Copyright
©2010 Baishideng Publishing Group Co.
World J Clin Oncol. Nov 10, 2010; 1(1): 12-17
Published online Nov 10, 2010. doi: 10.5306/wjco.v1.i1.12
Published online Nov 10, 2010. doi: 10.5306/wjco.v1.i1.12
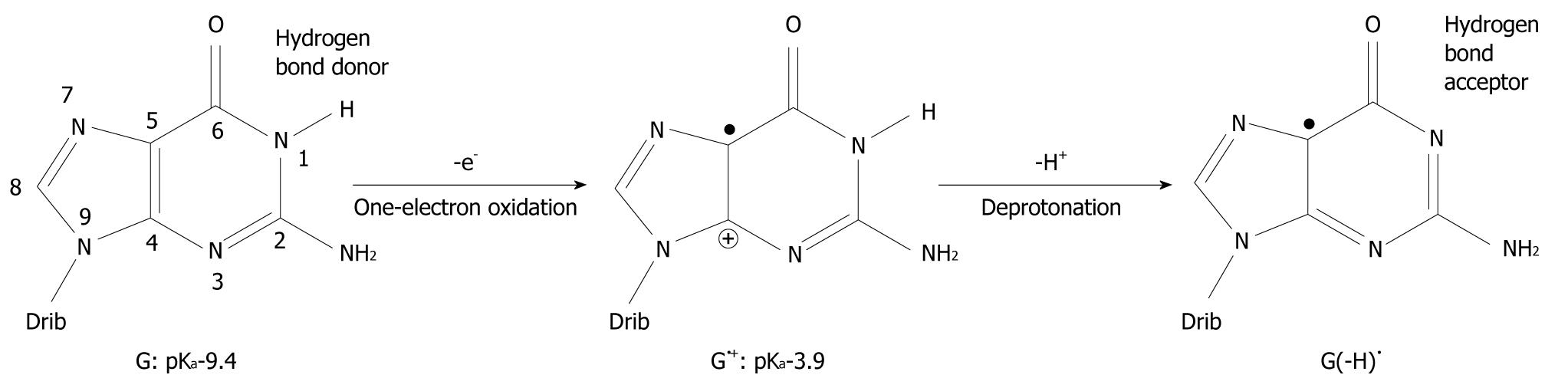
Figure 1 Oxidation of guanine (G) and deprotonation of its radical cation (G•+).
The pKa-value of G is drastically lowered upon one-electron oxidation and subsequent