contexts.
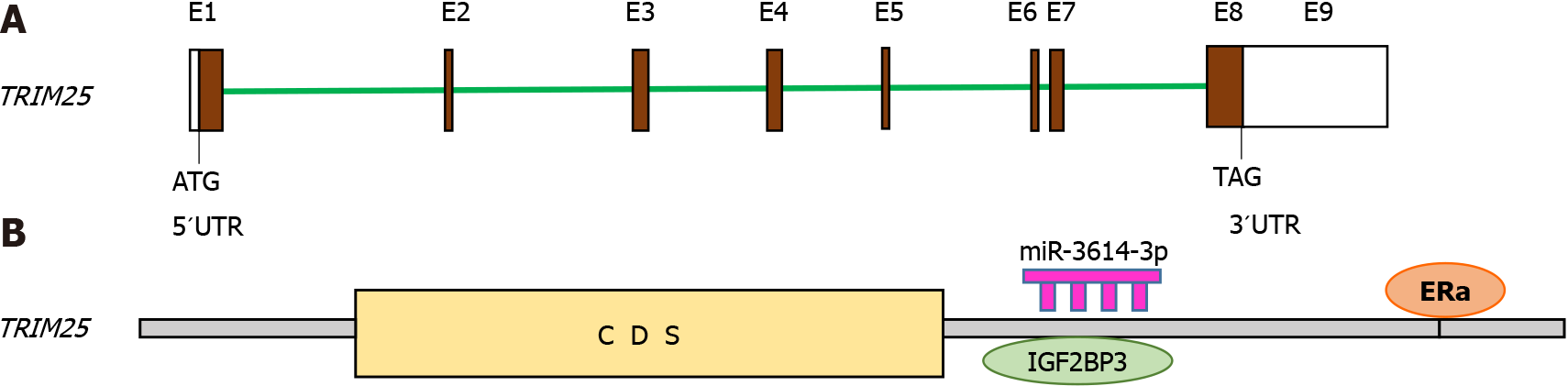
The human TRIM25 gene covers approximately 25 kb of genomic DNA and is organized in nine exons (Figure 3A). TRIM25 is also named as EFP, since its expression in breast cancer cells is induced by estrogens within 30 min. At the mechanistic level, it has been demonstrated that the TRIM25 3´UTR has one estrogen responsive element (ERE). Interestingly, this region of TRIM25 3´UTR possesses an enhancer activity that acts in an estrogen-dependent manner, increasing TRIM25 expression in breast cancer cells[41]. Additionally, Urano et al[42] have reported that TRIM25 is an estrogen-responsive E3-Ub ligase. They found that implantation of transfected TRIM25-MCF7 cells into ovariectomized mice resulted in the formation of prominent tumors, while implantation of vector-transfected MCF7 cells did not result in the formation of tumors.
Figure 3 Structure of TRIM25 gene and its regulation.
The structure of the TRIM25 gene is shown in (A) Regulation of TRIM25 expression by ERα, (B) miRNA-3614-3p, and IGF2BP3 through its 3´UTR region.
Furthermore, TRIM25 expression can also be modulated by miRNA-3614-3p and proteins associated with mRNAs, such as IGF2BP3 (Figure 3B). The TRIM25 3´-UTR region contains the pri-miRNA-3614-3p, as well as a binding site for miRNA-3614-3p. Hence, miRNA-3614-3p prevents the expression of its own host gene, TRIM25, via miR-3614-mediated degradation, after its binding to the TRIM25 3´-UTR. Furthermore, IGF2BP3 protein can bind to the TRIM25 3´UTR to inhibit miRNA-3614 maturation. Breast cancer cell growth is blocked by TRIM25 downregulation through two events: (1) miRNA-3614-3p overexpression or (2) IGF2BP3 depletion[43].
