Copyright
©The Author(s) 2024.
World J Clin Oncol. Aug 24, 2024; 15(8): 1061-1077
Published online Aug 24, 2024. doi: 10.5306/wjco.v15.i8.1061
Published online Aug 24, 2024. doi: 10.5306/wjco.v15.i8.1061
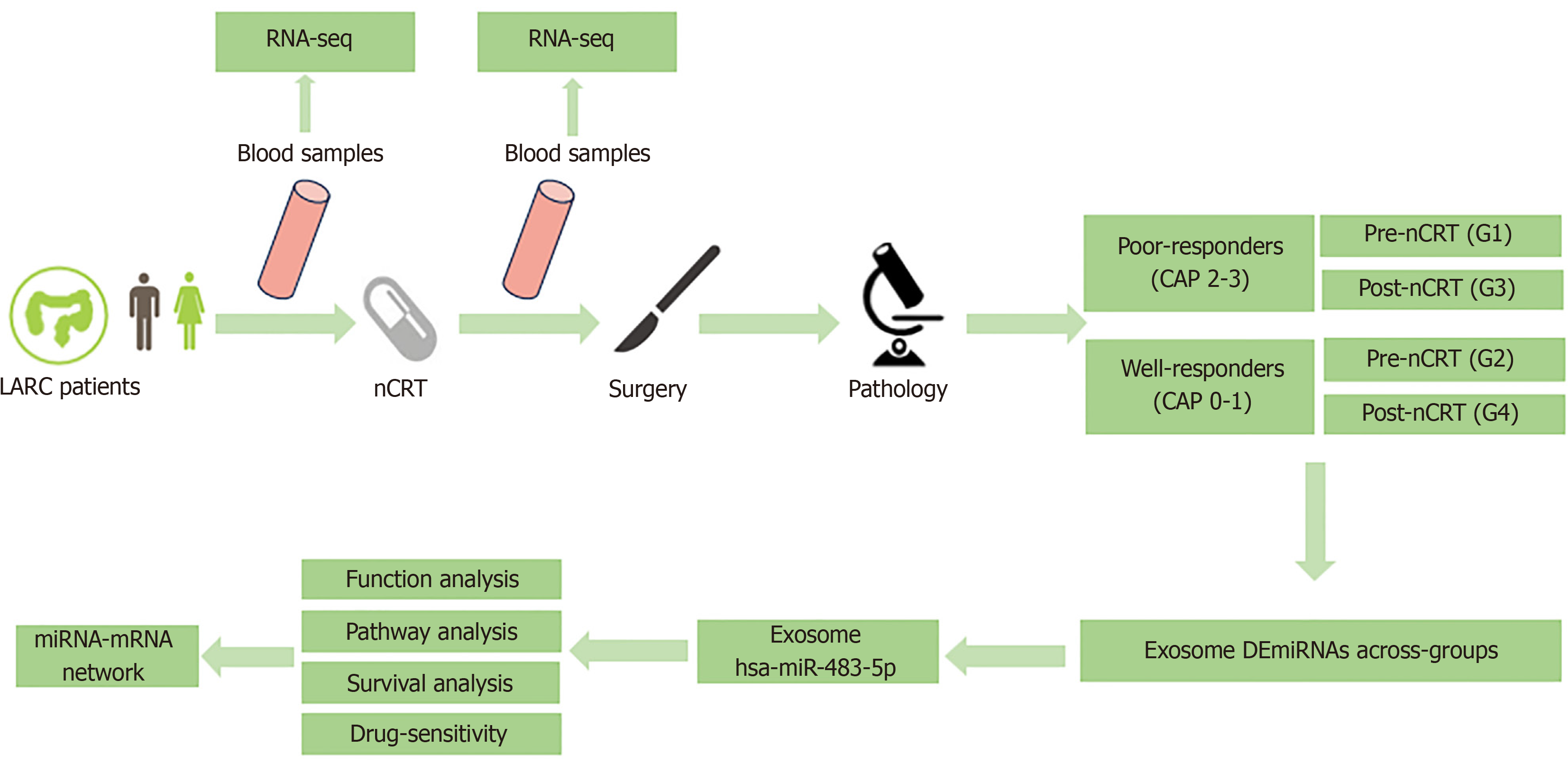
Figure 1 The flow diagram of study design.
nCRT: Neoadjuvant chemoradiotherapy; RNA seq: RNA sequencing; LARC: Locally advanced rectal cancer; CAP: College of American Pathologists; DE: Differentially expressed; miRNA: MicroRNA.
