Published online Nov 5, 2024. doi: 10.4292/wjgpt.v15.i6.98146
Revised: August 6, 2024
Accepted: September 10, 2024
Published online: November 5, 2024
Processing time: 128 Days and 4.7 Hours
With the rising prevalence of chronic liver diseases worldwide, there exists a need to diversify our artillery to incorporate a plethora of diagnostic and therapeutic methods to combat this disease. Currently, the most common causes of liver disease are non-alcoholic fatty liver disease, hepatitis, and alcoholic liver disease. Some of these chronic diseases have the potential to transform into hepatocellular carcinoma with advancing fibrosis. In this review, we analyse the relationship between the gut and liver and their significance in liver disease. This two-way relationship has interesting effects on each other in liver diseases. The gut microbiota, through its metabolites, influences the metabolism in numerous ways. Careful manipulation of its composition can lead to the discovery of numerous therapeutic potentials that can be applied in the treatment of various liver diseases. Numerous cohort studies with a pan-omics approach are required to understand the association between the gut microbiome and hepatic disease progression through which we can identify effective ways to deal with this issue.
Core Tip: We explore the bidirectional impact of gut-liver interactions on liver disease, highlighting how gut microbiota metabolites affect metabolism. It suggests that altering gut microbiota composition could unveil new treatments for liver ailments. Future cohort studies using pan-omics will be crucial in understanding gut microbiome links to liver disease progression and finding effective interventions.
- Citation: Jeyaraman N, Jeyaraman M, Mariappan T, Muthu S, Ramasubramanian S, Sharma S, Santos GS, da Fonseca LF, Lana JF. Insights of gut-liver axis in hepatic diseases: Mechanisms, clinical implications, and therapeutic potentials. World J Gastrointest Pharmacol Ther 2024; 15(6): 98146
- URL: https://www.wjgnet.com/2150-5349/full/v15/i6/98146.htm
- DOI: https://dx.doi.org/10.4292/wjgpt.v15.i6.98146
The liver and gut microbiota exhibit a complex, bidirectional relationship essential for maintaining metabolic equilibrium. Metabolic byproducts from the gut microbiome are transported to the liver via the portal vein, while the liver contributes to gut health by secreting bile and immunoglobulins into the intestinal tract[1]. This physiological exchange is crucial for sustaining a well-balanced metabolic state[2]. In various hepatic disorders, there is a notable perturbation of this equilibrium, a condition known as dysbiosis. Such imbalance is characterized by a decrease in microbial diversity and proliferation of pathogenic bacteria within the gut[3,4]. This altered microbial landscape is indicative of the significant role that the gut microbiota plays in the pathology of liver diseases. Dysbiosis is influenced by a confluence of genetic factors, environmental exposures, and lifestyle choices, which collectively contribute to the progression of liver diseases.
The mechanisms through which dysbiosis exacerbates liver disease are multifaceted. Primarily, it leads to immune dysregulation, which allows for the unchecked progression of disease. Additionally, alterations in energy utilization occur, and there is an increase in intestinal permeability. This heightened permeability facilitates the translocation of toxic metabolites from the gut into the liver. Once in the liver, these toxic substances trigger a pro-inflammatory response. This inflammatory state not only worsens liver function but also promotes the progression of liver disease, establishing a deleterious cycle that further impairs both liver and gut health[4-7]. Thus, understanding the interplay between the gut microbiota and liver function is critical for identifying potential therapeutic targets aimed at restoring this crucial phy
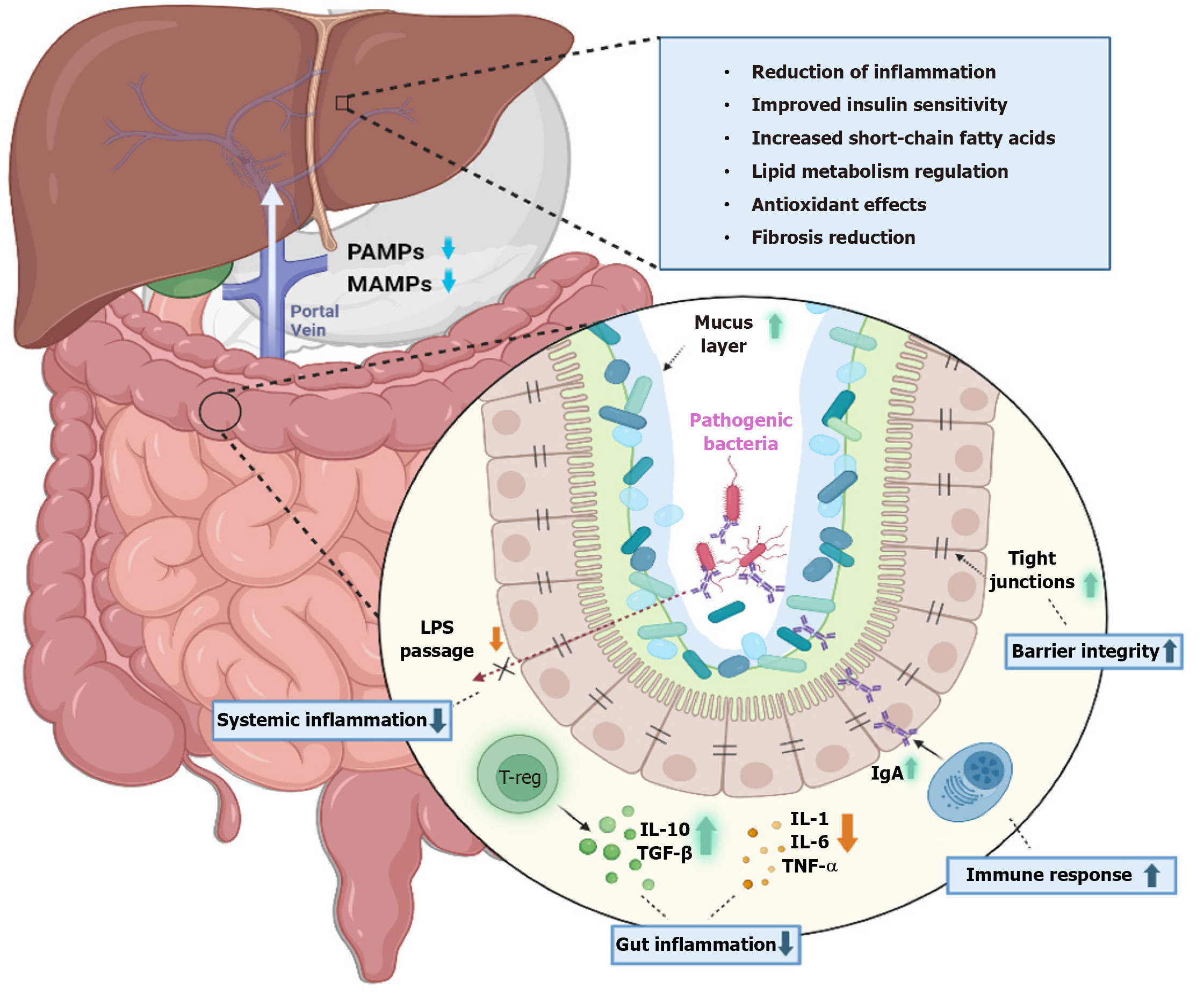
The gut and liver are interlinked majorly through the portal circulation[8-10]. This acts as a medium through which gut metabolites reach the liver. In between there exists a selectively permeable barrier through which nutrients and essential microbial products are translocated. It also acts as a barrier to harmful bacterial products and microbes. This function is achieved through tight junctions between enterocytes which predominantly consist of desmosomes, claudins, occludins, E-cadherins, and adhesion proteins. The short-chain fatty acids produced by the microbiota by the breakdown of dietary fibres have diverse roles such as energy production for intestinal cells, regulating motility of the gut, immune regulation, absorption of nutrients and anti-inflammatory products, and more importantly, altering carbohydrate and lipid meta
The human gut is host to 2172 taxonomically distinct species, predominantly composed of the phyla Firmicutes, Bacte
In alcoholic individuals, an increase in Proteobacteria, Streptococci, and Enterobacteria, and a decrease in Bacteroi
| Ref. | Population | Disease focus | Key findings | Implications |
| Chen et al[87], 2011 | 36 cirrhosis patients; 24 healthy controls | Cirrhosis | ↑ Proteobacteria; ↑ Fusobacteria; ↑ Enterobacteriacea; ↑ Veillonellacea; ↑ Streptococcaceae; ↓ Bacteroidetes; ↓ Lachnospiraceae | Dysbiosis due to increased Enterobacteriaceae and Streptococcaceae may affect the prognosis of cirrhosis patients |