The regulation of TRIM25 expression has an impact on other important proteins in breast cancer such as ERα. It has been demonstrated that ZBTB7A binds to regulatory regions of the gene that encodes for ERα (ESR1) to induce its expression in breast cancer cells, and regulates other ERα-dependent genes, facilitating breast cancer progression. Additionally, TRIM25 seems to be regulated by ZBTB7A since when ZBTB7A is inhibited, TRIM25 is upregulated. TRIM25 enhances ERα degradation via the UPS system, maintaining a positive correlation between ZBTB7A and ERα in breast cancer[44].
With respect to TRIM25 expression in breast tissue, a study detected TRIM25 in the cytoplasm of normal breast tissue cells, and in cell culture media, as a secreted factor. Also, high levels of TRIM25 have been detected in lactating breast, suggesting that TRIM25 may play a role in mammary gland differentiation. In a study low levels of TRIM25 were found by immunohistochemistry in breast cancer[45]. Nevertheless, in another study, TRIM25 immunoreactivity was positively correlated with poor prognosis of breast cancer[46].
TRIM25 AND PROTEIN UBIQUITINATION
Previous studies have shown that the Capicua (CIC) protein has an important role as a prognostic factor in different types of cancer, and this role is related to changes in the expression of targets downstream of the MAPK signaling cascade[47]. However, a MAPK-independent mechanism has been proposed where the interaction of CIC with ATXN1L prevents CIC degradation by TRIM25[48]. Whereas TRIM25 repression increases CIC levels and its tumor suppressor function, TRIM25 induction decreases it, indicating that CIC degradation is mediated by TRIM25 via the UPS. TRIM25 is enriched in both ATXN1L knockout and knockdown cell systems[48]. Another CIC interacting protein is 14-3-3 sigma, which has been shown to act as a tumor suppressor by negatively regulating the cell cycle and causing G2/M arrest. In breast cancer, 14-3-3 sigma expression is downregulated through epigenetic silencing by hypermethylation, and this inactivation has been suggested to be an early event in breast carcinogenesis[49]. A second mechanism for 14-3-3 sigma downregulation involves its interaction with TRIM25[42,50]. It has been shown that degradation of 14-3-3 sigma promoted breast tumor growth in a TRIM25 dose-dependent manner[42,50]. Overexpression of TRIM25 has been shown to drastically reduce the levels of 14-3-3 protein, which are recovered by MG132 (a potent inhibitor of proteasome function), suggesting that TRIM25 directly downregulates 14-3-3 sigma through a proteasome-dependent mechanism[42]. Several studies suggest that in human breast cancer, the regulation of 14-3-3 sigma may involve both mechanisms[49].
Another player in estrogen-dependent breast development and carcinogenesis is AT motif-binding factor 1 (ATBF1), which has been described as a potential tumor suppressor and is upregulated by direct binding of ERα to its promoter. TRIM25 also interacts with ATBF1, mediating its degradation by the UPS. Thus, ATBF1 antagonizes TRIM25-mediated cell proliferation[51]. It is also known that the transcription factor Kruppel-like factor 5 (KLF5), is a multifunctional transcription factor that inhibits estrogen-induced gene expression and cell proliferation. In breast cancer cells, estrogen-induced TRIM25 causes KLF5 protein degradation by the UPS. At the transcriptional level, estrogen downregulates KLF5, however, it takes a longer period of time than its effect at the protein level. It has been proposed that KLF5 and estrogen signaling regulate each other through a feedback mechanism, where high levels of KLF5 inhibit the function of estrogen by interacting with the ERα. Moreover, TRIM25 overexpression prevents KLF5 ubiquitination and enhances its own ubiquitination. Ubiquitinated TRIM25 is unable to interact with KLF5, whereas TRIM25 without ubiquitination marks can associate with KLF5 in breast cancer cells[52].