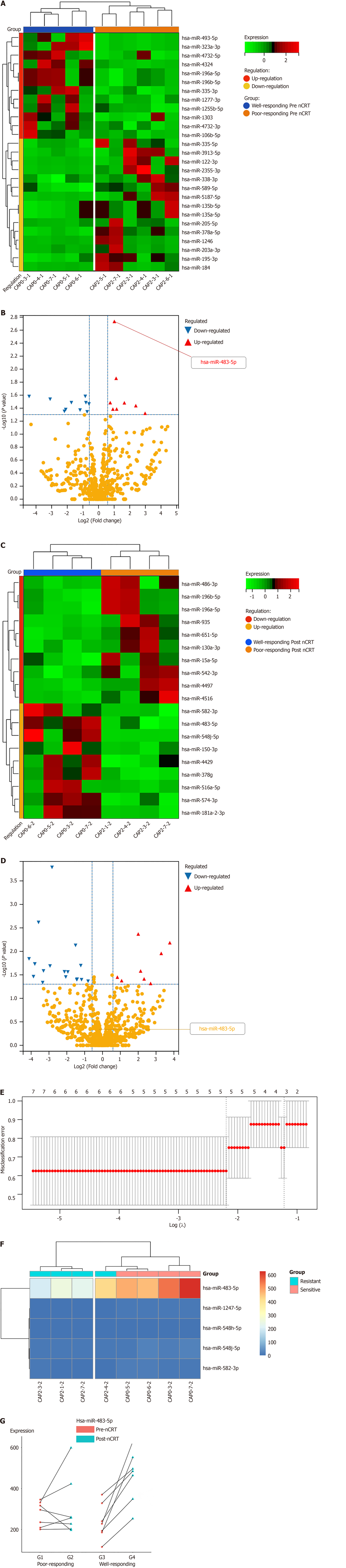
Figure 2 The expression profiles of exosome microRNAs and the selection of interest microRNAs.
A-D: The heatmaps and volcano plots of exosome differentially expressed (DE) microRNAs (miRNAs) in poor-responders and good responders pre- neoadjuvant chemotherapy (nCRT) (A and B) and post-nCRT (C and D); E and F: Showed the results of least absolute shrinkage and selection operator regression analysis to identify the exosome hsa-miR-483-5p as candidates for further analysis; G: Shows the cross-group comparison of the expression profiles of hsa-miR-483-5p among the four group. DE: Differentially expressed: NCRT: Neoadjuvant chemotherapy; CAP: College of American Pathologists.