deprotonation of the N1 proton changes it from a hydrogen bond donor to a hydrogen bond acceptor. The number of atoms constituting G is shown. Drib: 2’-deoxyribose moiety.
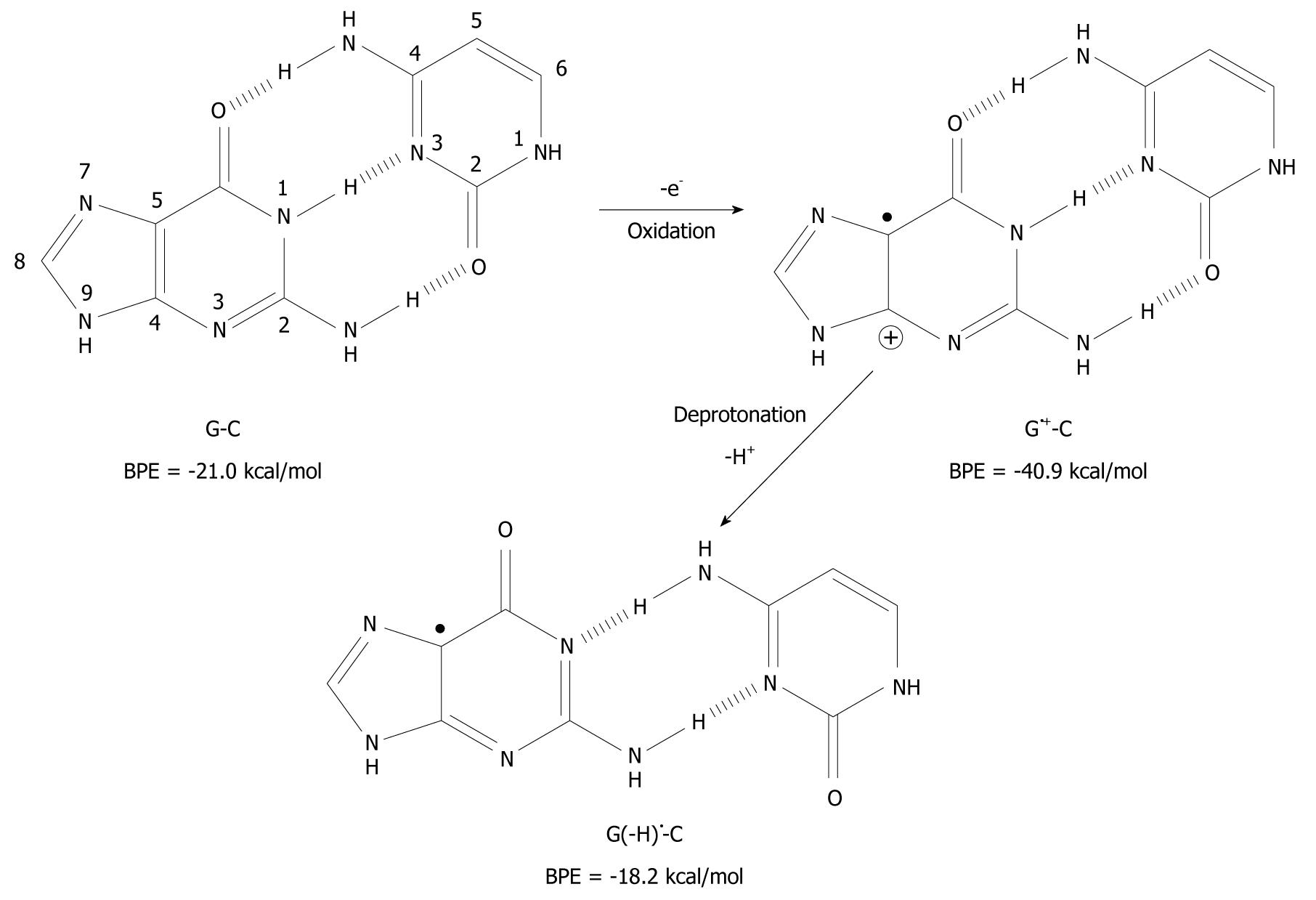
Figure 2 Deprotonation-induced structural change of the G-C base pair initiated by one-electron oxidation leading to the shifted base pair G(-H)• - C.
BPE: Base pairing energy.
