Copyright
©The Author(s) 2022.
World J Clin Oncol. Nov 24, 2022; 13(11): 866-879
Published online Nov 24, 2022. doi: 10.5306/wjco.v13.i11.866
Published online Nov 24, 2022. doi: 10.5306/wjco.v13.i11.866
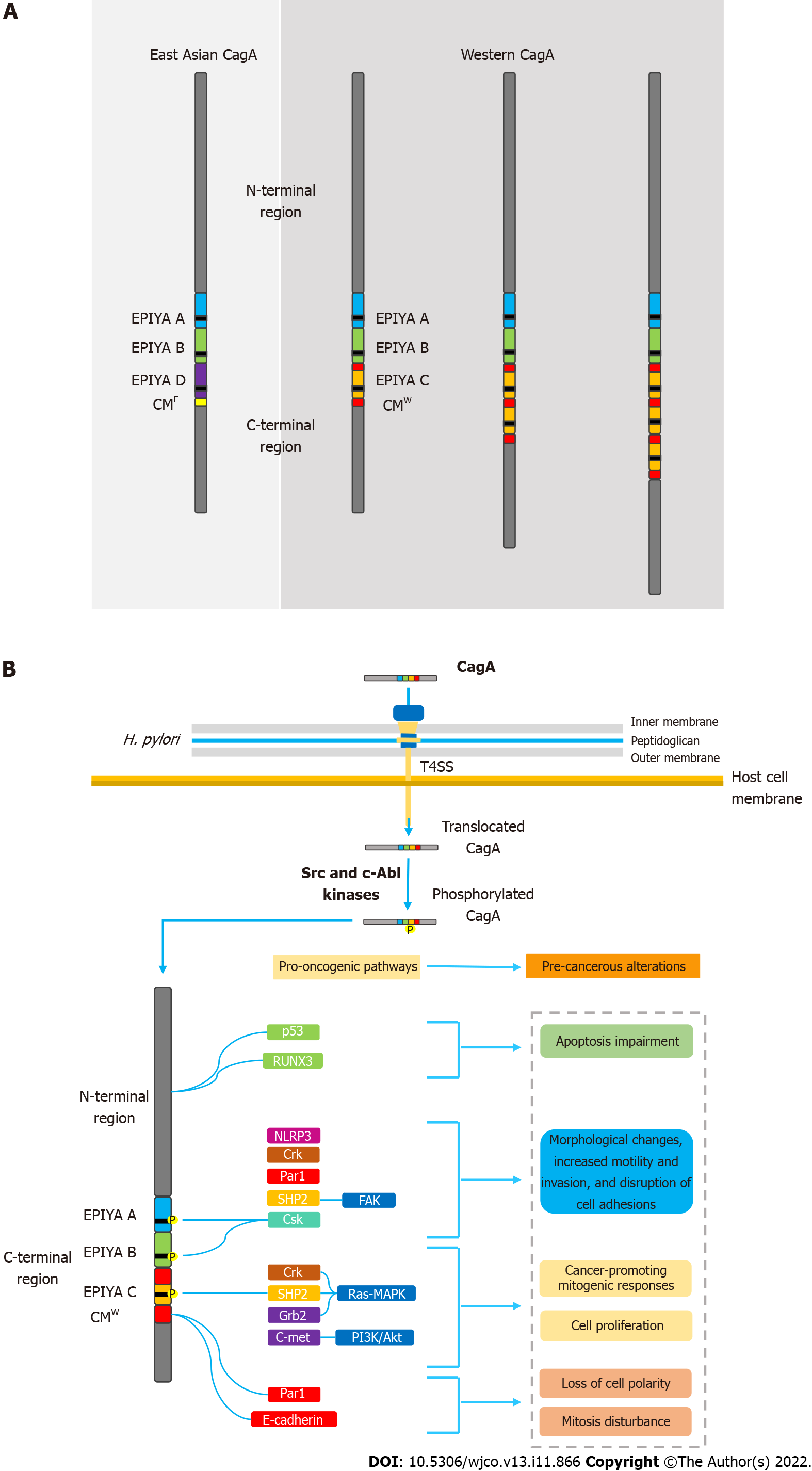
Figure 1 Helicobacter pylori oncoprotein: molecular structure and CagA-mediated carcinogenesis underlying mechanisms.
A: Structural domain differences between East Asian and western CagA. CagA tertiary structure is characterized by a structured N-terminal region and an unstructured C-terminal tail. The oncoprotein contains repetitive sequences in its C-terminal polymorphic region, known as the EPIYA motifs and CM motif. EPIYA-A and EPIYA-B motifs were identified in almost all CagA-positive H. pylori strains, followed by one, two, or three EPIYA-C sequences in western strains, or an EPIYA-D segment in East Asian strains. The CM motif, although highly conserved, possesses a 5-amino-acid difference between East Asian and western strains, hence distinguishing East Asian and Western CagA; B: Molecular mechanisms of CagA-mediated carcinogenesis. Upon translocation, CagA EPIYA motifs are tyrosine-phosphorylated by Src family or c-Abl kinases of the host cell. After phosphorylation, CagA localizes to the inner leaflet of the plasmatic membrane and acts as a promiscuous scaffold or hub protein that simultaneously disturbs multiple host signaling pathways, involved in regulation of a large range of cellular processes, including proliferation, differentiation and apoptosis. Ultimately, the disharmonic interaction between CagA and host proteins leads to pro-oncogenic cellular alterations. CM: CagA-multimerization; CMW: western CagA.