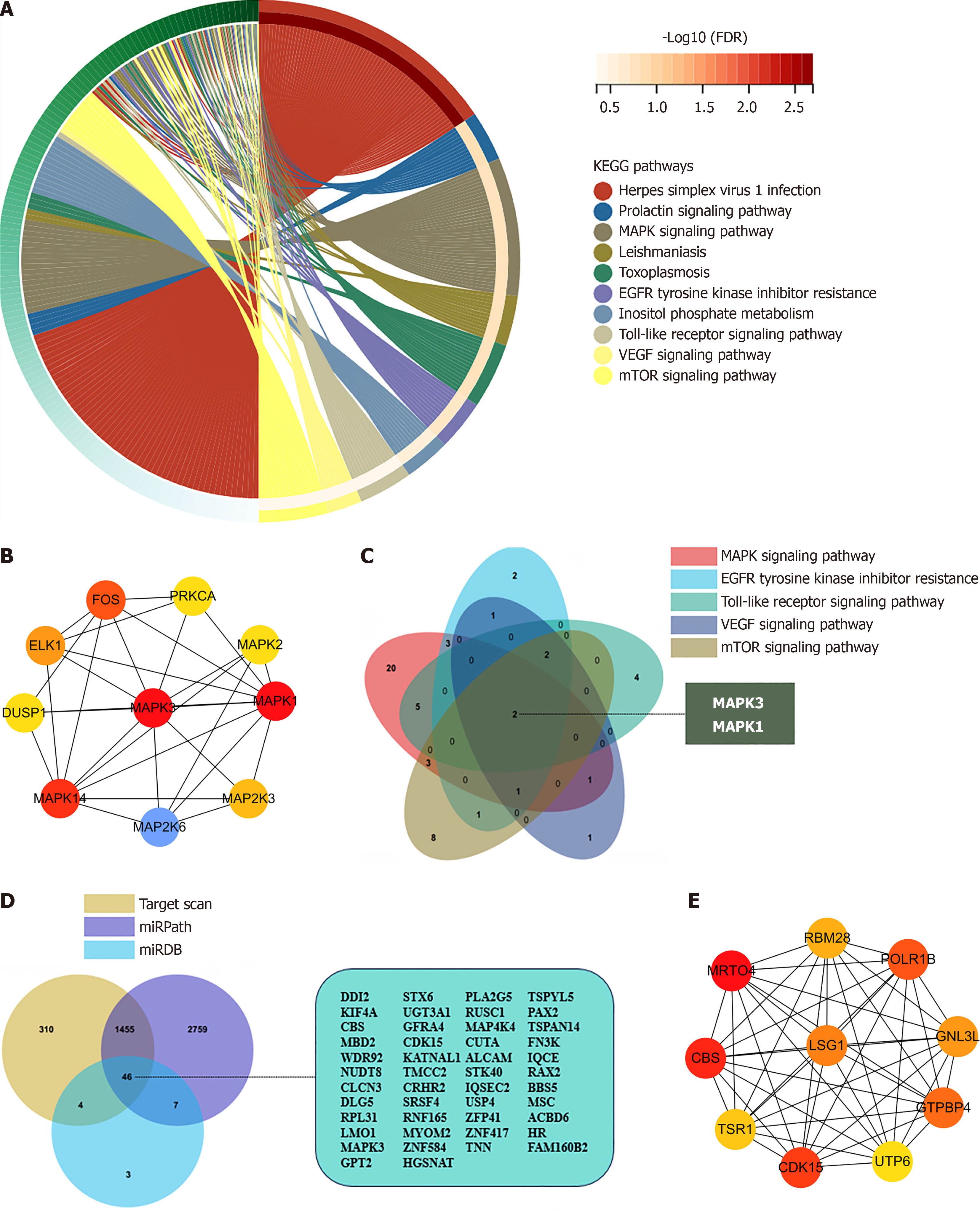
Figure 3 The bioinformatics analysis of the target genes of exosome hsa-miR-483-5p.
A: Represents the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment results of target genes; B: The protein-protein interaction (PPI) network and the hub genes selected in the enriched five KEGG pathways; C: The selected common target genes of hsa-miR-483-5p enriched in the five KEGG pathways, with two identified hub genes-MAPK1 and MAPK3; D: The PPI network and the hub genes selected from the target genes of exosome hsa-miR-483-5p; E: The selection of