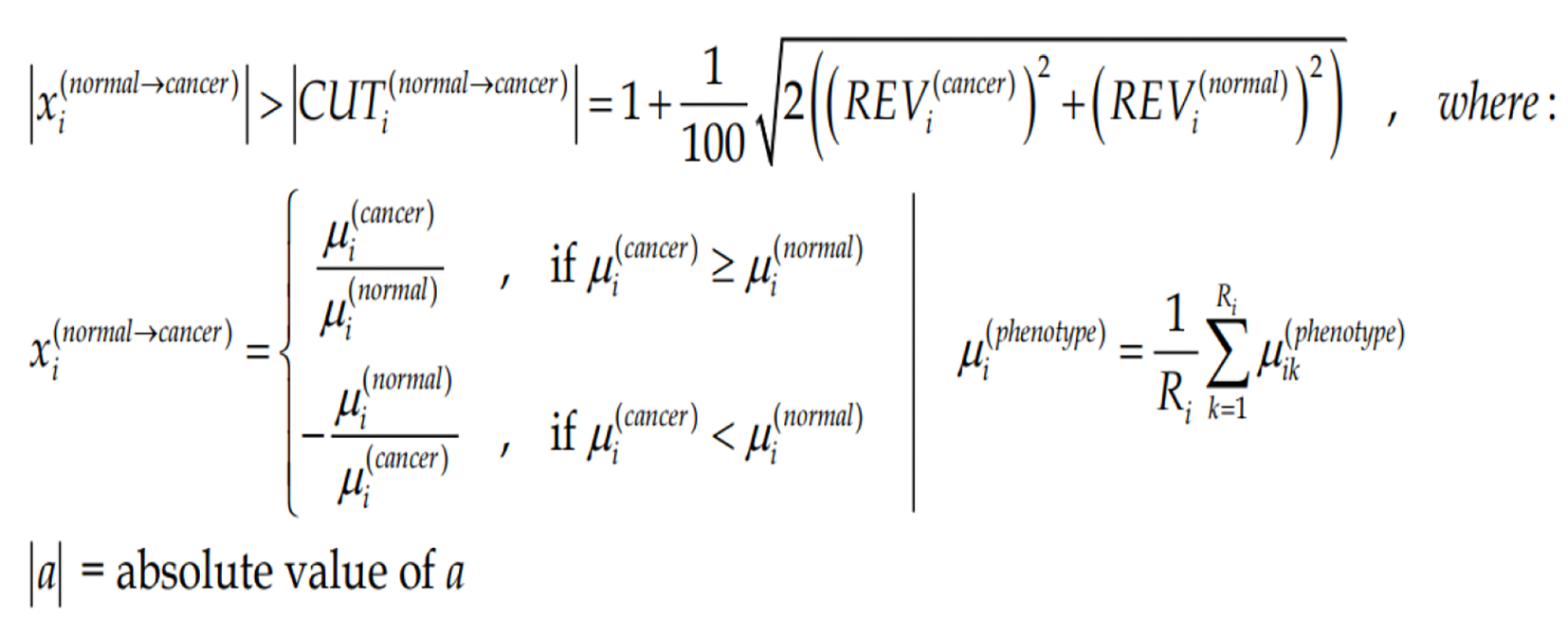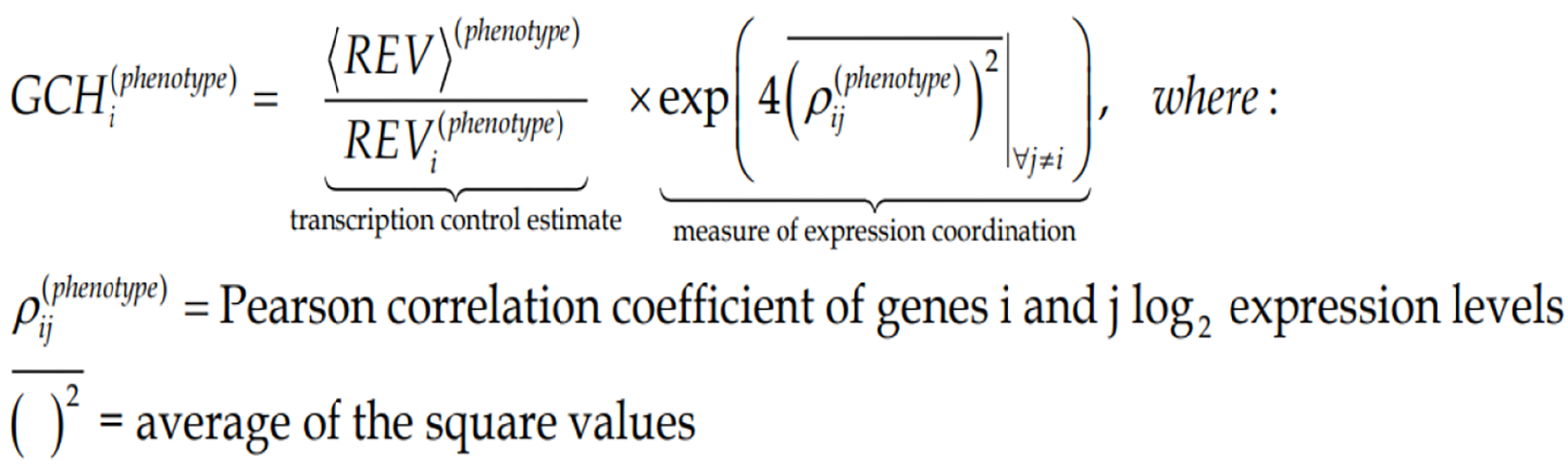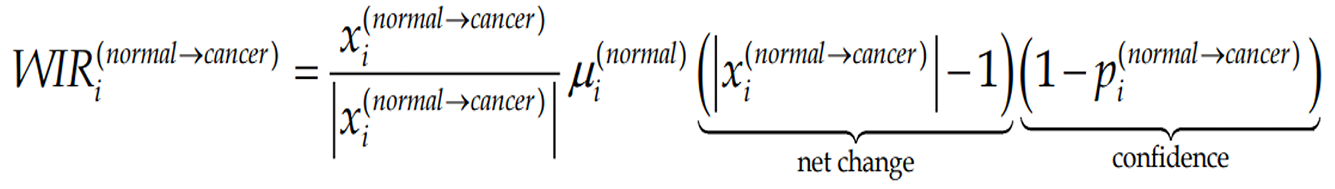Copyright
©The Author(s) 2020.