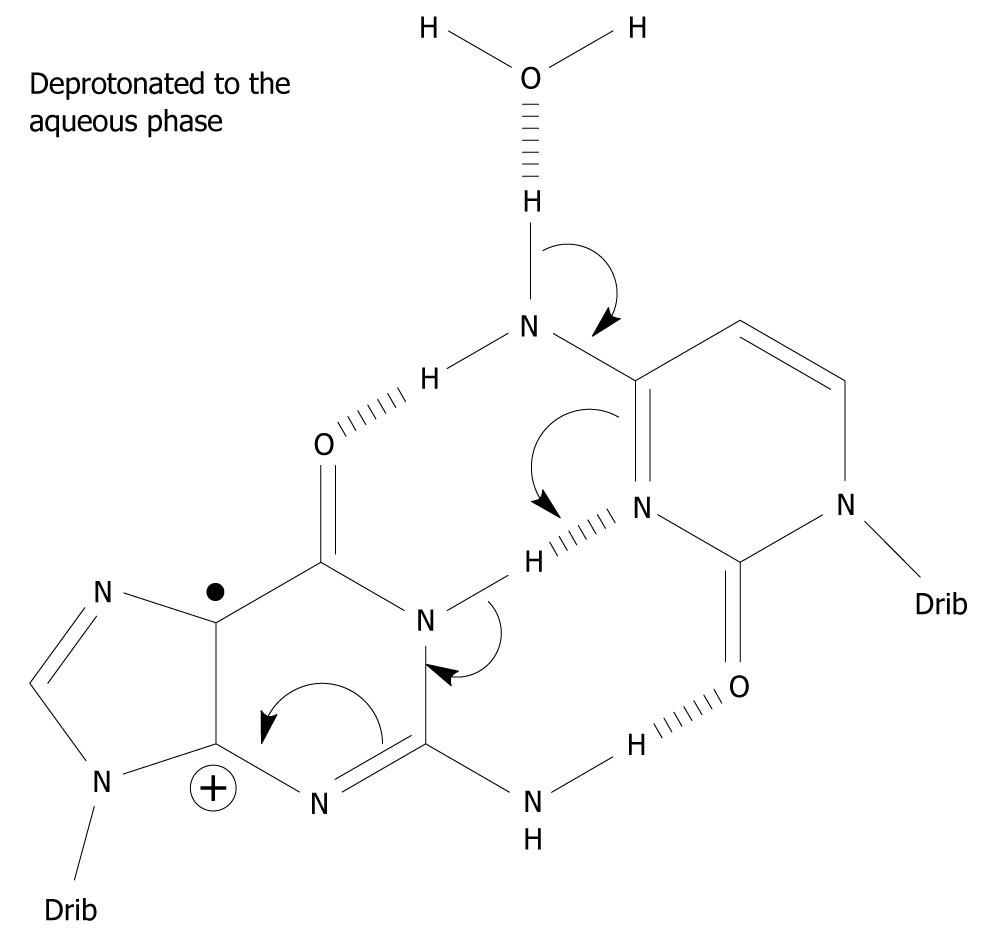
Figure 3 A possible mechanism which involves the exocyclic amine moiety on C as the proton donor of the one-electron oxidized base pair in which the initial charge sits on G, i.
e. in the complementary strand. Spin-charge separation between G and C plays a crucial role in the reaction cascade. The depicted deprotonation can lead to the formation of G(-H)•-C[34].
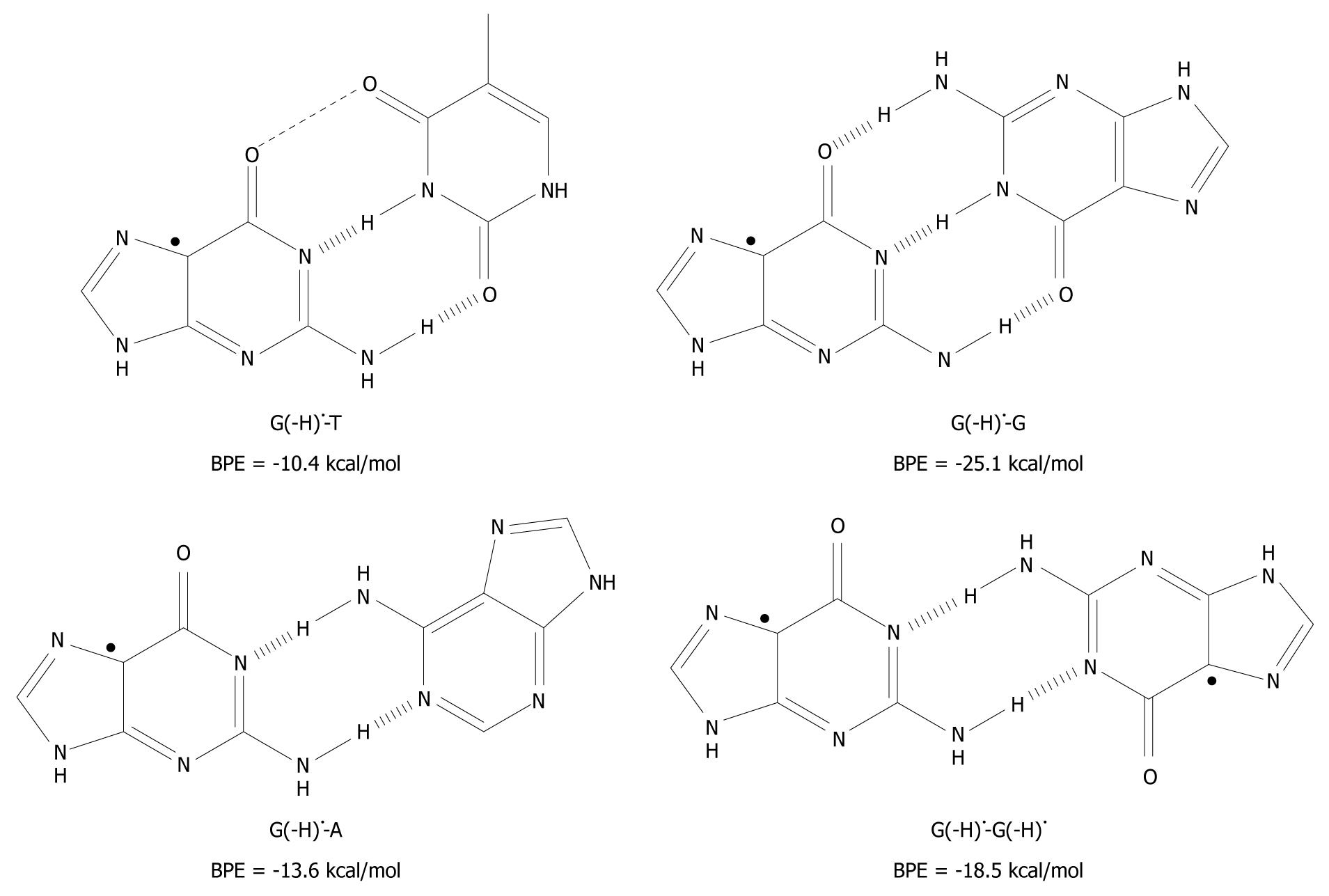