target genes of hsa-miR-483-5p from the three miRNA-associated databases, with chosen 46 common genes. KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: Protein-protein interaction.
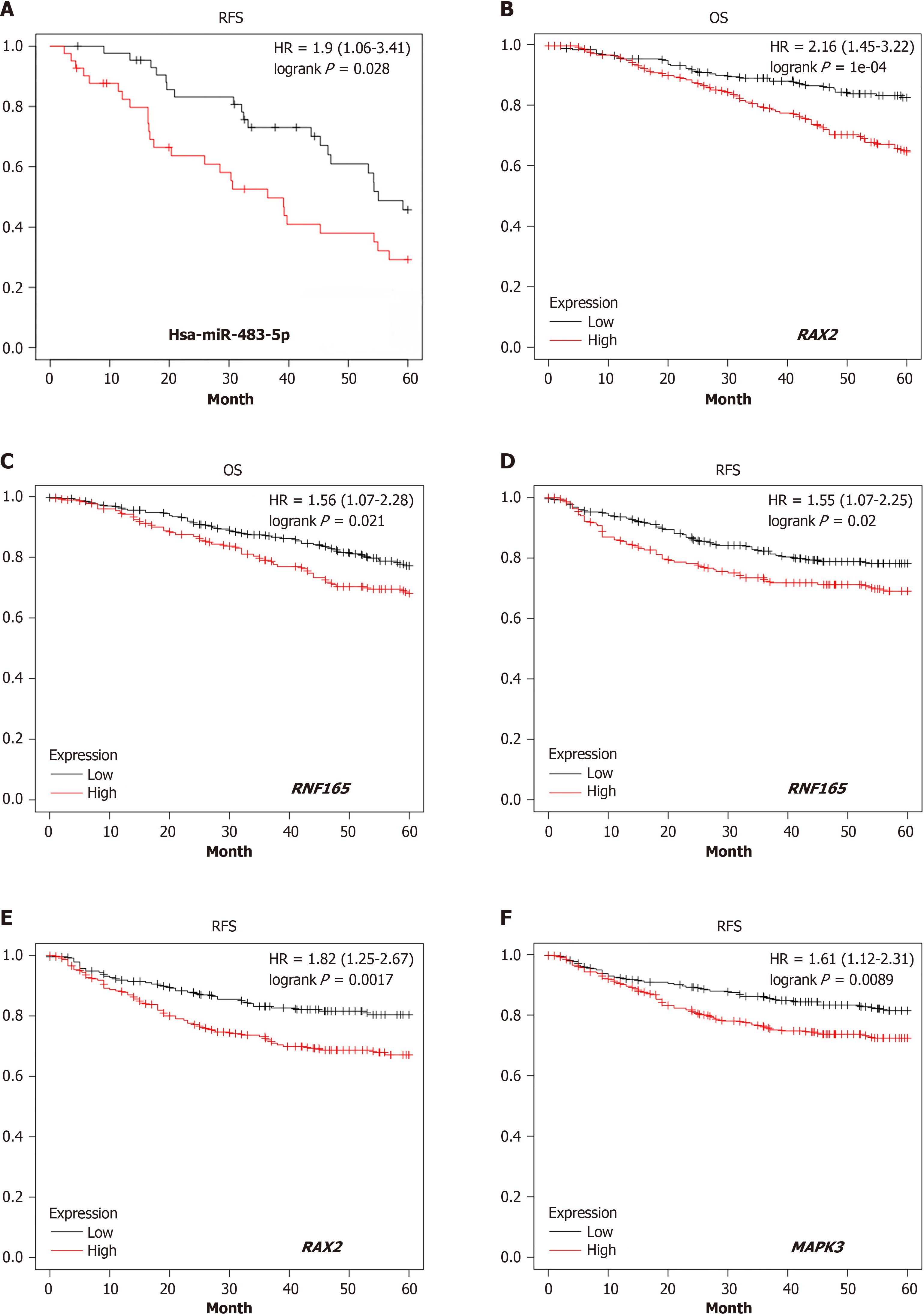
Figure 4 The survival analysis of target genes of hsa-miR-483-5p using TCGA database.
A: The up-regulation of hsa-miR-483-5p is associated with a relatively poor survival outcomes in breast cancer patients, suggesting its role in the pathogenesis of tumors; B and C: Demonstrated that the low-expression of target genes of RAX2 and RNF165 are correlated with better 5 years-overall survival in colorectal cancer patients; D-F: Represents that the low-expression of target genes of RAX2, RNF165, and MAPK3 are positively associated with superior 5 years-recurrence-free survival, correspondingly. OS: Overall survival; RFS: Recurrence-free survival; CRC: Colorectal cancer; HR: Hazard ratio.
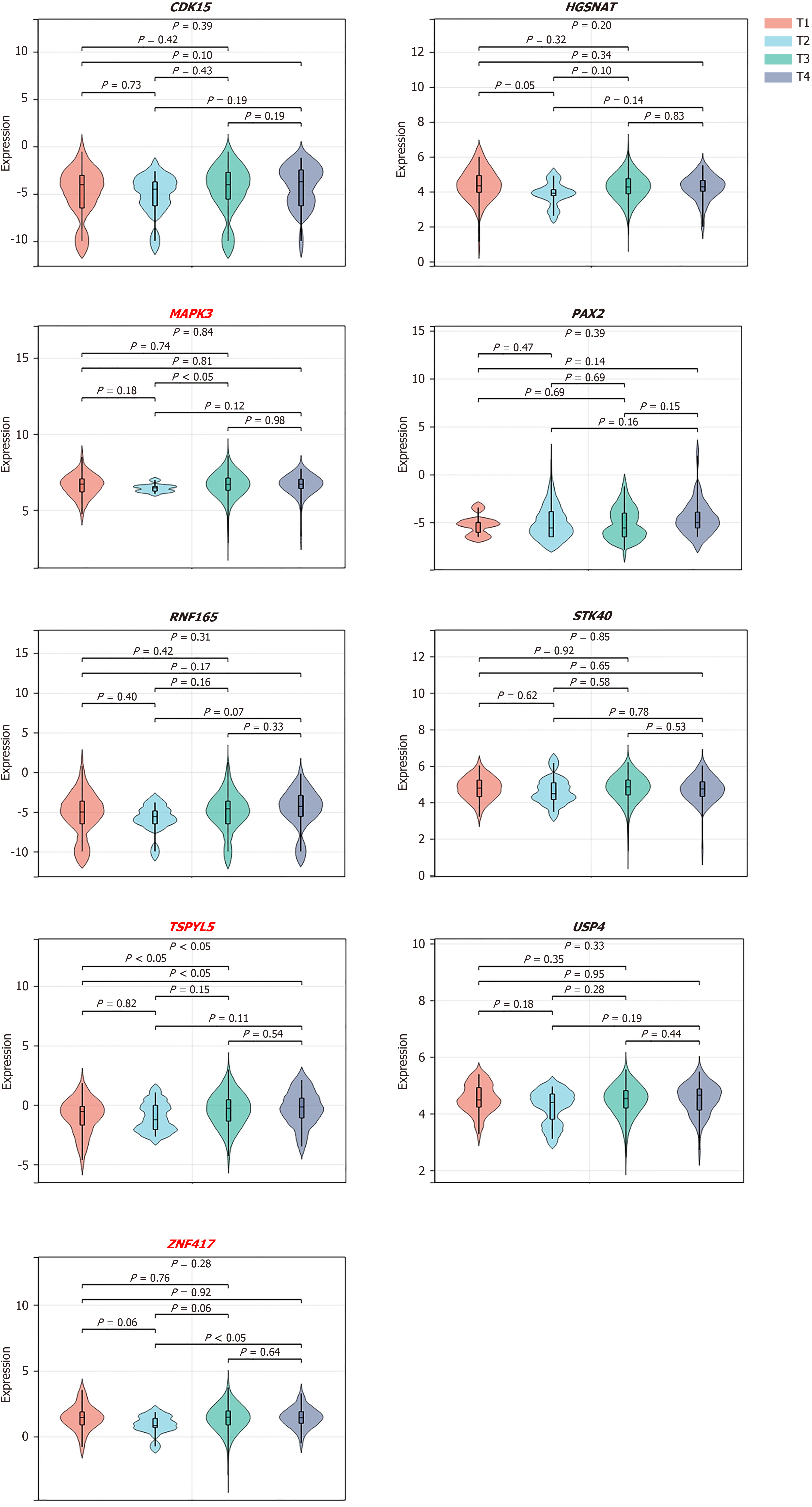
Figure 5 The association of the expression level of 9 target genes and the tumor stage.
The results revealed that the expression levels of target genes of MAPK3, TSPYL5, and ZNF417 are negatively correlated with the tumor stages.
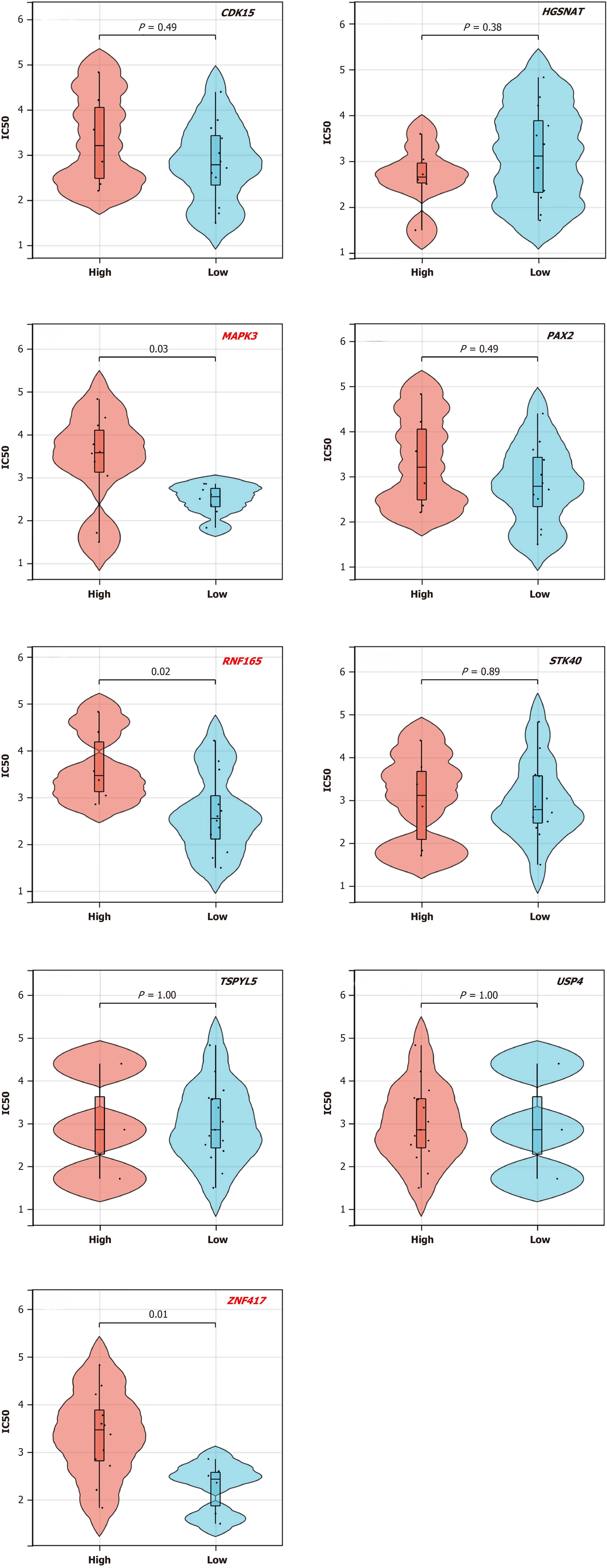
Figure 6 The relationship of the 9 target genes and drug-sensitivity of oxaliplatin.