| Liu et al[88], 2012 | Cirrhosis patients vs healthy controls | Cirrhosis | ↓ Bifidobacterium; ↓ Bacteroidetes; ↑ Proteobacteria; ↑ Fusobacteria; ↑ Enterobacteriaceae; ↑ Enterococcus | On releasing endotoxin by enterobacteriaceae, intestinal permeability is increased |
| Bajaj et al[89], 2012 | 25 cirrhosis patients: 17 with HE and 8 without HE; 10 healthy controls | Cirrhosis | ↑ Bacteroidetes; ↑ Veillonellaceae in HE; ↑ Enterobacteriacea; ↑ Alcaligeneceae; ↑ Porphyromonadacea; ↑ Fusobacteriaceae; ↓ Ruminococcaceae; ↓ Lachnospiraceae | Dysbiosis was found in patients with HE compared to healthy individuals; endotoxemia, impaired cognition, and inflammation in the liver were seen in patients with HE |
| Mutlu et al[26], 2012 | ALD patients vs healthy control | ALD | ↑ Proteobacteria; ↓ Bacteroidetes; ↓ Firmicutes; ↑ Enterobacteriaceae; ↓ Bacteroidetes; ↓ Lactobacillus | Decreased beneficial bacteria and increased intestinal permeability result in systemic endotoxemia |
| Zhang et al[90], 2013 | 26 cirrhosis patients with HE; 25 cirrhosis patients without HE; 26 healthy controls | Cirrhosis | ↑ Streptococcus salivarius in HE; ↑ Streptococcaceae; ↑ Veillonellaceae | Streptococcus salivarius was found in patients with HE due to increased ammonia |
| Wong et al[91], 2013 | NASH patients and healthy controls | NASH | ↓ Firmicutes; ↓ Clostridiales (Faecalibacterium & Anaerosporobacte); ↑ Bacteroidetes (Parabacteroides & Allisonella) | |
| Mouzaki et al[92], 2013 | 33 NAFLD patients; 11 steatosis patients; 22 NASH patients; 17 normal controls | NAFLD; NASH; steatosis | ↑ C. Coccoides in NASH; ↓ Bacteroidetes in NASH | The relationship between Bacteroidetes and liver disease state was independent of increase in BMI |
| Zhu et al[51], 2013 | 22 NASH patients; 25 obese people; 16 healthy controls | NASH | ↑ Bacteroides (Prevotella); ↑ Proteobacteria (Escherichia); ↓ Firmicutes; ↓ Actinobacteria | Increased population of ethanol producing bacteria in patients with NASH contributed to disease progression; increased ethanol-producing bacteria (Escherichia) was due to the use of antibiotics |
| Raman et al[30], 2013 | 30 NAFLD patients; 30 healthy controls | NAFLD | ↑ Proteobacteria; ↑ Firmicutes; ↓ Bacteroidetes | Faecal ester volatile organic compounds could negatively influence the microbiome composition of patients with NAFLD |
| Kakiyama et al[93], 2013 | 47 cirrhosis patients; 14 healthy controls | Cirrhosis | ↑ Staphylococcaeae; ↑ Enterobacteriaceae; ↑ Enterococcaceae; ↓ Lachnospiraceae; ↓ Ruminococcaceae; ↓ Clostridiales XIV; ↓ Blautia | Increased pathogenic bacteria as a result of gut dysbiosis in cirrhotic patients with altered bile acid composition |
| Qin et al[94], 2014 | 98 cirrhosis patients; 83 controls | Cirrhosis | ↑ Proteobacteria; ↑ Veillonella; ↑ Streptococcus; ↓ Bacteroidetes; ↓ Lachnospiraceae; ↓ Ruminococcaceae; ↓ Blautia | Oral commensals were found in the gut of cirrhotic patients |
| Bajaj et al[4,95,96], 2014, 2016, and 2019 | HE patients vs healthy control | HE due to cirrhosis | ↑ Megasphaera; ↑ Enterococcus; ↑ Burkholderia; ↑ Veillonellaceae; ↓ Fecalibacterium; ↓ Blautia; ↓ Roseburia; ↓ Dorea | Increased pathogenic bacteria are linked with poor cognition and inflammation |
| Bajaj et al[97], 2014 | Cirrhosis patients vs healthy controls | Cirrhosis | ↑ Veillonella spp.; ↑ Streptococcus spp.; ↓ Bacteroidetes; ↓ Firmicutes | |
| Grat et al[98], 2016 | 15 HCC patients; 5 patients without HCC; all participants with cirrhosis underwent liver transplantation | HCC | ↑ E. coli; ↑ Enterobacteriaceae; ↑ Enterococcus; ↑ Lactobacillus; ↑ H2O2-producing Lactobacillus species | Increased faecal counts of E. coli were noted in the cirrhotic-HCC group, indicating its association with HCC development |
| Llopis et al[27], 2016 | Severe AH patients vs healthy control | Alcoholic hepatitis | ↑ Bifidobacteria; ↑ Streptococci; ↑ Enterobacteria; ↓ Clostridium leptum; ↓ Faecalibacterium prausnitziithan | Decreased anti-inflammatory bacteria and enhanced intestinal dysbiosis result in gut permeability which facilitates microbiota translocation |
| Chen et al[99], 2016 | 30 cirrhosis patients; 28 healthy controls | Cirrhosis | ↑ Veillonella; ↑ Megasphaera; ↑ Dialister; ↑ Atopobium; ↑ Prevotella; ↑ Firmicutes | Raised oral commensal bacteria were found in duodenal mucosal microbiota of cirrhotic patients |
| Ahluwalia et al[100], 2016 | 87 patients with HE; 40 healthy controls | Cirrhosis | ↑ Enterobacteriaceae; ↓ Lachnospiraceae; ↓ Ruminococcaceae | Specific bacterial families were associated with astrocytic and neuronal MRI changes; gut dysbiosis in cirrhosis was linked with systemic inflammation, elevated ammonia levels, and neuronal dysfunction |
| Yang et al[101], 2017 | ALD patients vs healthy controls | ALD | ↑ Candida; ↓ Epicoccum; ↓ Galactomyces | |
| Dubinkina et al[102], 2017 | ALD patients vs healthy controls | ALD | ↑ Bifidobacterium; ↑ Streptococcus spp; ↑ Lactobacillus spp; ↓ Prevotella; ↓ Paraprevotella; ↓ Alistipes | |