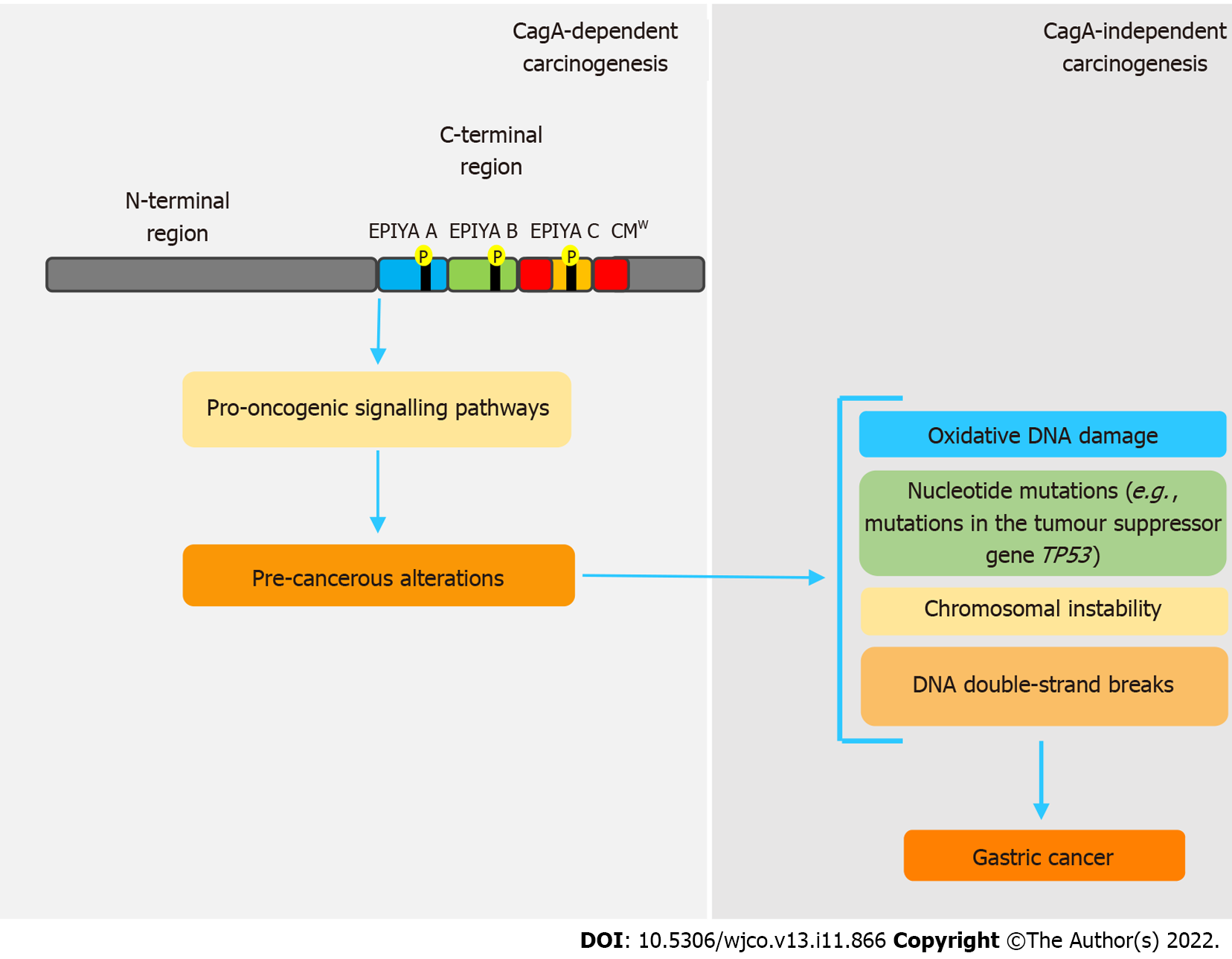
Figure 2 Simplified model of CagA-mediated hit-and-run carcinogenesis.
The pro-oncogenic properties of CagA are already well established. However, genetic and epigenetic alterations caused by this oncoprotein provide a favorable environment for carcinogenesis, independently of its presence, in a hit-and-run carcinogenesis mechanism. In this sense, through interaction with various host proteins, CagA leads to chromosomal instability, double-strand breaks and repeated nucleotide mutations, which are correlated to gastric cancer development. CMW: western CagA.