The results indicated that the lower expression of target genes of RNF165, ZNF417, and MAPK3 are prone to exhibit relatively lower IC50 value.
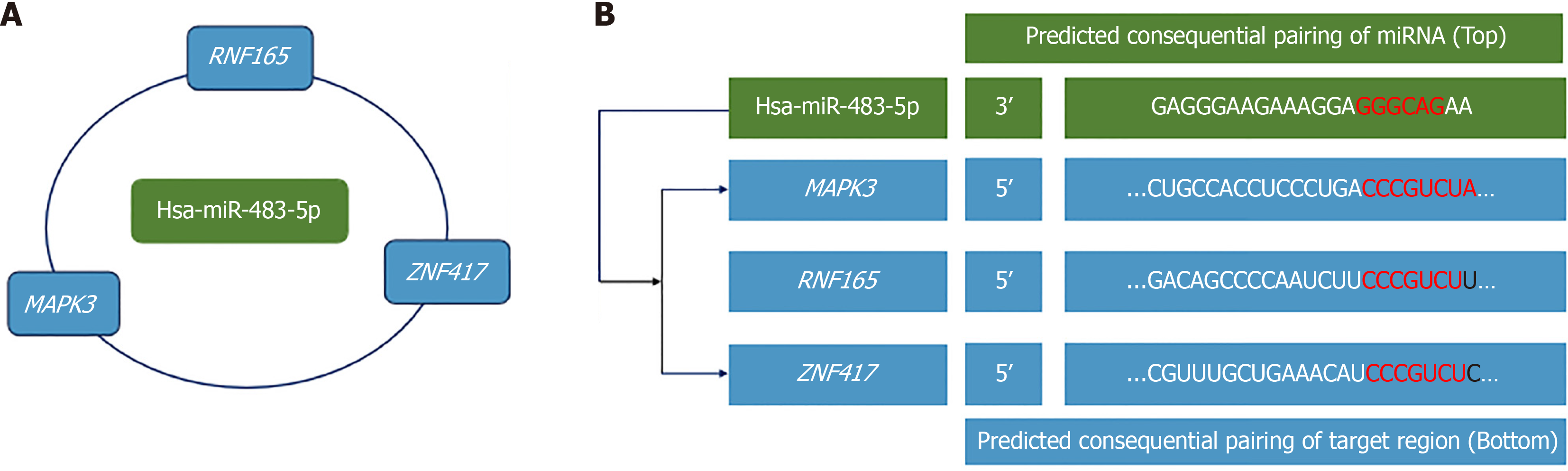
Figure 7 The construction of microRNAs-mRNA network.
A: The construction of “hsa-miR-483-5p/RNF165”, “hsa-miR-483-5p/ZNF417”, and “hsa-miR-483-5p/MAPK3” network; B: The potential binding sites of hsa-miR-483-5p and its target genes predicted from Target Scan database. miRNA: MicroRNA.
- Citation: Li GB, Shi WK, Zhang X, Qiu XY, Lin GL. Hsa-miR-483-5p/mRNA network that regulates chemotherapy resistance in locally advanced rectal cancer identified through plasma exosome transcriptomics. World J Clin Oncol 2024; 15(8): 1061-1077
- URL: https://www.wjgnet.com/2218-4333/full/v15/i8/1061.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i8.1061