| Chierico et al[29], 2017 | 61 NASH/NAFLD patients; 54 healthy controls | NAFLD; NASH | ↑ Actinobacteria; ↑ Bradyrhizobium; ↑ Anaerococcus; ↑ Peptoniphilus; ↑ P.acnes; ↑ Enterobacteriaceae (Escherichia coli); ↑ Dorea; ↑ Ruminococcus; ↓ Bacteroidetes; ↓ Oscillospira; ↓ Rikenellaceae | Increased microbial diversity in NASH/NAFLD; decreased Bacteroidaceae and Bacteroides were observed in NAFLD and NASH, while they were increased in obese patients compared to controls; increased ethanol-producing bacteria (Enterobacteriaceae) in NAFL/NASH compared to controls |
| Loomba et al[103], 2017 | NAFLD patients and healthy controls | NAFLD | ↑ Escherichia coli; ↑ Bacteriodes vulgatus; ↓ Ruminococcus spp.; ↓ Eubacterium rectale; ↓ Faecalibacterium prausnitzii | |
| Liu et al[104], 2018 | 36 cirrhosis patients; 20 healthy controls | Cirrhosis | ↑ Firmicutes; ↓ Bacteroidetes | Microbial dysbiosis in cirrhotic patients with Child-Pugh scores > 5 led to decreased gut motility |
| Ren et al[105], 2019 | 75 early HCC patients; 40 Liver cirrhosis patients; 75 healthy controls | HCC | ↑ Actinobacteria; ↑ Gemmiger; ↑ Parabacteroides; ↑ Paraprevotella; ↑ Klebsiella; ↑ Haemophilus; ↓ Verrucomicrobia; ↓ Alistipes; ↓ Phascolarctobacterium; ↓ Ruminococcus; ↓ Oscillibacter; ↓ Faecalibacterium; ↓ Clostridium IV; ↓ Coprococcus | Decreased butyrate-producing bacteria and increased LPS-producing bacteria observed in early HCC |
| Ponziani et al[106], 2019 | 21 NAFLD-related cirrhosis patients with HCC; 20 NAFLD related cirrhosis patients without HCC; 20 healthy controls | HCC | ↑ Bacteroides; ↓ Ruminococcaceae; ↑ Bifidobacterium | Increased faecal calprotectin in HCC patients is an indicator of inflammatory state |
| Piñero et al[107], 2019 | 407 cirrhosis patients: 25 with HCC; 25 without HCC; 25 healthy controls | HCC | ↑ Erysipelotrichaceae; ↑ Odoribacter; ↑ Butyricimonas; ↓ Leuconostocaceae; ↓ Fusobacterium; ↓ Lachnospiraceae | Decreased Prevotella in cirrhotic patients with HCC, is associated with activation of several inflammatory pathways |
| Ni et al[108], 2019 | 68 primary HCC patients: (23 Stage I, 13 Stage II, 30 Stage III, 2 Stage IV); 18 healthy controls | HCC | ↑ Dysbiosis index Proteobacteria (Enterobacter, Haemophilus); ↑ Desulfococcus; ↑ Prevotella; ↑ Veillonella; ↓ Cetobacterium | Dysbiosis is seen in patients with primary HCC when compared to healthy controls |
| Liu et al[69], 2019 | 57 HCC patients (35 with HBV related HCC, 22 with non-HBV non-HCV related HCC); 33 healthy controls | HCC | ↑ Bifidobacterium; ↑ Lactobacillus; ↓ Proteobacteria; ↓ Firmicutes | Decreased anti-inflammatory and increased pro-inflammatory bacteria in non-HBC non-HCV related HCC patients are positively correlated with alcohol consumption |
| Schwimmer et al[109], 2019 | 87 NAFLD patients; 37 healthy controls | NAFLD | ↑ Bacteroidetes; ↑ Proteobacteria; ↓ Firmicutes | Decreased α-diversity in NAFLD was associated with differences in bacterial abundance rather than an increase in specific phyla or genus; increased bacterial pro-inflammatory products (LPS) were seen in patients with NAFLD |
| Duarte et al[110], 2019 | NASH patients; healthy controls | NASH | ↑ Bacteroides; ↑ Proteobacteria; ↑ Enterobacteriaceae; ↑ Escherichia; ↓ Firmicutes; ↓ Actinobacteria; ↑ Klebsiella pneumoniae | Increased alcohol-producing bacteria supply a constant source of ROS which results in liver inflammation |
| Kravetz et al[111], 2020 | 44 NAFLD patients; 29 healthy controls | NAFLD | ↓ Bacteroidetes; ↓ Prevotella; ↓ Gemmiger; ↓ Oscillospira | Decreased bacterial diversity in patients with NAFLD is associated with an increase in the rate of inflammation in NAFLD |
| Lang et al[65], 2020 | NAFLD patients and healthy controls | NAFLD | ↓ Virus and bacteriophage diversity; ↑ Escherichia; ↑ Enterobacteria; ↑ Lactobacillus phage | |
| Lang et al[112], 2021 | NAFLD patients and healthy controls | NAFLD | ↑ Gemmiger; ↓ Faecalibacterium; ↓ Bacteroides; ↓ Prevotella | |
| Behary et al[113], 2021 | 32 NAFLD-HCC patients; 28 NAFLD-cirrhosis patients; 30 non-NAFLD controls | HCC | ↑ Proteobacteria; ↑ Enterobacteriaceae; ↑ Bacteroides xylanisolvens; ↑ B. caecimuris; ↑ Ruminococcus gnavus; ↑ Clostridium bolteae; ↑ Veillonella parvula; ↑ Bacteroides caecimuris; ↑ Veillonella parvula; ↑ Clostridium bolteae; ↑ Ruminococcus gnavus; ↓ Oscillospiraceae; ↓ Erysipelotrichaceae; ↓ Eubacteriaceae | Increased B. caecimuris and Veillonella parvula distinguish NAFLD-HCC from NAFLD-cirrhosis and non-NAFLD controls; decreased gut microbial α-diversity and increased SCFAs serum levels in NAFLD-HCC result in immunosuppression |
| Trebicka et al[114], 2021 | Cirrhosis patients vs healthy controls | Cirrhosis | ↑ Enterobacteriaceae; ↑ Alcaligenaceae; ↑ Streptococcaceae; ↑ Veillonellaceae; ↑ Fusobacteriaceae; ↓ Bacteroidetes; ↓ Ruminococcaceae; ↓ Lachnospiraceae | Pathogenic organisms' overgrowth results in accelerated disease progression and endotoxemia which results in reduction of organisms that can produce SCFAs and anti-bacterial peptides |
| Solé et al[115], 2021 | 182 cirrhosis patients | Cirrhosis | ↑ Enterococcus; ↑ Streptococcus in ACLF; ↑ Faecalibacterium; ↑ Ruminococcus; ↑ Eubacterium in decompensated patients | As cirrhosis progressed from compensated to uncompensated to ACLF, there was a marked reduction in metagenomic richness |
As cirrhosis progresses from compensated to uncompensated to acute-on-chronic liver failure, there is a marked reduction in metagenomic richness.