World J Clin Oncol. Sep 24, 2020; 11(9): 679-704
Published online Sep 24, 2020. doi: 10.5306/wjco.v11.i9.679
Published online Sep 24, 2020. doi: 10.5306/wjco.v11.i9.679
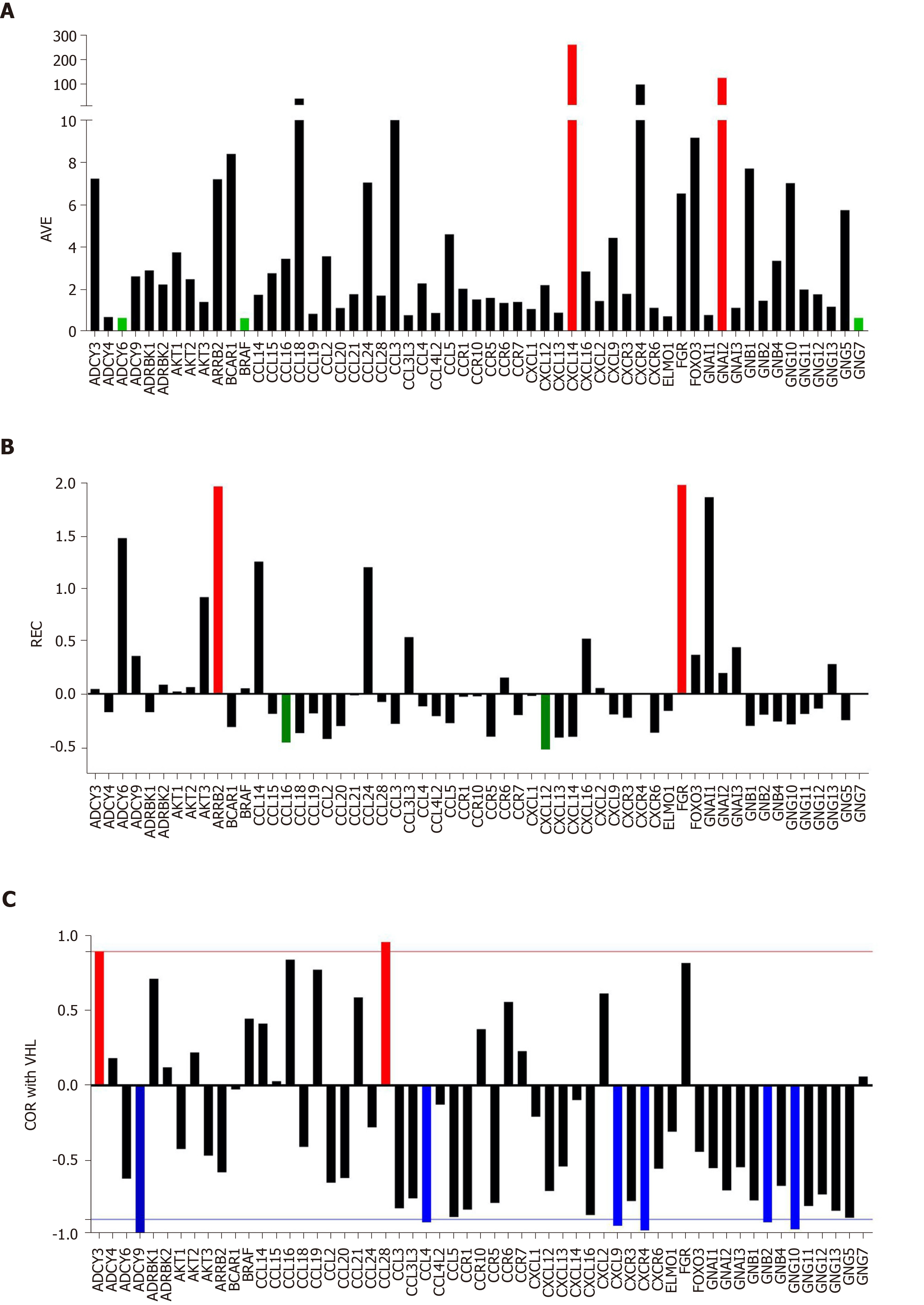
Figure 1 Individual gene quantifiers in the normal kidney tissue (NOR) from a 74-year-old clear cell renal cell carcinoma patient.
A: Average expression levels of the first alphabetically ordered 60 genes involved in the chemokine signaling. Brown columns indicate the most highly expressed genes and the blue ones the least expressed genes; B: Relative expression variability of the first alphabetically ordered 60 genes involved in the chemokine signaling. Brown columns indicate the most controlled genes and the blue ones the least controlled genes; C: Pearson product-moment correlation coefficient (COR) of VHT with the first alphabetically ordered 60 genes involved in the chemokine signaling. Brown columns indicate the significantly synergistically expressed partners of VHL and the blue ones the significantly antagonistically expressed partners. Note the independence of the three features. COR: Pearson product-moment correlation coefficient of VHT; REV: Relative expression variability.
