Copyright
©The Author(s) 2020.
World J Clin Oncol. Aug 24, 2020; 11(8): 563-572
Published online Aug 24, 2020. doi: 10.5306/wjco.v11.i8.563
Published online Aug 24, 2020. doi: 10.5306/wjco.v11.i8.563
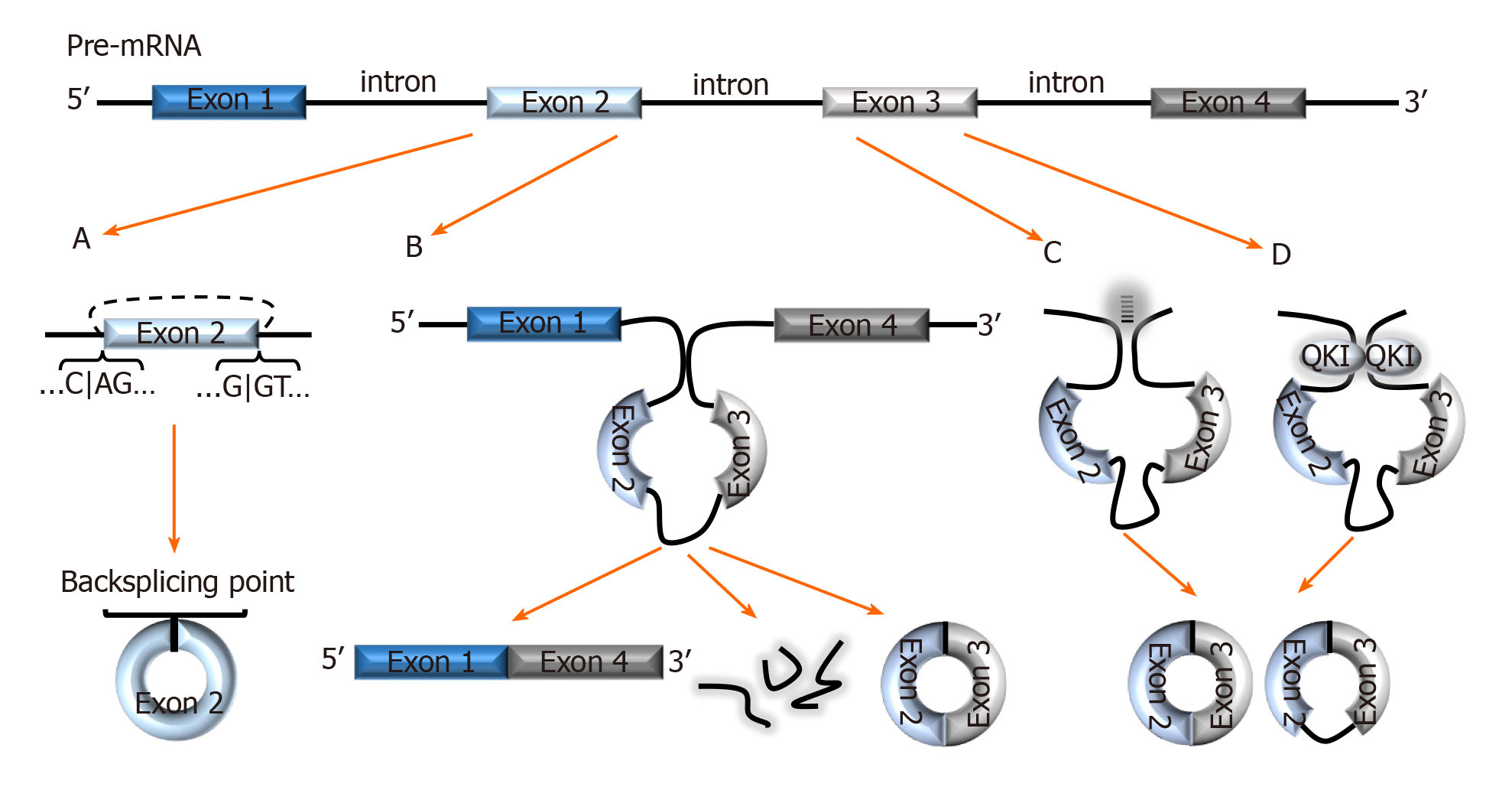
Figure 1 Biogenesis of circRNAs.
A: In backsplicing, circRNAs are usually flanked by the canonical splicing motifs, AG-GT, and covalently fuse the 5′ site of an upstream exon (acceptor) with the 3′ end of a downstream exon (donor); B: In the exon skipping model, an unstable intermediate lariat consisting of introns and skipped exons are generated after splicing. The intermediate lariat is then spliced to produce circRNA; C: Flanking introns containing complementary sequences (Alu repeats) bind and increase the possibility of backsplicing; D: RNA-binding proteins, such as Quaking can bind to flanking introns and dimerize to create a closed RNA loop which facilitates backsplicing. QKI: Quaking.
- Citation: Tucker D, Zheng W, Zhang DH, Dong X. Circular RNA and its potential as prostate cancer biomarkers. World J Clin Oncol 2020; 11(8): 563-572
- URL: https://www.wjgnet.com/2218-4333/full/v11/i8/563.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i8.563