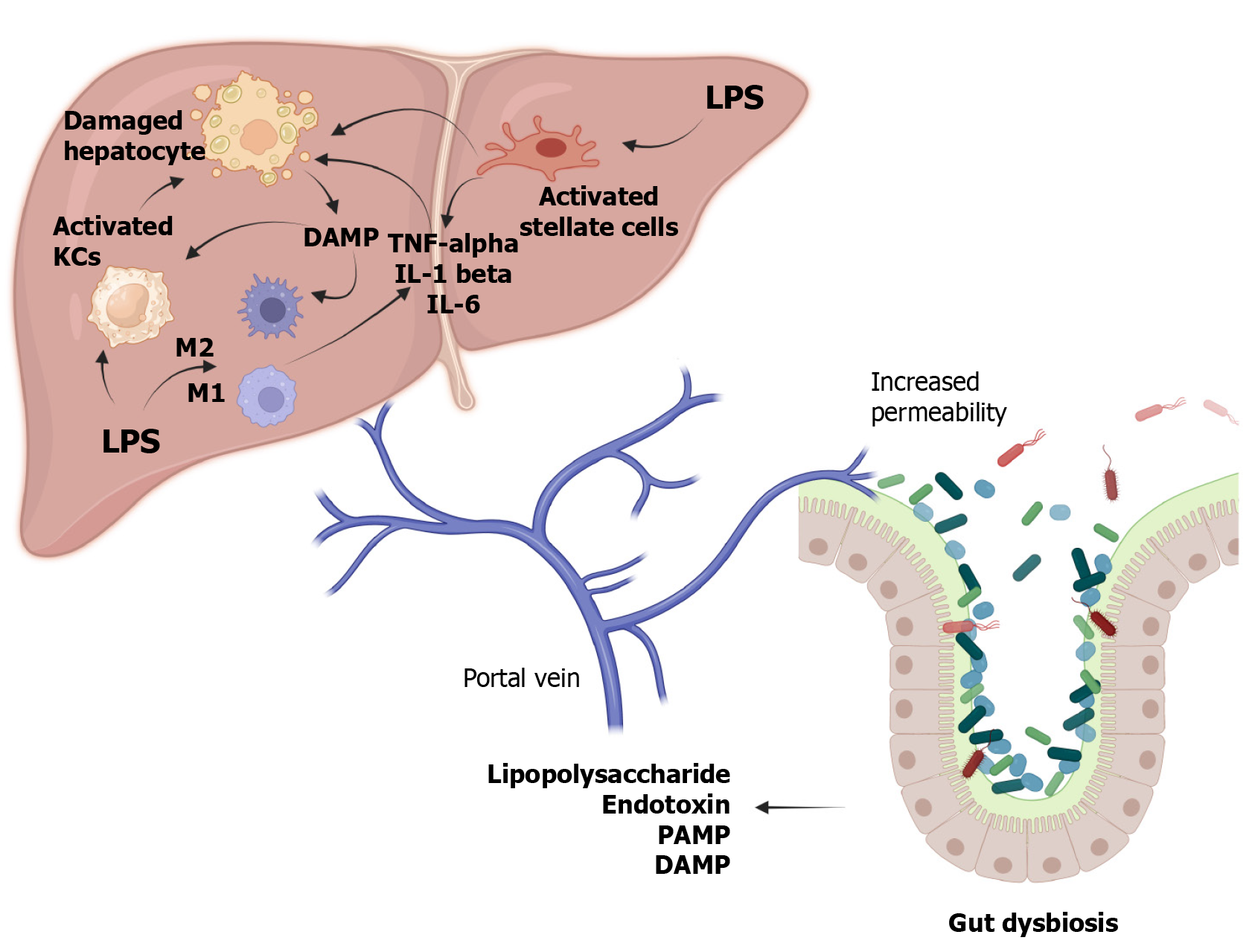
The onset of liver pathology is often precipitated by dysbiosis, which leads to enhanced intestinal permeability. Various factors, including diet, environment, lifestyle, medications, age, and gender, can alter the gut microbiome[34]. This alteration facilitates the release of lipopolysaccharide (LPS), endotoxins, pathogen-associated molecular patterns, damage-associated molecular patterns, and other gut-derived metabolites into the bloodstream. Once in circulation, LPS interacts with Toll-like receptor 4 (TLR4) on endothelial cells, Kupffer cells, and hematopoietic stem cells, and with TLR9 on dendritic cells. Activation of TLR4 also stimulates liver stellate cells, initiating fibrogenesis and the release of pro-inflammatory and profibrotic mediators like TNF-α, IL-1β, and interleukin (IL)-6, along with chemokines such as CCL2, CXCL2, and CXCL10[35,36]. These inflammatory responses and metabolic disruptions elevate serum-free fatty acid and triglyceride levels, leading to their accumulation in the liver and further inflammatory changes[37]. LPS also affects the secretion of adipokines such as adiponectin, IL-6, and leptin, which enhance hepatic inflammation[38,39]. Moreover, LPS reduces adrenergic stimulation, diminishes the protective effects of IL-10, and decreases reactive oxygen species (ROS) production[40-42]. Enhanced TLR signalling in the colonic mucosa also increases the expression of the inflammasome nucleotide-binding oligomerization domain-like receptor family pyrin domain containing 3 in patients with NASH rather than simple steatosis[43].
Another pathway of liver injury involves alterations in bile acid metabolism. Normally, PBAs are converted into secondary bile acids such as lithocholic acid and deoxycholic acid (DCA) through 7α-dehydroxylation by bacteria like Lachnospiraceae and Blautia[9,44]. In chronic liver conditions, the inflammatory mediators released inhibit PBA synthesis via CYP7A1, creating a conducive environment for pathogenic bacteria such as Enterobacteriaceae and Porphyromonadaceae due to the reduced production of antimicrobial agents typically stimulated by PBAs[45]. Alternatively, activation of sterol 27-hydroxylase (CYP27A1) results in the production of chenodeoxycholic acid but not cholic acid (CA). This decrease in CA leads to reduced DCA levels, which otherwise inhibit bacterial overgrowth by displaying potent antimicrobial activity[46-48]. Another study observed a reduction in the secondary to primary bile acids (BAs) ratio and a decrease in total faecal BA concentration in the terminal stages of cirrhosis[49]. These shifts result in diminished FXR activation and increased damage mediated by ROS[50]. Consequently, these changes foster bacterial overgrowth and dysbiosis, perpetuating the vicious cycle of increased permeability, immune dysregulation, metabolic imbalance, and hepatocellular damage. Figure 2 illustrates the complex interactions within the gut-liver axis and the mechanisms of its failure, leading to liver injury.