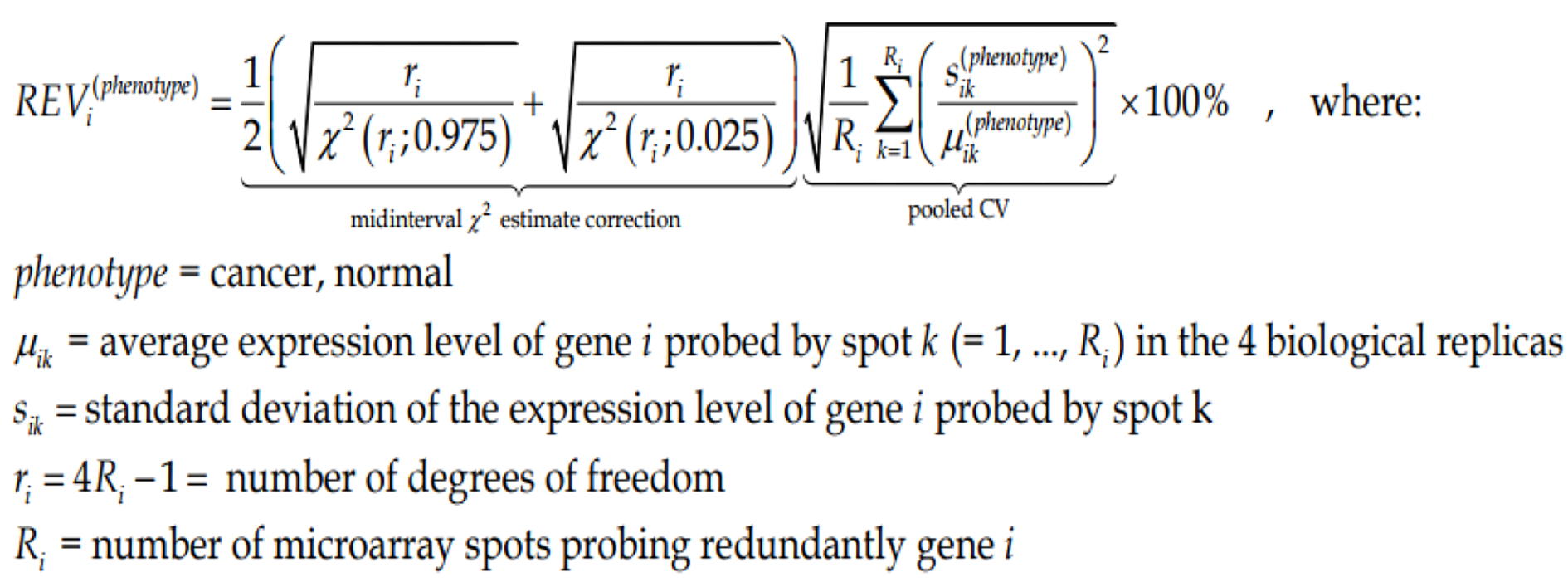
Figure Formula 1
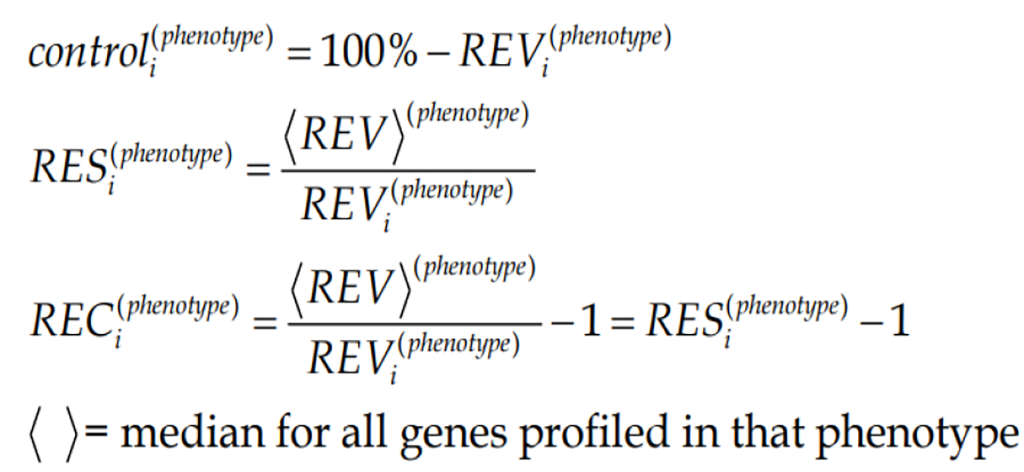
Figure Formula 2
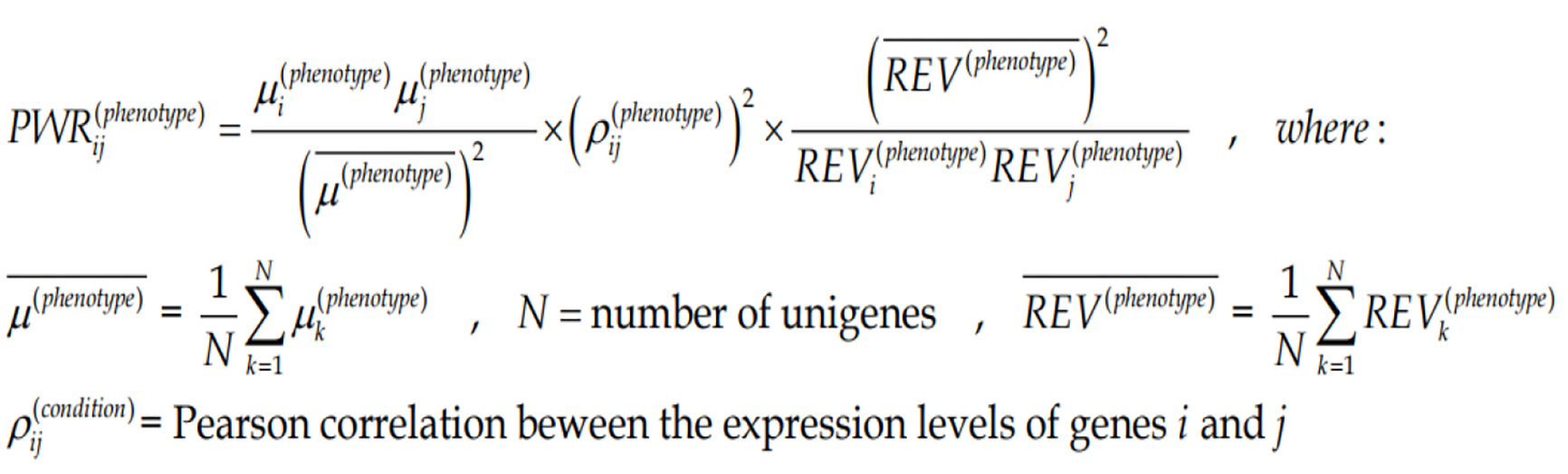
Figure Formula 3
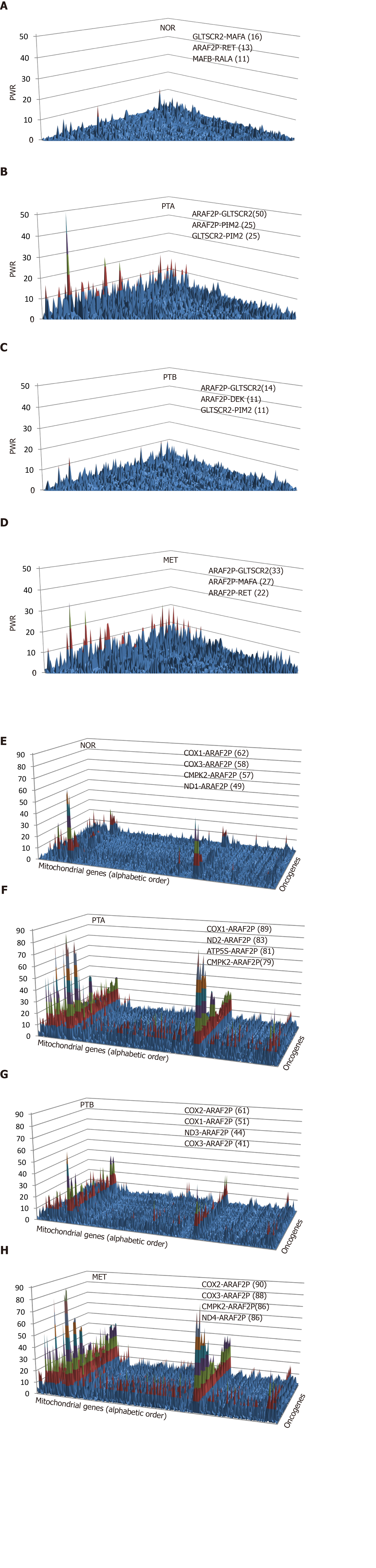
Figure 2 Pair-Wise Relevance Analysis of the oncogenes and interplay of oncogenes with mitochondrial genes in the normal tissue (NOR) from the right kidney, two primary cancer nodules (PTA, PTB) from the right kidney and the metastasis region in the chest wall of a clear cell renal cell carcinoma patient.
A-D: Pair-Wise Relevance Analysis of the oncogenes in the normal tissue (NOR) from the right kidney, two primary cancer nodules (PTA, PTB) from the right kidney and the metastasis region in the chest wall of a clear cell renal cell carcinoma patient; E-H: Interplay of oncogenes with mitochondrial genes in the normal tissue (NOR) from the right kidney, two primary cancer nodules (PTA, PTB) from the right kidney and the metastasis region in the chest wall of a clear cell renal cell carcinoma patient.
