Copyright
©The Author(s) 2017.
World J Clin Oncol. Feb 10, 2017; 8(1): 67-74
Published online Feb 10, 2017. doi: 10.5306/wjco.v8.i1.67
Published online Feb 10, 2017. doi: 10.5306/wjco.v8.i1.67
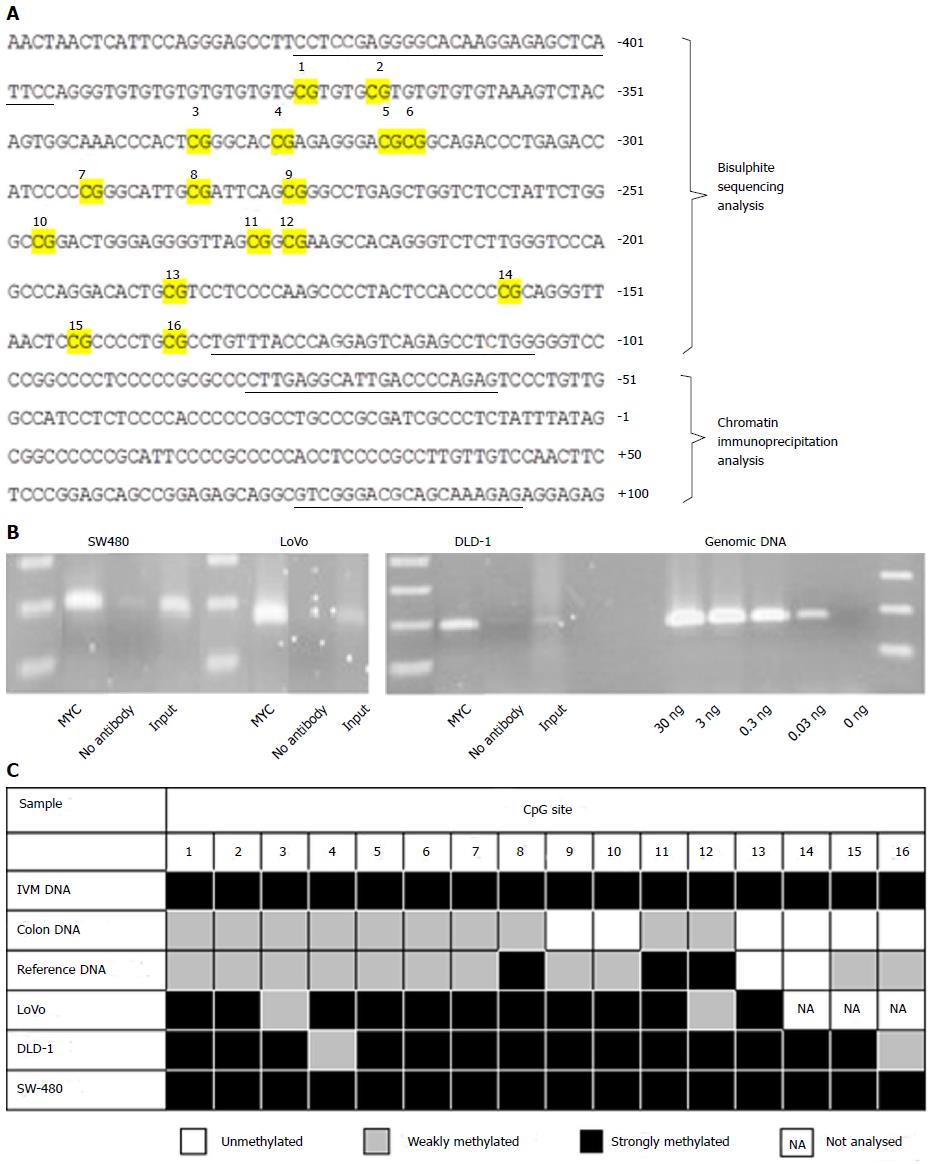
Figure 1 Epigenetic and chromatin immunoprecipitation analysis of the NDRG2 promoter in three colorectal cancer cell lines.
A: The NDRG2 gene sequence around the transcriptional start site at +1. Primer-binding regions for PCR are underlined and CpG sites subjected to methylation analysis are numbered 1 to 16; B: Endogenous MYC interacts with the NDRG2 core promoter. ChIP analysis was carried out on SW-480, LoVo and DLD-1 cell extracts using antibody against the transcription factor MYC. “No antibody” was without antibody and “input” served as a positive control. Genomic DNA was used as positive control for the PCR reaction; C: The NDRG2 promoter is hypermethylated in three colorectal cancer cell lines. Bisulfite sequencing was carried out on human genomic DNA from LoVo, DLD-1 and SW-480 cell lines, normal colonic DNA, reference DNA and in vitro SssI-methylated (IVM) DNA. Each CpG site was rated as unmethylated, weakly methylated (≤ 50% methylated), or strongly methylated (> 50% methylated).
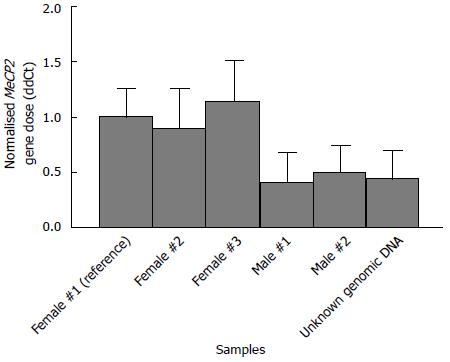
Figure 2 Validation of the gene copy number experimental setup.
Bar diagram showing the calculated delta delta Ct values (ddCt) of the X-linked MeCP2 gene normalised to GFAP, giving the expected result (one copy in males and two copies in females). A ddCt value of 1.00 in the reference female genomic sample represents the normal two alleles. Data are presented as mean (filled bars) and SD (whiskers).
- Citation: Lorentzen A, Mitchelmore C. NDRG2 gene copy number is not altered in colorectal carcinoma. World J Clin Oncol 2017; 8(1): 67-74
- URL: https://www.wjgnet.com/2218-4333/full/v8/i1/67.htm
- DOI: https://dx.doi.org/10.5306/wjco.v8.i1.67