NAFLD has been linked to gut dysbiosis influenced by dietary and lifestyle factors. Elevated levels of Proteobacteria, Enterobacteriaceae, and Escherichia, which are known alcohol-producing bacteria, have been observed in patients with NASH[51]. These bacteria may cause liver damage by enabling the translocation of toxins through the portal circulation. In cases of NAFLD or NASH, there is an increase in Bacteroidetes, Proteobacteria, and Actinobacteria[24,28,29,52-54]. Conversely, other studies indicate an increase in Firmicutes and a decrease in Bacteroidetes, Proteobacteria, and Actino
Alcohol consumption disrupts the gut microbiota. Research involving ethanol-fed mice showed intestinal cell death, which increases permeability due to the deterioration of tight junctions[56]. A significant rise in endotoxemia was observed in alcoholics, patients with alcoholic hepatitis, and those with cirrhosis compared to the general population[57]. Mutlu et al[26], in 2012, noted a reduction in Bacteroidetes and Firmicutes and an increase in Proteobacteria. Severe alcoholic liver disease is associated with a higher proportion of Streptococci, Bifidobacteria, and Enterobacteria, and a reduction in anti-inflammatory microorganisms like Faecalibacterium prausnitzii[27]. Furthermore, Parasutterella excrementihominis, absent in alcoholic mouse microbiota but present in non-alcoholic ones, suggests a protective role for this bacterium.
Cirrhotic patients exhibit a decline in Lachnospiraceae, Clostridia, Ruminococcaceae [Firmicutes phylum], and Bacte
The LPS-TLR4 axis is implicated in promoting carcinogenesis via activation of stellate and Kupffer cells, chronic inflammation, and fibrosis, though it does not initiate carcinogenesis[64]. Another pathway involves the activation of the nuclear factor-κB pathway through TLR-4, which stimulates the release of inflammatory cytokines such as IL-1B and IL-18[65,66]. LPS can also trigger epithelial-mesenchymal transition[67]. A common alteration in the gut microbiota in hepatocellular carcinoma (HCC) is an increased Firmicutes/Bacteroidetes ratio[68]. There is also an elevation in inflammatory bacteria like Enterococcus, Escherichia, and Shigella, alongside a reduction in Faecalibacterium, Ruminococcus, and Ruminoclostridium in HCC patients[69]. Additionally, Zheng et al[66] found a decrease in butyrate-producing bacteria such as Clostridium, Coprococcus, and Ruminococcus, and an increase in LPS-producing bacteria like Neisseria, Enterobacteriaceae, and Veillonella in patients with HCC and cirrhosis. These microbial shifts could serve as potential biomarkers for HCC.
The pathogenesis of this condition is primarily dependent on the interplay between genetic and environmental factors. It has been found that genetically susceptible individuals has HLA-DRB1 0301 and HLA-DRB1 0401 genotypes which, on interaction with environmental factors such as viruses (cytomegalovirus, hepatitis A, B, C, and E viruses, and Ebstein-Barr virus) or drugs (minocycline), leads to a dysregulated pro-inflammatory response where the antigen presenting cells set off a cascade of events where helper T cells get activated. Activated helper T cells release a stream of cytokines which in turn activate cytotoxic T cells to release a group of cytokines resulting in an antibody mediated cell toxicity, eventually leading to hepatocellular injury.
Another mechanism in the development of autoimmune hepatitis is through molecular mimicry where the antibodies directed against environmental antigens become self-directed to self-antigens due to similarities of environmental antigens with self-antigens as per Floreani et al[67]. In individuals with AIH, there were significant reductions in species of Bifidobacterium and Lactobacillus, which resulted in increased gut permeability and enhanced translocation of bacteria indicated by increased lipopolysaccharide levels that were positively correlated with the disease severity as per Lin et al[70].
These pathologies show an increased association between disease progression and dysbiosis. A decrease in Bacteroides, Lactobacillus, Bifidobacterium and an increase in Enterococcus and Enterobacteriaceae, which resulted in altered gut microbiome, were seen in chronic hepatitis B. With limited studies on hepatitis C and gut dysbiosis, it was found that there was a reduction in alpha-diversity and altered gut microbiome. One of the reasons for altered microbiome is that a reduction in bile production leads to an increase in pathogenic species in the gut[71-73].
Another interesting correlation was observed between primary sclerosing cholangitis (PSC) and gut dysbiosis. According to Bajer et al[74], in patients with PSC and PSC-IBD there was an increase in Veillonella, Enterococcus, Clostridium, Streptococcus, Rothia, and Hemophilus and a decrease in Coprococcus. This observation is explained by the fact that the pro-inflammatory state set by PSC leads to increased gut permeability and the products of bacteria such as SCFAs and bile acids leads to disease progression[74].
In primary biliary cholangitis (PBC), a study by Lv et al[75] stated that there was an increase in Veillonella, Bifidobacterium, Neisseria, and Klebsiella and a decrease in Ruminococcus, Bacteroides eggerthii, and Hallella. But further studies are required for establishing treatments that alter the gut microbiome in patients with PBC and PSC/PSC-IBD.
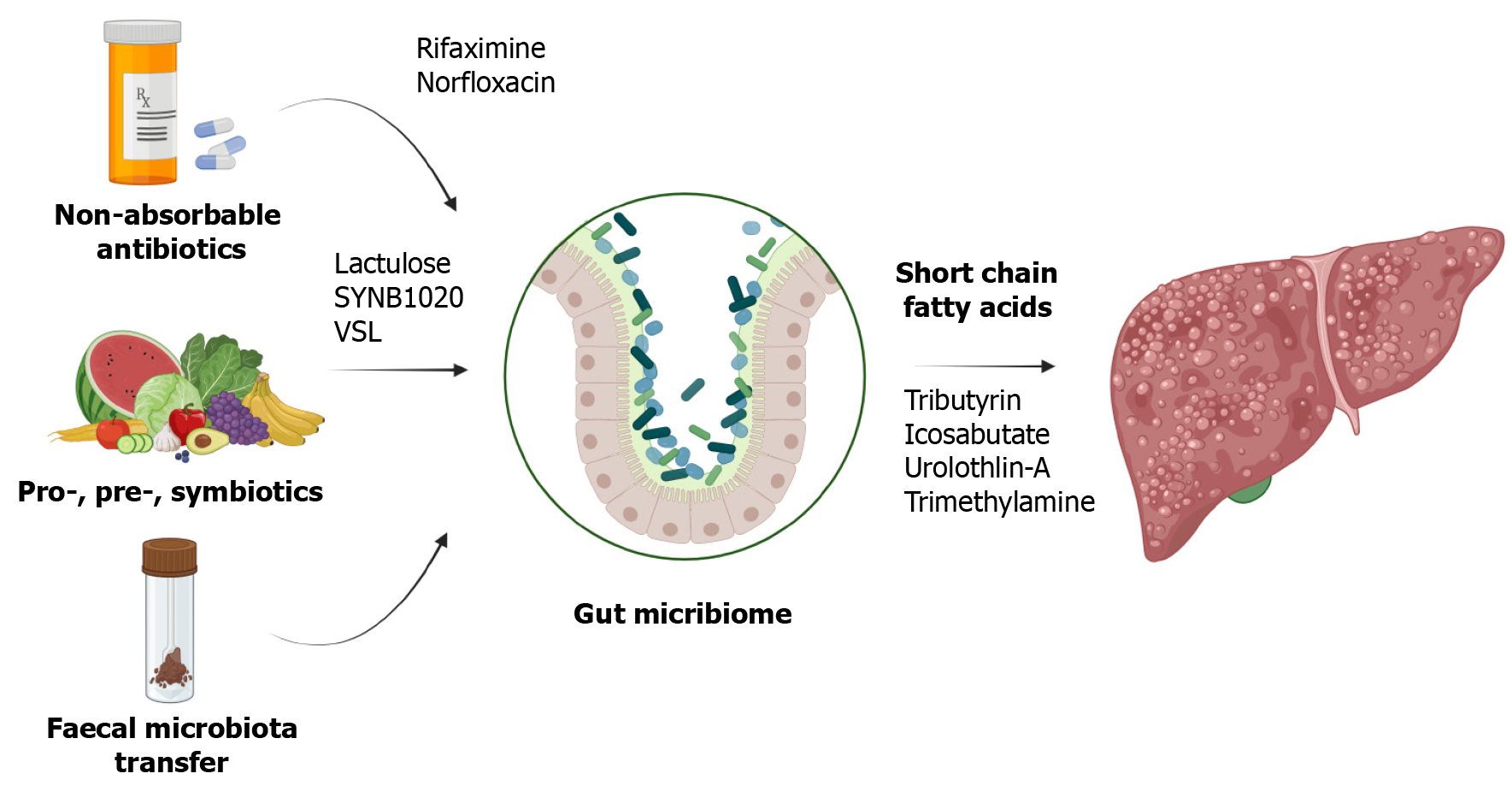
Targeted changes in human microbiota are achieved through probiotics, prebiotics, and synbiotics as shown in Figure 3.