Figure Formula 4
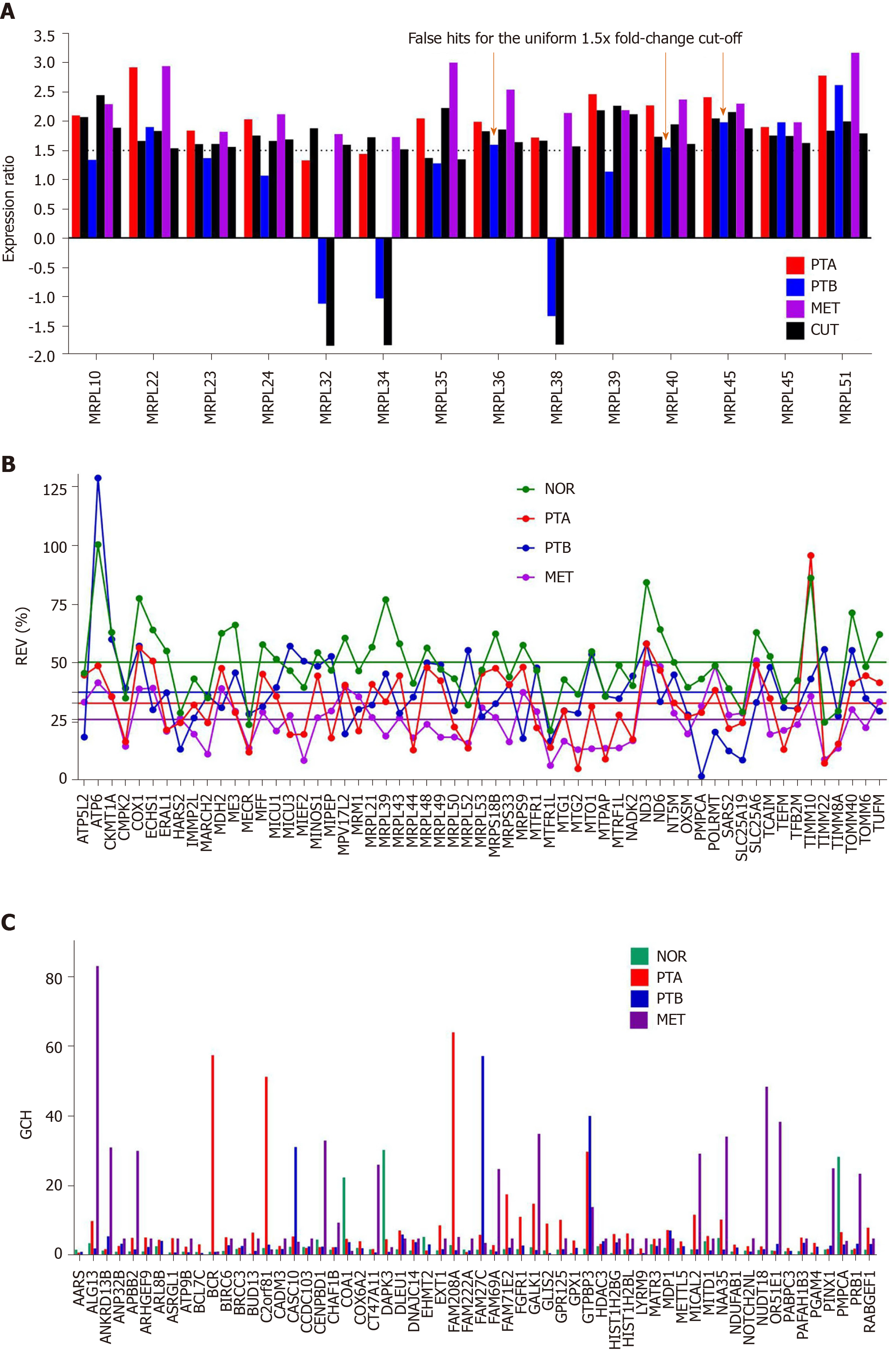
Figure 3 Comparing individual gene quantifiers in the four frozenly preserved specimens of a clear cell renal cell carcinoma 74 years old man.
A: Expression ratios (negative for down-regulation) of 14 MRPL genes in the three cancer samples (PTA, PTB, MET) with respect to the normal tissue (NOR) from the studied clear cell renal cell carcinoma case. The arrows indicate the genes which would be false hits in the standard uniform 1.5× absolute fold-change cut. Note that regulation is higher for most genes in MET than in PTA which at its turn is higher than in PTB; B: Relative expression variability (REV) of 59 randomly selected mitochondrial genes. Note the significantly lower averages for the cancer regions with respect to the control (healthy) tissue; C: GCH