TRIM25 MEDIATES ISGYLATION
Under DNA damage, a p53-dependent pathway induces 14-3-3 sigma expression, which sequesters cyclin B1-CDC2 complexes outside the nucleus, favoring G2 arrest. However, in breast cancer, the downregulation of 14-3-3 sigma is common due to CpG methylation, p53 inactivation, and degradation via the UPS. TRIM25 acts as an E3-Ub ligase, mediating 14-3-3 sigma degradation (as it was mentioned previously). Intriguingly, TRIM25 and the E2-Ub conjugating enzyme UBC8 participate in the posttranslational modification of 14-3-3 sigma via ISGylation[53]. In addition to the identification of TRIM25 as an E3 Ligase for ISG15, it has also been shown that this activity is RING domain-dependent[19]. Interestingly, TRIM25 is a target of ISGylation through its own E3 ISG15 Ligase activity. The ring-finger domain of TRIM25 is implicated in its auto-ISGylation, and the TRIM25 Lysine modified by ISG15 is K117. TRIM25 carrying a mutation in this residue was resistant to its own ISGylation. However, this mutant EFP-K117R enhances the ISGylation of its target, 14-3-3 sigma. All data suggest that autoISGylation of TRIM25 inhibits its function as an ISG15 E3 Ligase and the ISGylation of its protein target, 14-3-3 sigma[54].
Moreover, the carboxyl terminus of Hsp70-interacting protein (CHIP) is an E3-Ub ligase that is a target of ISGylation in HEK293 cells upon stimulation with type I IFN. The ISGylation target residues of CHIP are Lys143/144/145 and Lys287, and as a result of this modification, the activity of CHIP as an E3 ubiquitin ligase is increased, affecting targets such as c-Myc. Hence, the ISGylation confers an antitumor activity to CHIP, and this modification can be mediated by HERC5 and TRIM25[55].
TRIM25 IN BREAST CANCER: FUTURE CHALLENGES
TRIM25 is a common component in the ubiquitination and ISGylation pathways. However, the mechanisms that define participation in one or the other system remain to be elucidated. Similarly, whether there is a competition between the TRIM25-dependent substrate ubiquitination or ISGylation should be evaluated. It is not completely known the effect of these modifications on the stability, localization, interaction profile, and the specific functions of their substrates in breast cancer. Furthermore, many other signaling pathways with cancer and anti-cancer activities may affect the expression and abundance of TRIM25 in mammary tumors.
TRIM25 is an ERα-regulated gene[41], and consequently, mammary tumors that do not express this receptor may have a different pattern of expression, regulation, and activity of TRIM25. In addition, the TRIM25-regulated proteome may be altered by treatment with anti-estrogens commonly used in endocrine therapy, such as tamoxifen (a type of SERM) and fulvestrant (a type of SERD). All these aspects related to TRIM25 may have impact in the detection, prognosis, and treatment of breast tumors. To date, the studies indicate that TRIM25 is a key element involved in molecular pathways that differentially modify the proteome in mammary tumors in comparison with normal mammary tissue.
TRIM25 is also involved in other cancer types. For example, it has been proposed that TRIM25 may be a biomarker and target for therapies against endometrial[56] and lung cancer[57]. Nevertheless, its ability to be induced in response to estrogens and its relationship with ERα-associated pathways indicate its relevance in breast cancer. Additional studies have shown that deregulation of TRIM25 is associated with metastasis and poor survival outcome of patients with breast cancer[30]. Moreover, it has been proposed that TRIM25 can modulate gene signatures at the transcriptional and post-transcriptional levels, in addition to its effect on the proteome in breast cancer. Therefore, the identification of proteins modified by TRIM25 may help to further establish its functions and relationships with other molecular mechanisms, and modifications in this disease.
CONCLUSION
In conclusion, the data presented here suggest that TRIM25 may be a biomarker in breast cancer. Although more studies are required to define the molecular mechanisms of regulation and action of TRIM25, the investigations performed so far suggest that TRIM25 may be a potential target for new therapeutic strategies to control breast cancer progression.
ACKNOWLEDGEMENTS
We thank Cerecedo CR for her comments.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Huo Q, Riis M S-Editor: Wu YXJ L-Editor: A P-Editor: Yuan YY