Probiotics, which are live organisms, are administered as supplements to supplant pathogenic bacteria. Research has demonstrated that a mixture of Lactobacillus, Streptococcus thermophilus, and Bifidobacteria ameliorated steatosis in mice induced by a high-fat diet[76]. Lactobacillus GG was shown to mitigate intestinal oxidative stress, leakage, and liver damage in rat models of alcoholic steatohepatitis[77]. Additionally, combinations of probiotics have been effective in slowing the progression of HCC in mice by reducing TH17 cells[78]. Akkermansia muciniphila has been noted to strengthen tight junctions and maintain intestinal permeability in alcoholic steatohepatitis models[79]. There is also evidence suggesting that probiotic therapy can enhance the efficacy of immunotherapy in cancer patients[80].
Studies have shown that pectin can modify the intestinal microbiota in mice, prevent steatosis, and decrease inflammation. Common prebiotics include oligosaccharides, polyunsaturated fatty acids, and polyphenols[81]. A meta-analysis involving 1309 patients with NAFLD reported significant reductions in body mass index, liver enzymes, serum chole
The combined use of prebiotics and probiotics, known as synbiotics, has shown enhanced benefits. According to Hadi et al[83], synbiotic consumption led to improvements in lipid profiles and metabolic hepatic steatosis. Malaguarnera et al[84] found that after 6 mo of administering Bifidobacterium longum and Fructo-oligosaccharide to 66 patients with NASH, there were significant reductions in serum AST, LPS, inflammatory mediators, fat denaturation, and the NASH activity index.
Faecal microbiota transplantation (FMT) aims to replace the intestinal flora with a healthier one, improving gut permeability and reducing endotoxemia and inflammatory molecules through the increased production of anti-microbial peptides. In a study by Ferrere et al[85], fecal bacteria from alcohol-resistant mice were transplanted to alcohol-sensitive receptor mice, effectively preventing alcohol-induced intestinal disorders and fatty liver hepatitis. This treatment altered the bacterial composition, decreasing Bacteroides and increasing Actinobacteria and Firmicutes. In a recent randomized, double-blind trial, patients with alcohol-related liver disease and cirrhosis received FMT from a donor with a Lachnospiraceae and Ruminococcaceae rich microbiota. Results from this trial showed reductions in IL-6 and LPS-binding protein levels and an increase in butyrate/isobutyrate levels on day 15 in the FMT group, in contrast to the control and placebo groups[86](Table 2).
| Interventions | Mechanism of action | Targeted disease | Clinical outcomes | Ref. |
| Prebiotics (pectin) | Restore Bacteroides level | Alcoholic liver disease | Control dysbiosis | Ferrere et al[85], 2017 |
| Prebiotics (Fructo-oligosaccharide) | Promote fatty acid oxidation | NAFLD | Reduced hepatocyte damage and inflammation | Matsumoto et al[116], 2017 |
| Probiotics (E. coli Nissle strain) | ↑ Lactobacillus species; ↑ Bifidobacterium species; ↓ Proteus hauseri; ↓ Citrobacter species; ↓ Morganella species | Cirrhosis (humans) | Significant improvement in gut microbiome with decreased endotoxemia, bilirubin, and ascites | Lata et al[117], 2007 |
| Probiotics (Lactobacillus reuteri GMNL-263) | ↑ Bifidobacteria; ↑ Lactobacilli; ↓ Clostridia | Hepatic steatosis (rats) | ↓ Blood glucose levels, TNF-α and IL-6 production by adipose tissue | Hsieh et al[118], 2013 |
| Probiotics | ↑ Parabacteroide; ↑ Allisonella; ↓ Faecalibacterium; ↓ Anaerosporobacter | NASH (humans) | ↑ Bacteroidetes ↓ Firmicutes | Wong et al[91], 2013 |
| Probiotic: VSL#3 (8 probiotic mixture) | GLP-1 | NAFLD | Decrease BMI and increase GLP-1 and activated GLP1 | Alisi et al[119], 2014 |
| Probiotics (VSL #3) | ↑ Lactobacillus species | Cirrhosis (humans) | Reduced hospitalization due to HE with daily intake of probiotic for 6 mo | Dhiman et al[120], 2014 |
| Probiotics (Lactobacillus GG) | ↑ Firmicutes species; ↓ Enterobacteriaceae; ↓ Porphyromon adacea; | Cirrhosis (humans) | ↓ Endotoxemia and TNF-α after 8 wk; ↓ dysbiosis due to decreased Enterobacteriaceae and increased Firmicutes species | Bajaj et al[95], 2014 |
| Probiotics (cholesterol lowering probiotics and anthraquinone from Cassia obtusifolia L) | ↑ Bacteroides; ↑ Lactobacillus P; ↑ Arabacteroides; ↓ Oscillospira | NAFLD (rats) | Improve intestinal barrier and decrease endotoxemia and inflammatory cytokines | Mei et al[121], 2015 |
| Probiotics (Prohep: Lactobacillus rhamnosus GG (LGG), viable Escherichia coli Nissle 1917 (EcN), and heat-inactivated VSL#3) | ↑ Alistipes; ↑ Butyricimonas; ↑ Mucispirillum; ↑ Oscillibacter; ↑ Parabacteroides; ↑ Paraprevotella; ↑ Prevotella; ↑ Bacteroidetes; ↓ Firmicutes; ↓ Proteobacteria | HCC (mice) | ↑ Anti-inflammatory bacteria; ↓ Th17-inducing bacteria and segmented filamentous pro inflammatory bacteria | Li et al[77], 2016 |
| Probiotics | ↑ Ruminococcus; ↑ Saccharibacteria (TM7 phylum); ↓ Verrucomicrobia; ↓ Veillonella | NAFLD (rats) | ↓ TC, TG, lipid deposition, and inflammation | Liang et al[122], 2019 |
| Six probiotic mixtures | Gut microbiota | NAFLD | Reduce intrahepatic fat and body weight | Ahn et al[123], 2019 |
| Probiotics (multispecies strain) | ↑ Lactobacillus (brevis, salivarius, lactis); ↑ Faecalibacterium prausnitzii; ↑ Syntrophococcus sucromutans; ↑ Alistipes shahii; ↑ Bacteroides vulgatus; ↑ Prevotella | Cirrhosis (humans) | Gut microbiome enrichment in compensated cirrhosis patients and improved gut barrier function | Horvath et al[124], 2020 |
| Probiotics (Bifidobacterium animalis spp. Lactis 420) | ↑ Lactobacillus; ↑ Alistipes; ↑ Rikenella; ↑ Clostridia; ↓ Bacteroides; ↓ Ruminococcus | HCC (Mice) | Reduced liver injury and improved immune homeostasis via: Increment in tight junction proteins; ↓ Serum endotoxin levels; ↑ fecal SCFAs; ↑ α-diversity regulation of pro-inflammatory cytokines; (-) RIP3-MLKL signalling pathway of liver macrophages | Zhang et al[125], 2020 |
| Probiotics (Bifidobacterium and Lactobacillus) | ↑ Bacteroidetes; ↑ Bifidobacterium; ↑ Bacteroides; ↑ Clostridium; ↑ Ruminococcus; ↑ Anaerostipes; ↑ Blautia; ↓ Firmicutes; ↓ Faecalibacterium; ↓ Helicobacter; ↓ Staphylococcus | HCC (Mice) | ↑ Treg cell differentiation; ↑ SCFAs; ↓ infiltration of inflammatory cells in the liver; ↓ ALT, AST; ↓ Th1, Th17 cells; (-) LPS translocation to the liver; (-) activation of the TLR/NF-kB pathway | Liu et al[126], 2021 |
| Probiotic | Gut barrier | NAFLD | Mohamad et al[127], 2021 | |
| FMT | ↑ Lactobacillaceae; ↑ Bifidobacteriaceae; ↑ Bacteroidetes; ↑ Firmicutes | HE | Improves dysbiosis and SCFAs | Bajaj et al[86], (2017) |
| FMT | Gut microbiota | Cirrhosis | Reduced systemic inflammation | Bajaj et al[128], 2019 |
| FMT | Allogenic FMT: ↑ Ruminococcus ↑ Eubacterium hallii; ↑ Faecalibacterium; ↑ Prevotella copri; Autologous FMT: ↑ Lachnospiraceae | NAFLD | Decreased steatosis and liver inflammation and enhanced liver endothelial function | Witjes et al[129], 2020 |
| FMT | Gut microbiota | NAFLD | Reduced intestinal permeability | Craven et al[130], 2020 |