of the top 3 genes in each condition plus 48 randomly selected ones. NOR: Normal kidney tissue; PTA: Cancer nodule (primary tumor) A; PTB: Cancer nodule (primary tumor) B; MET: Metastatic chest wall. Colored continues lines are the average REVs in each condition. Note that the GCHs of the same gene are region dependent.
Figure Formula 5
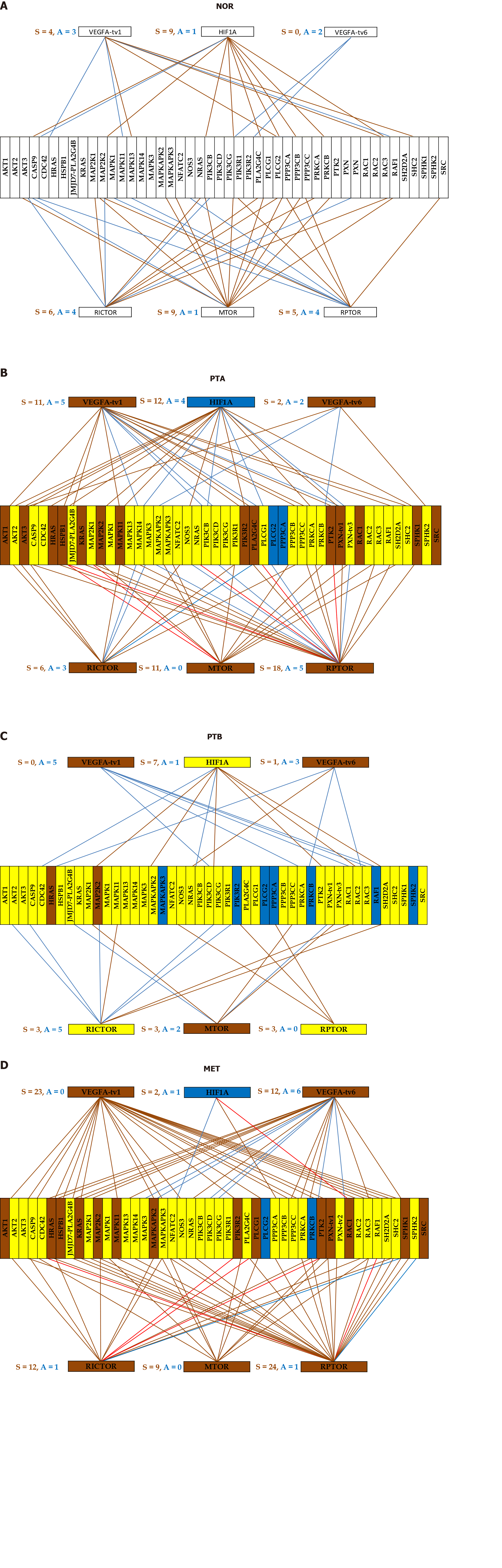
Figure 4 Gene networking depends on phenotype and may include different coordination for alternative transcripts.
A-D: A brown/blue line indicates that the connected genes are (P < 0.05) significantly synergistically/antagonistically expressed. Brown/blue background of the gene symbol denotes significant up-/down regulation, while the yellow background stands for not statistically significant expression change. NOR: Normal kidney tissue; PTA: Cancer nodule (primary tumor) A; PTB: Cancer nodule (primary tumor) B; MET: Metastatic chest wall.
