Copyright
©2012 Baishideng Publishing Group Co.
World J Clin Oncol. May 10, 2012; 3(5): 67-79
Published online May 10, 2012. doi: 10.5306/wjco.v3.i5.67
Published online May 10, 2012. doi: 10.5306/wjco.v3.i5.67
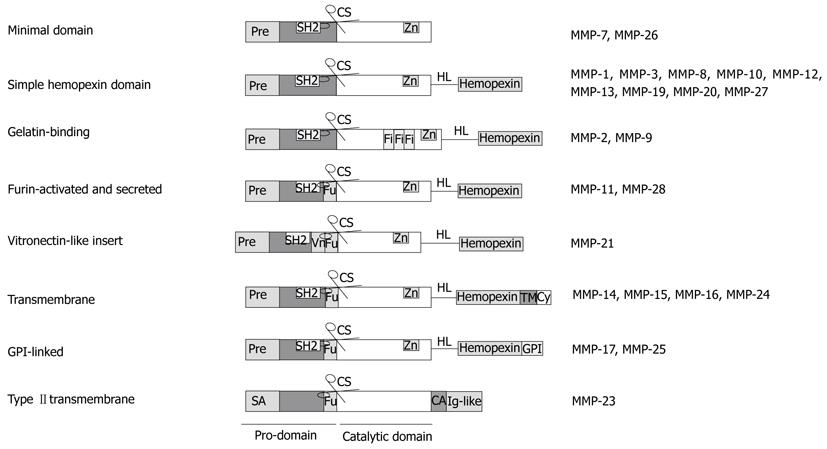
Figure 1 Structural groups of matrix metalloproteinases and their domain composition.
Pre: Amino-terminal signal sequence, directing matrix metalloproteinases (MMPs) to the endoplasmic reticulum; SA: Signal anchor for cell membrane targeting; Fu: Recognition motif for intracellular furin-like serine proteinases, allowing intracellular activation of MMPs by cutting off the pro-domain at the cleavage site; Vn: Vitronectin-like insert; Fi: Collagen-binding type II repeats of fibronectin, HL: hinge linker, connecting the catalytic domain with the hemopexin domain, which mediates interaction with tissue inhibitors of metalloproteinases (TIMPs), cell-surface molecules and proteolytic substrates; CA: Cysteine array; TM: Single-span transmembrane domain; Cy: Cytoplasmic domain.
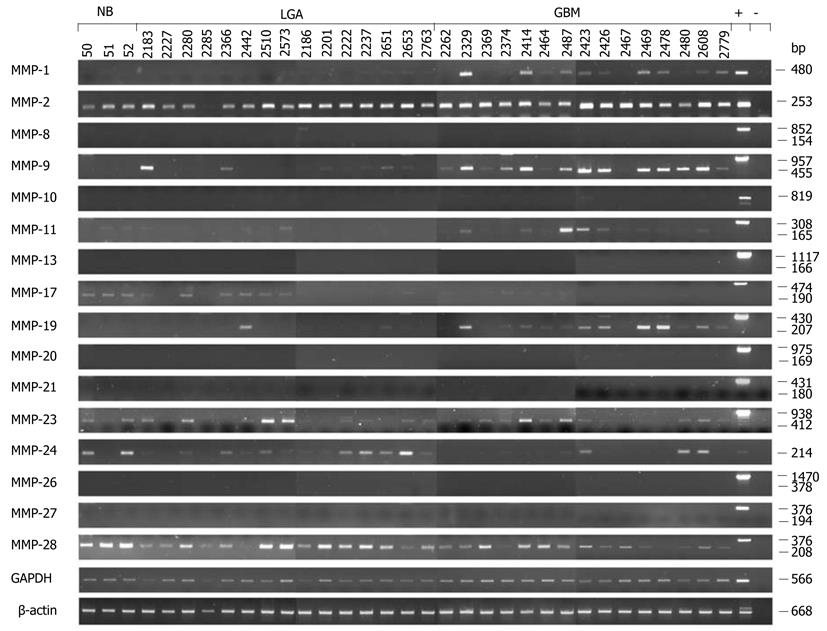
Figure 2 Expression analysis of matrix metalloproteinases in normal brain and tumor brain samples by semiquantitative real-time reverse transcription PCR.
Total RNA from normal brain (NB, lanes 1 to 3), low grade astrocytoma (LGA, lanes 4 to 18) and glioblastoma multiforme tissue samples (GBM, lanes 19 to 33) was used as a template for real-time reverse transcription PCR (RT-PCR) analysis. Primers, specific for each transcript, were designed in flanking exons, resulting in longer amplicons if human genomic DNA was amplified [positive control (+)] and in shorter amplicons representing cDNAs. In several cases (MMP-1, -10 and -24) HBMEC (human brain microvascular endothelial cell) cDNA was used as a positive control. The various cDNA concentrations were normalized to that of the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) which was used as an internal loading control. GAPDH transcripts were amplified in 20 cycles, whereas amplification of MMP transcripts was performed in 33 cycles. All technical procedures, PCR conditions and primer sequences have been published[77].
- Citation: Hagemann C, Anacker J, Ernestus RI, Vince GH. A complete compilation of matrix metalloproteinase expression in human malignant gliomas. World J Clin Oncol 2012; 3(5): 67-79
- URL: https://www.wjgnet.com/2218-4333/full/v3/i5/67.htm
- DOI: https://dx.doi.org/10.5306/wjco.v3.i5.67