| FMT | ↑ Bifidobacterium; ↑ Lactobacillus; ↓ Escherichia coli | HCC | Decreased AST, ALT, and serum IgG levels and prevented progression of alcohol induced hepatitis | Liang et al[131], 2021 |
| FMT | Gut microbiota | NAFLD | Reduces gut dysbiosis and decreases fat accumulation | Xue et al[132], 2022 |
| Synbiotics [Bifidobacterium longum and Fructo-oligosaccharide] | Gut microbiota | NASH | Reduced liver inflammation and hepatocyte damage | Malaguarnera et al[84], 2012 |
| Synbiotics [Bifidobacterium animalis and inulin] | Gut microbiota | NAFLD | Improved steatosis and liver enzyme levels | Lambert et al[133], 2015 |
| Synbiotics | Gut microbiota | NAFLD | Increased levels of Bifidobacterium and Faecalibacterium, and decreased Oscillibacter and Alistipes | Scorletti et al[134], 2020 |
The therapeutic landscape for liver diseases is complex, due to challenges such as tissue specificity, drug resistance, and selectivity, complicating the establishment of clear relationships between specific liver conditions and causative organisms. Current research often relies on animal models, primarily mice, which differ significantly in gut microbial diversity from humans. This variation limits the direct applicability of findings to human models. Although advan
The current understanding of how microbial metabolites affect liver pathology is limited, and further research is needed to identify key microbial strains or metabolites critical in disease progression. This could pave the way for targeted therapies. Moreover, there is a pressing need for non-invasive biomarkers that reflect the gut-liver axis accu
Our article highlights the potential of gut microbiome manipulation as a transformative approach to liver disease treatment, with fewer side effects and complications compared to traditional methods. Therapeutic strategies such as the administration of probiotics, prebiotics, synbiotics, and FMT have shown promise in modulating the gut microbiota to enhance liver health. As we move forward, the integration of these interventions into personalized medicine is essential, utilizing detailed individual microbiome profiles to tailor therapies. The future of liver disease management will be shaped by continued research and innovation. Longitudinal studies and clinical trials are imperative to validate the therapeutic potentials identified and to refine these strategies, propelling us into a new era of precision medicine in hepatology.
| 1. | Pabst O, Hornef MW, Schaap FG, Cerovic V, Clavel T, Bruns T. Gut-liver axis: barriers and functional circuits. Nat Rev Gastroenterol Hepatol. 2023;20:447-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 155] [Reference Citation Analysis (0)] |
| 2. | Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020;72:558-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 1260] [Article Influence: 252.0] [Reference Citation Analysis (1)] |
| 3. | Li R, Mao Z, Ye X, Zuo T. Human Gut Microbiome and Liver Diseases: From Correlation to Causation. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:235-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 469] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 5. | Fukui H. Role of Gut Dysbiosis in Liver Diseases: What Have We Learned So Far? Diseases. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 6. | Zheng Z, Wang B. The Gut-Liver Axis in Health and Disease: The Role of Gut Microbiota-Derived Signals in Liver Injury and Regeneration. Front Immunol. 2021;12:775526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 7. | Wang L, Cao ZM, Zhang LL, Li JM, Lv WL. The Role of Gut Microbiota in Some Liver Diseases: From an Immunological Perspective. Front Immunol. 2022;13:923599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 8. | Brandl K, Kumar V, Eckmann L. Gut-liver axis at the frontier of host-microbial interactions. Am J Physiol Gastrointest Liver Physiol. 2017;312:G413-G419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 131] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 9. | Fukui H. Leaky Gut and Gut-Liver Axis in Liver Cirrhosis: Clinical Studies Update. Gut Liver. 2021;15:666-676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 10. | Plaza-Díaz J, Solís-Urra P, Rodríguez-Rodríguez F, Olivares-Arancibia J, Navarro-Oliveros M, Abadía-Molina F, Álvarez-Mercado AI. The Gut Barrier, Intestinal Microbiota, and Liver Disease: Molecular Mechanisms and Strategies to Manage. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 11. | Anand S, Mande SS. Host-microbiome interactions: Gut-Liver axis and its connection with other organs. NPJ Biofilms Microbiomes. 2022;8:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 85] [Reference Citation Analysis (0)] |
| 12. | Rauf A, Khalil AA, Rahman UU, Khalid A, Naz S, Shariati MA, Rebezov M, Urtecho EZ, de Albuquerque RDDG, Anwar S, Alamri A, Saini RK, Rengasamy KRR. Recent advances in the therapeutic application of short-chain fatty acids (SCFAs): An updated review. Crit Rev Food Sci Nutr. 2022;62:6034-6054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 94] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 13. | Zhou D, Chen YW, Zhao ZH, Yang RX, Xin FZ, Liu XL, Pan Q, Zhou H, Fan JG. Sodium butyrate reduces high-fat diet-induced non-alcoholic steatohepatitis through upregulation of hepatic GLP-1R expression. Exp Mol Med. 2018;50:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 14. | Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020;11:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 780] [Cited by in RCA: 1608] [Article Influence: 321.6] [Reference Citation Analysis (0)] |
| 15. | Chandrasekaran P, Weiskirchen R. The Role of SCAP/SREBP as Central Regulators of Lipid Metabolism in Hepatic Steatosis. Int J Mol Sci. 2024;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 16. | Liu W, Luo X, Tang J, Mo Q, Zhong H, Zhang H, Feng F. A bridge for short-chain fatty acids to affect inflammatory bowel disease, type 1 diabetes, and non-alcoholic fatty liver disease positively: by changing gut barrier. Eur J Nutr. 2021;60:2317-2330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 17. | Fiorucci S, Distrutti E. The Pharmacology of Bile Acids and Their Receptors. Handb Exp Pharmacol. 2019;256:3-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 18. | Yang ZX, Shen W, Sun H. Effects of nuclear receptor FXR on the regulation of liver lipid metabolism in patients with non-alcoholic fatty liver disease. Hepatol Int. 2010;4:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 19. | Ramires LC, Santos GS, Ramires RP, da Fonseca LF, Jeyaraman M, Muthu S, Lana AV, Azzini G, Smith CS, Lana JF. The Association between Gut Microbiota and Osteoarthritis: Does the Disease Begin in the Gut? Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 969] [Cited by in RCA: 2077] [Article Influence: 346.2] [Reference Citation Analysis (0)] |