Figure Formula 6
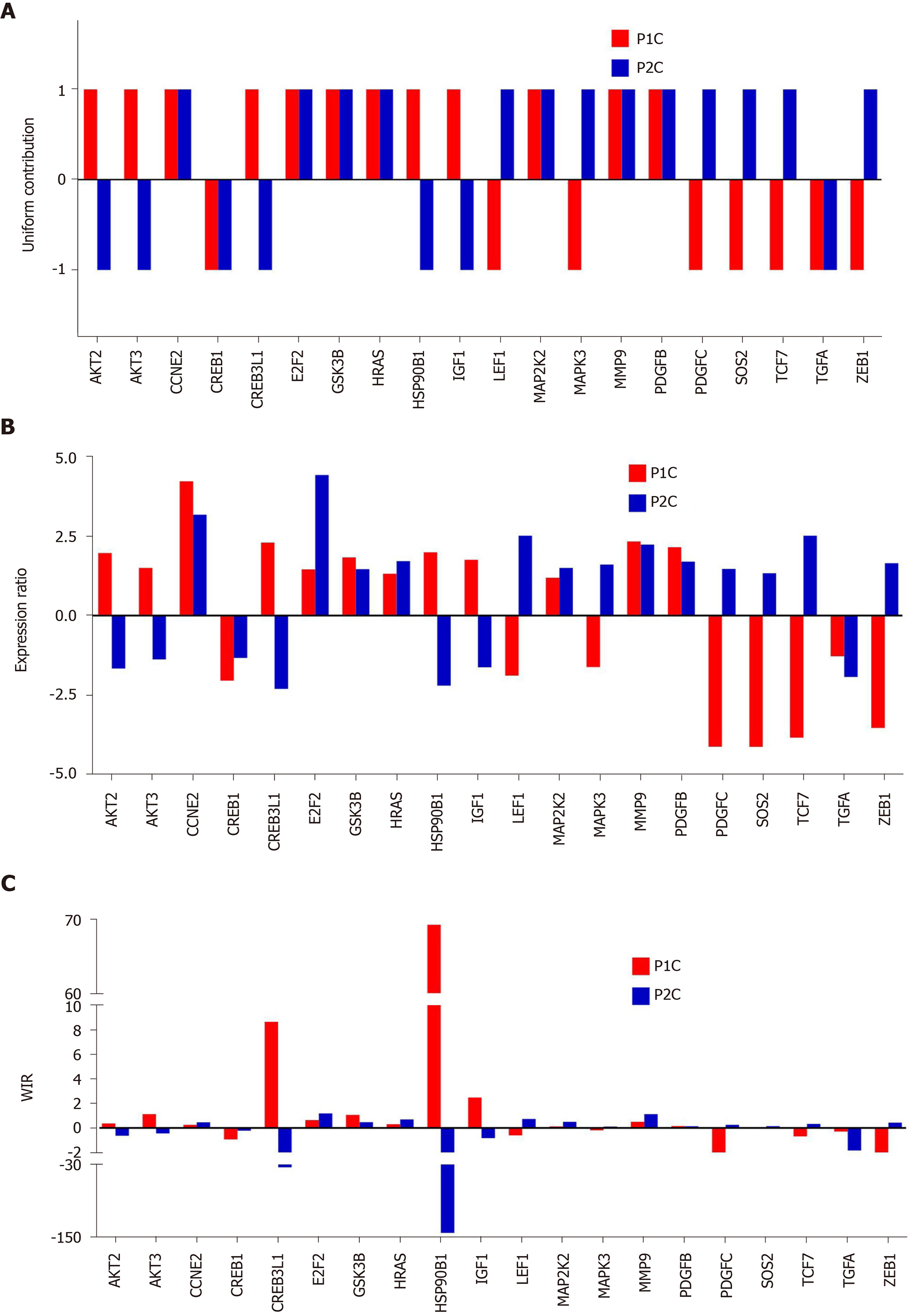
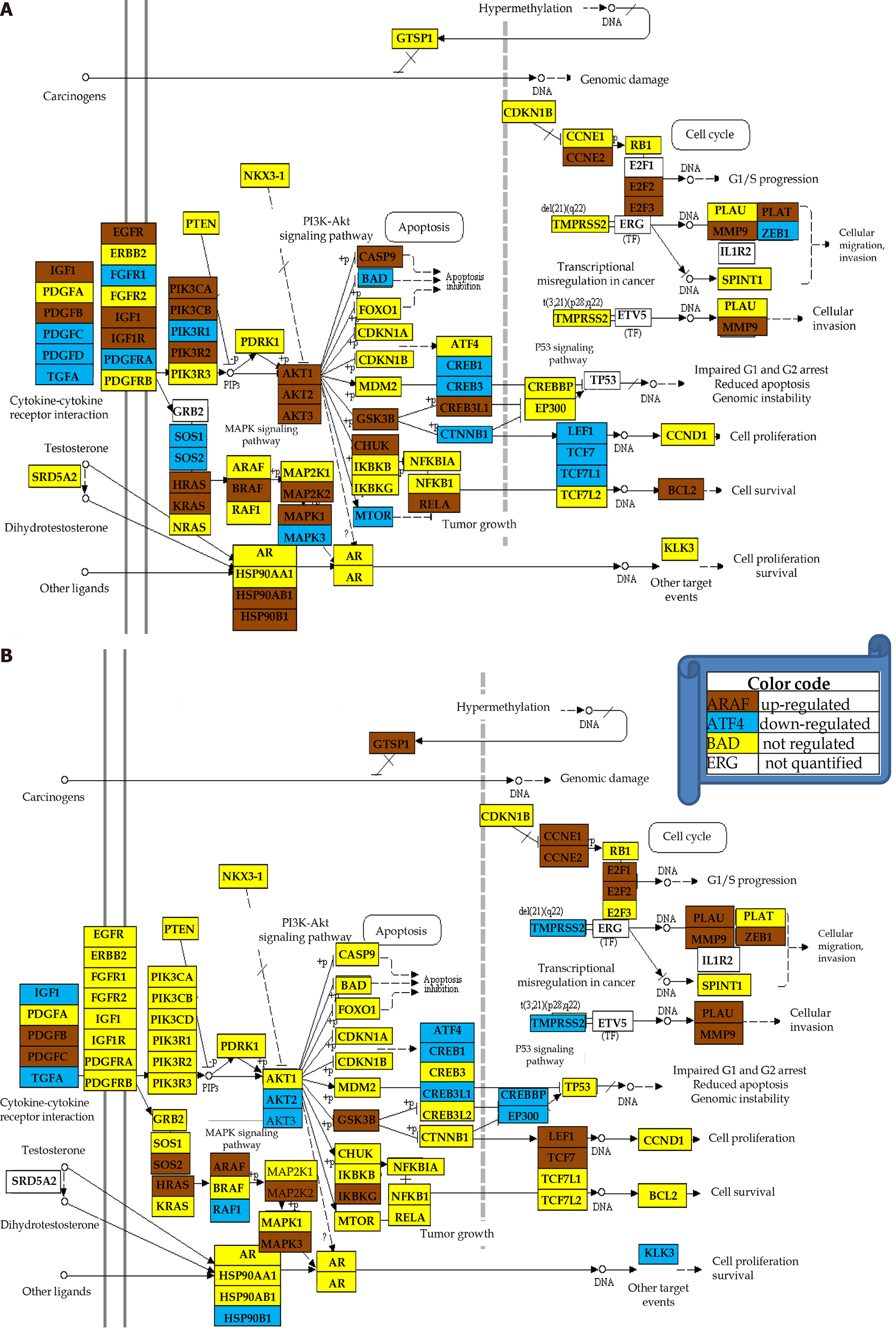
Figure 5 Quantification of expression regulation of prostate cancer pathway genes in two patients.
A: Uniform (unit) contribution; B: Expression ratio; C: Weighted individual (gene) regulations. Only the values of the significantly regulated genes in both patients were plotted. WIR: Weighted individual (gene) regulation.