Figure 4 The unnatural base pairs between G(-H)• and the other bases[19].
The substantial base pairing energy (BPE) for the non-classical complexes depicted leads to the conclusion that G(-H)• does not have any specificity for C.
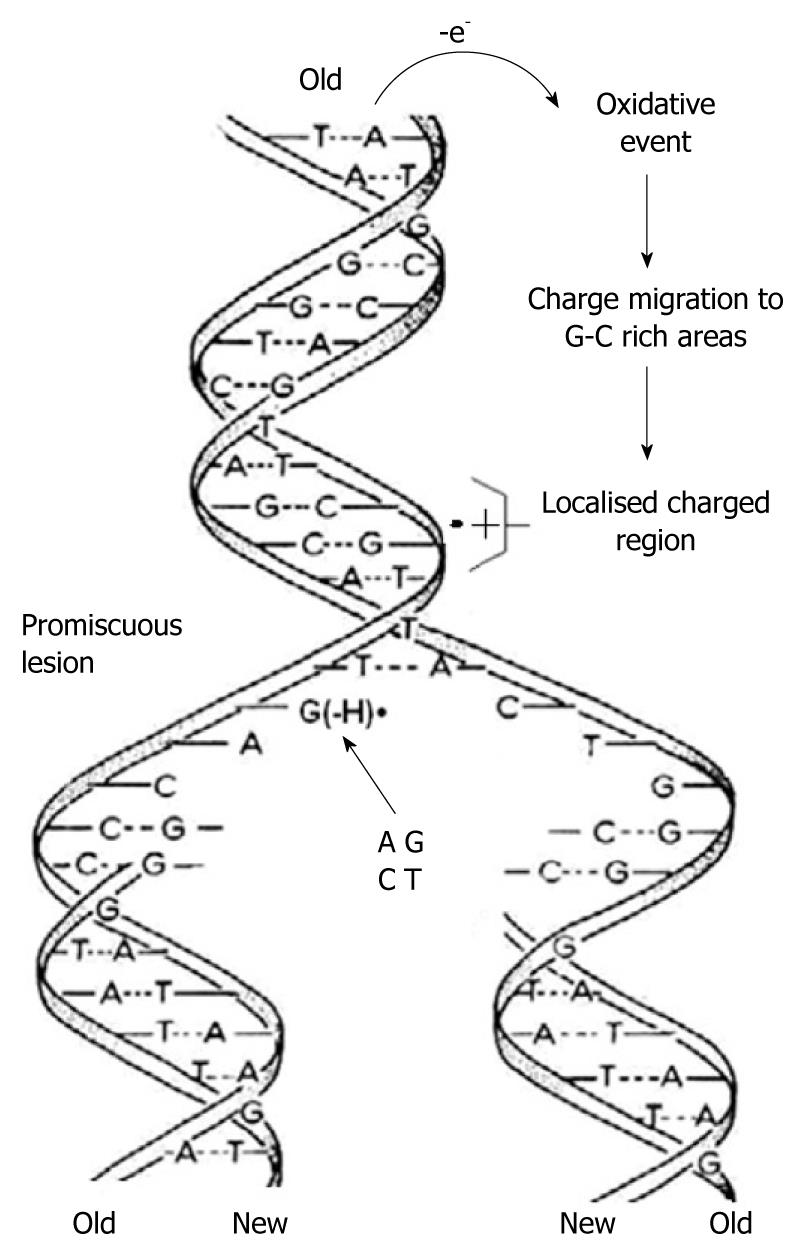
Figure 5 As the two strands of the double helix unwind, each pairs up with the appropriate bases to form a new double helix.