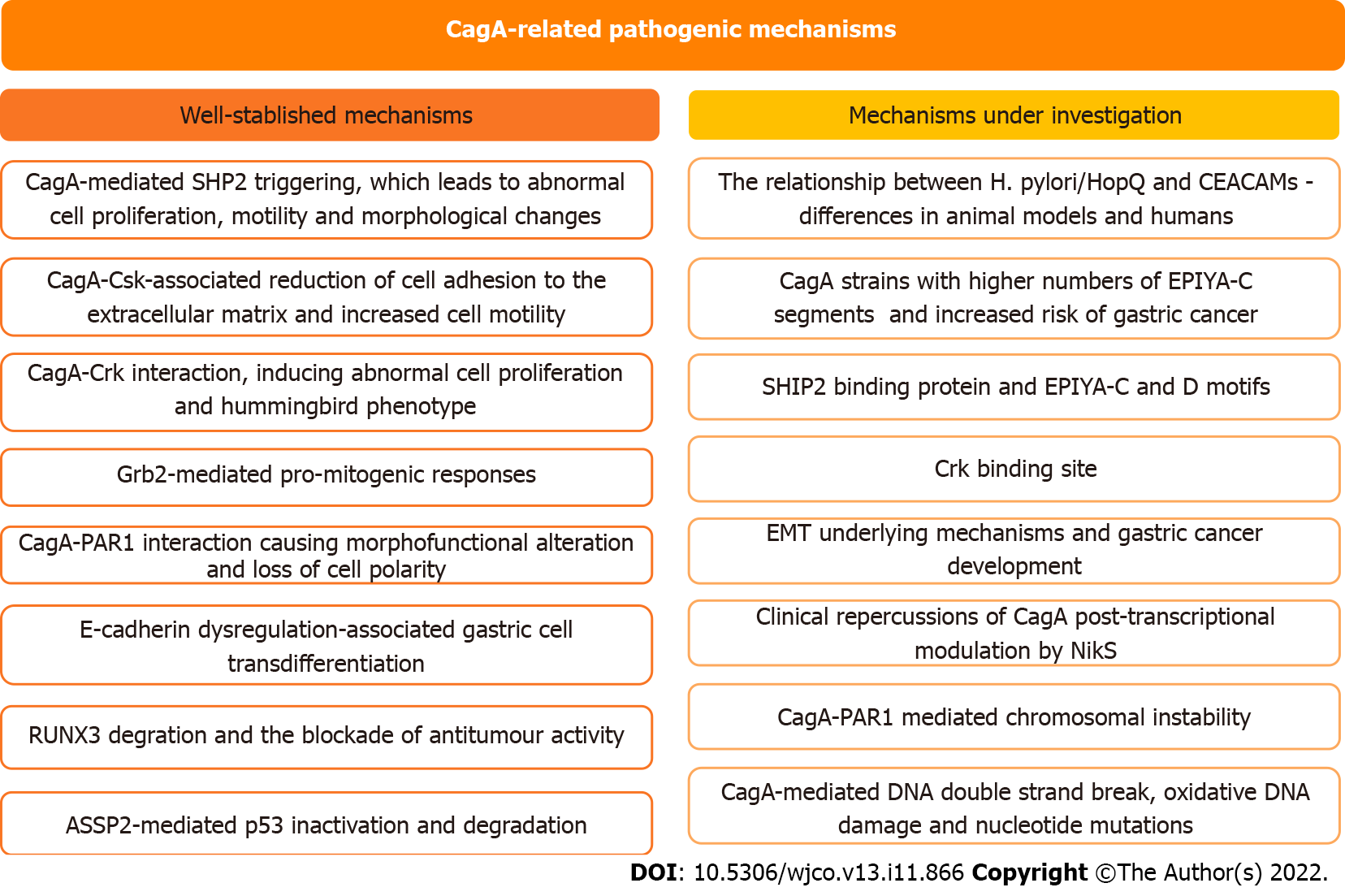
Figure 3 Status of the current understanding regarding CagA-related pathogenic mechanisms.
EMT: Epithelial to mesenchymal transition; CEACAMs: Carcinoembryonic antigen-related cell adhesion molecules; Grb2: Growth factor receptor-bound protein 2; ASPP2: Apoptosis-stimulating protein of p53 2; SHP2: Domain-containing protein tyrosine phosphatase 2; SHIP2: Src homology 2 domain-containing inositol 5'-phosphatase 2; RUNX3: Runt-related transcription factor 3; PAR1: Partitioning-defective 1; NikS: Nickel-regulated sRNA.
- Citation: Freire de Melo F, Marques HS, Rocha Pinheiro SL, Lemos FFB, Silva Luz M, Nayara Teixeira K, Souza CL, Oliveira MV. Influence of Helicobacter pylori oncoprotein CagA in gastric cancer: A critical-reflective analysis. World J Clin Oncol 2022; 13(11): 866-879
- URL: https://www.wjgnet.com/2218-4333/full/v13/i11/866.htm
- DOI: https://dx.doi.org/10.5306/wjco.v13.i11.866