Figure Formula 7
Figure Formula 8
Figure 6 Regulation of the prostate cancer pathway.
A: Patient 1; B: Patient 2. Note the different regulations of the pathways although the two patients have the same Gleason score.
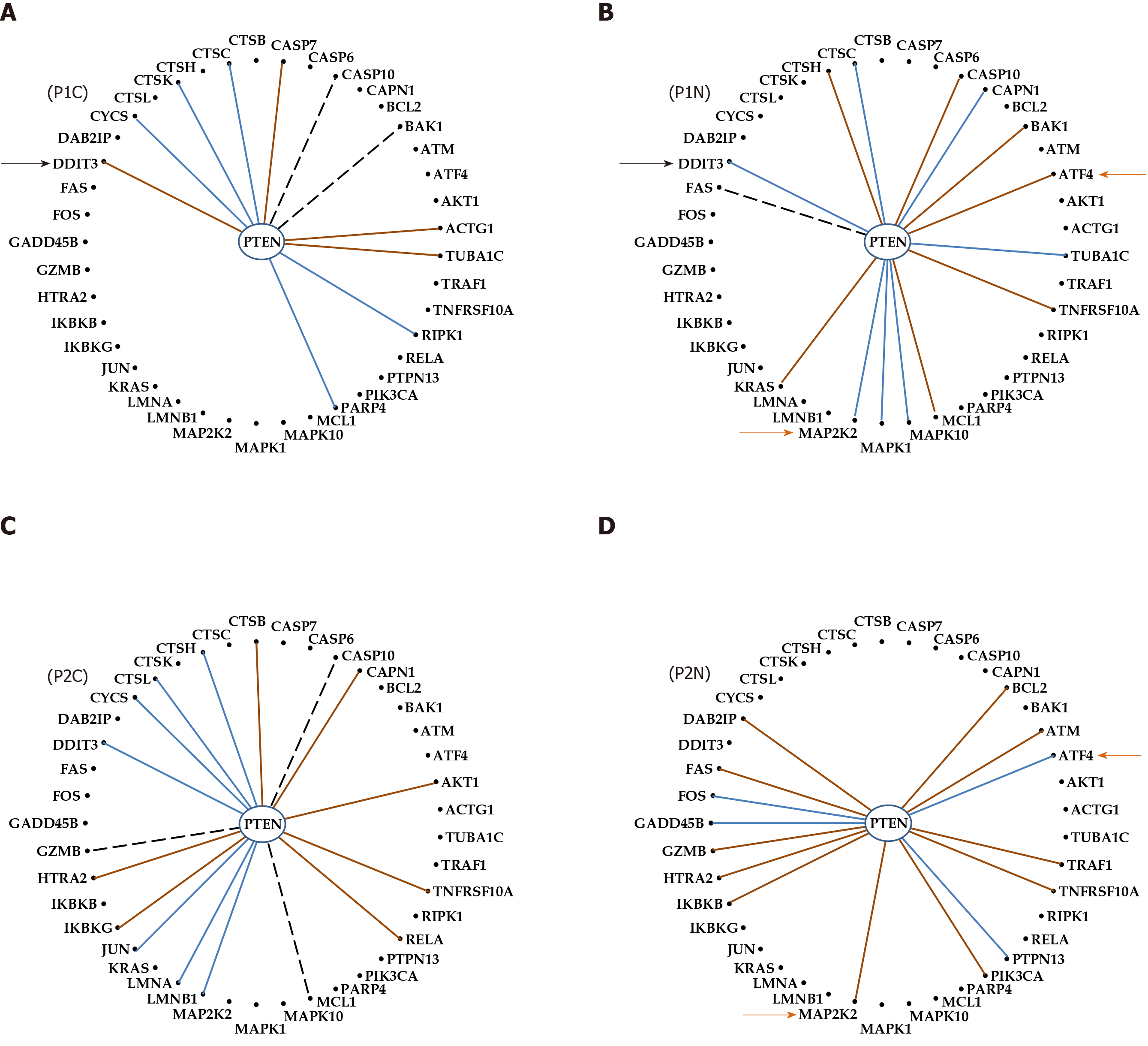
Figure 7 Even with the same phenotype and Gleason score, the genes are networked differently in the prostates of distinct persons.