The two new helices are identical to each other and to the original. This process is compromised by one-electron oxidation of the π-DNA stack, deprotonation from G•+ and the subsequent formation of G(-H)•, which is promiscuous with regard to base pairing.
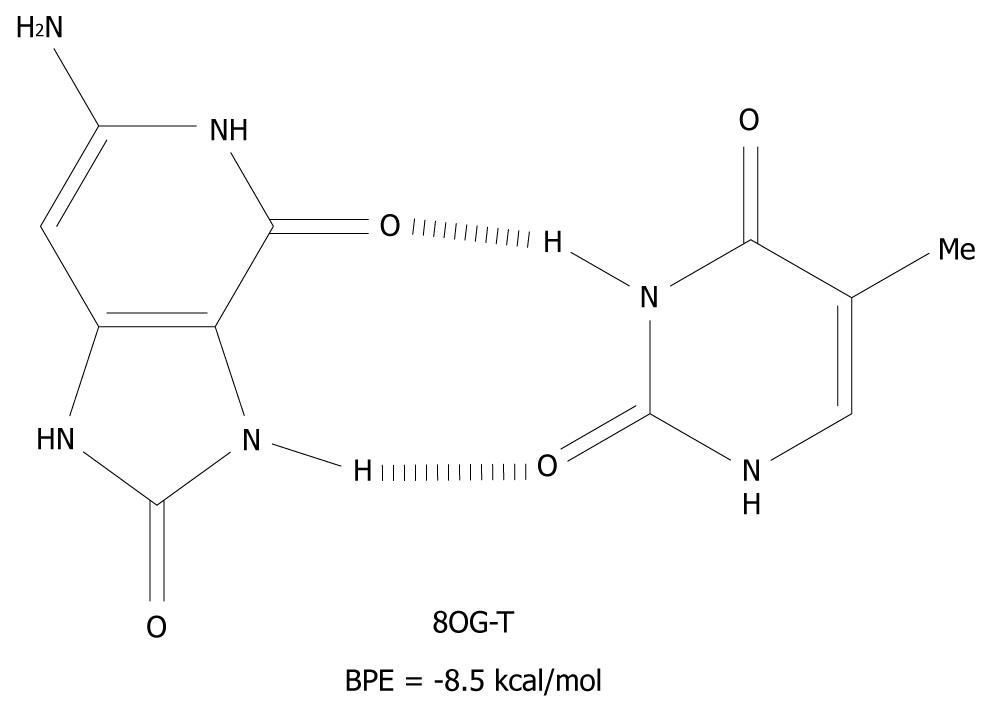
Figure 6 The syn-anti base pair of 8-oxoguanine-T.
BPE: Base pairing energy; 8OG: 8-oxoguanine.
- Citation: Reynisson J. Molecular mechanism of base pairing infidelity during DNA duplication upon one-electron oxidation. World J Clin Oncol 2010; 1(1): 12-17
- URL: https://www.wjgnet.com/2218-4333/full/v1/i1/12.htm
- DOI: https://dx.doi.org/10.5306/wjco.v1.i1.12