A: Cancer of patient P1; B: Normal tissue of patient P1; C: Cancer of patient P2; D: Normal tissue of patient P2; Continuous brown/blue lines indicate statistically (P < 0.05) significant synergistic/antagonistic pairing of the connected genes, while dashed black ones that the two genes are independently expressed. Black arrows point to a gene whose coordination with PTEN was switched from negative in the normal tissue to positive in the corresponding cancer nodule. Brown arrows mark the opposite coordination when comparing the normal tissues of the two patients.
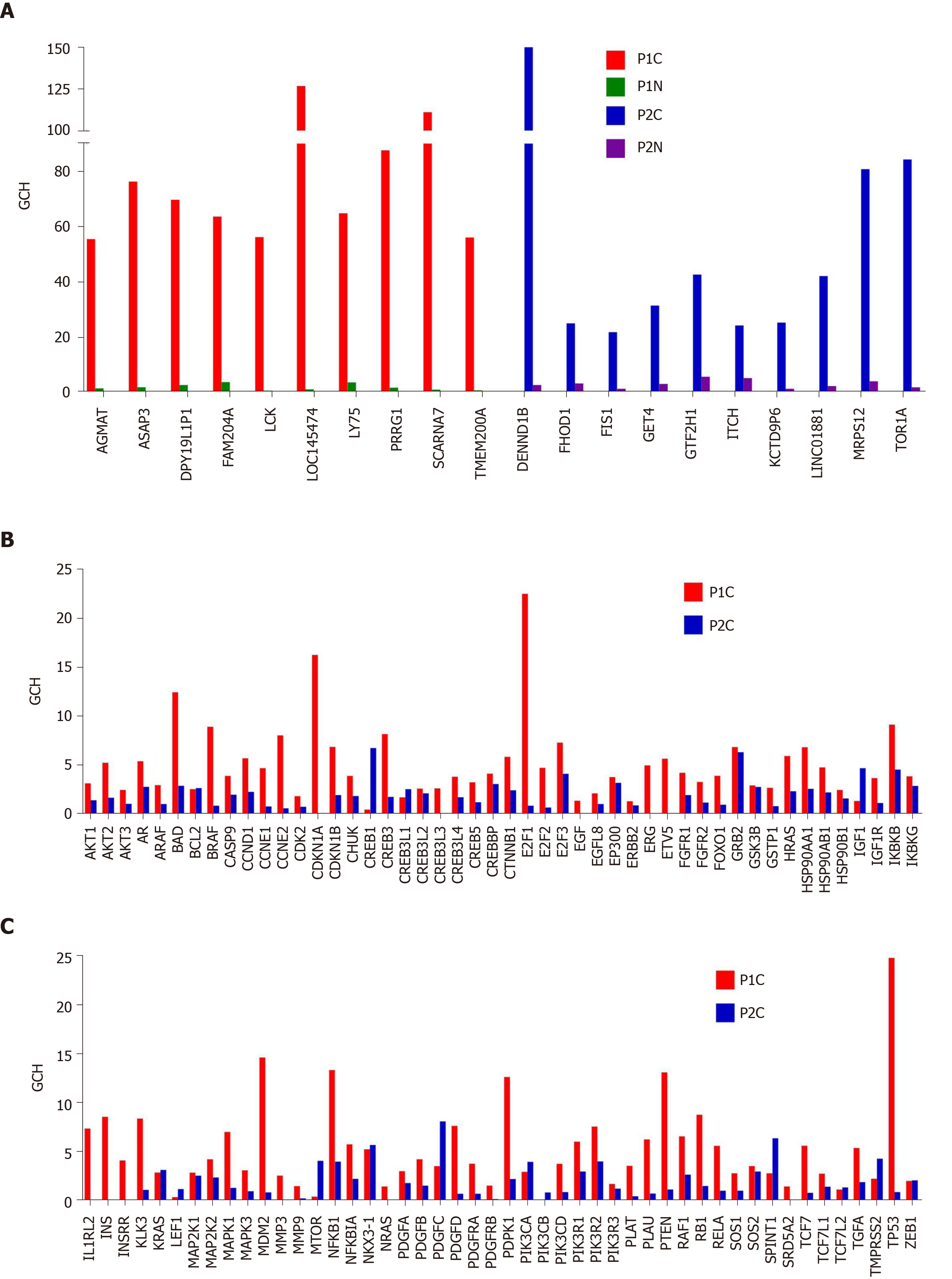
Figure 8 Biomarkers are not the most important genes for the cell phenotype.
A: Top 10 genes in the prostate cancer nodules of the two patients and their scores in the corresponding normal prostate tissues; B and C: Gene commanding height scores of the known prostate cancer biomarkers in the cancer nodules of the two pattients. GCH: Gene commanding height.
- Citation: Iacobas DA. Powerful quantifiers for cancer transcriptomics. World J Clin Oncol 2020; 11(9): 679-704
- URL: https://www.wjgnet.com/2218-4333/full/v11/i9/679.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i9.679