| 21. | Jeyaraman M, Nallakumarasamy A, Jain VK. Gut Microbiome - Should we treat the gut and not the bones? J Clin Orthop Trauma. 2023;39:102149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | NIH HMP Working Group; Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. The NIH Human Microbiome Project. Genome Res. 2009;19:2317-2323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1410] [Cited by in RCA: 1388] [Article Influence: 86.8] [Reference Citation Analysis (0)] |
| 23. | Birchenough GMH, Johansson MEV. Forming a mucus barrier along the colon. Science. 2020;370:402-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42:28-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 25. | Li F, Ye J, Shao C, Zhong B. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: a systematic review and Meta-analysis. Lipids Health Dis. 2021;20:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 26. | Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966-G978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 595] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 27. | Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, Lefèvre L, Langelier B, Cailleux F, González-Castro AM, Rabot S, Gaudin F, Agostini H, Prévot S, Berrebi D, Ciocan D, Jousse C, Naveau S, Gérard P, Perlemuter G. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 423] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 28. | Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, Cheng W, Li B, Li H, Lin W, Tian C, Zhao J, Han J, An D, Zhang Q, Wei H, Zheng M, Ma X, Li W, Chen X, Zhang Z, Zeng H, Ying S, Wu J, Yang R, Liu D. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab. 2019;30:675-688.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 322] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 29. | Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G, Dallapiccola B, Miccheli A, Alisi A, Putignani L. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 528] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 30. | Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868-75.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 520] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 31. | Li NN, Li W, Feng JX, Zhang WW, Zhang R, Du SH, Liu SY, Xue GH, Yan C, Cui JH, Zhao HQ, Feng YL, Gan L, Zhang Q, Chen C, Liu D, Yuan J. High alcohol-producing Klebsiella pneumoniae causes fatty liver disease through 2,3-butanediol fermentation pathway in vivo. Gut Microbes. 2021;13:1979883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 32. | Chen Z, Xie Y, Zhou F, Zhang B, Wu J, Yang L, Xu S, Stedtfeld R, Chen Q, Liu J, Zhang X, Xu H, Ren J. Featured Gut Microbiomes Associated With the Progression of Chronic Hepatitis B Disease. Front Microbiol. 2020;11:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 33. | Bajaj JS, Fagan A, Sikaroodi M, White MB, Sterling RK, Gilles H, Heuman D, Stravitz RT, Matherly SC, Siddiqui MS, Puri P, Sanyal AJ, Luketic V, John B, Fuchs M, Ahluwalia V, Gillevet PM. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 2017;23:907-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 34. | Stojic J, Kukla M, Grgurevic I. The Intestinal Microbiota in the Development of Chronic Liver Disease: Current Status. Diagnostics (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 35. | Arelaki S, Koletsa T, Sinakos E, Papadopoulos V, Arvanitakis K, Skendros P, Akriviadis E, Ritis K, Germanidis G, Hytiroglou P. Neutrophil extracellular traps enriched with IL-1β and IL-17A participate in the hepatic inflammatory process of patients with non-alcoholic steatohepatitis. Virchows Arch. 2022;481:455-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 36. | Han YH, Choi H, Kim HJ, Lee MO. Chemotactic cytokines secreted from Kupffer cells contribute to the sex-dependent susceptibility to non-alcoholic fatty liver diseases in mice. Life Sci. 2022;306:120846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 37. | Rennert C, Heil T, Schicht G, Stilkerich A, Seidemann L, Kegel-Hübner V, Seehofer D, Damm G. Prolonged Lipid Accumulation in Cultured Primary Human Hepatocytes Rather Leads to ER Stress than Oxidative Stress. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Arrese M, Cabrera D, Kalergis AM, Feldstein AE. Innate Immunity and Inflammation in NAFLD/NASH. Dig Dis Sci. 2016;61:1294-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 383] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 39. | Jennison E, Byrne CD. The role of the gut microbiome and diet in the pathogenesis of non-alcoholic fatty liver disease. Clin Mol Hepatol. 2021;27:22-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 40. | Thakur V, Pritchard MT, McMullen MR, Wang Q, Nagy LE. Chronic ethanol feeding increases activation of NADPH oxidase by lipopolysaccharide in rat Kupffer cells: role of increased reactive oxygen in LPS-stimulated ERK1/2 activation and TNF-alpha production. J Leukoc Biol. 2006;79:1348-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 170] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 41. | von Montfort C, Beier JI, Guo L, Kaiser JP, Arteel GE. Contribution of the sympathetic hormone epinephrine to the sensitizing effect of ethanol on LPS-induced liver damage in mice. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1227-G1234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 42. | Hill DB, Barve S, Joshi-Barve S, McClain C. Increased monocyte nuclear factor-kappaB activation and tumor necrosis factor production in alcoholic hepatitis. J Lab Clin Med. 2000;135:387-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 43. | Wree A, McGeough MD, Peña CA, Schlattjan M, Li H, Inzaugarat ME, Messer K, Canbay A, Hoffman HM, Feldstein AE. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med (Berl). 2014;92:1069-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 400] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 44. | Shen TD, Pyrsopoulos N, Rustgi VK. Microbiota and the liver. Liver Transpl. 2018;24:539-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4:382-387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 271] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 46. | Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1244] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 47. | Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1644] [Cited by in RCA: 2039] [Article Influence: 102.0] [Reference Citation Analysis (0)] |
| 48. | Ridlon JM, Kang DJ, Hylemon PB. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 49. | de Faria Ghetti F, Oliveira DG, de Oliveira JM, de Castro Ferreira LEVV, Cesar DE, Moreira APB. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr. 2018;57:861-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 50. | Fang J, Yu CH, Li XJ, Yao JM, Fang ZY, Yoon SH, Yu WY. Gut dysbiosis in nonalcoholic fatty liver disease: pathogenesis, diagnosis, and therapeutic implications. Front Cell Infect Microbiol. 2022;12:997018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 100] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 51. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1281] [Article Influence: 106.8] [Reference Citation Analysis (1)] |
| 52. | Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, Hunault G, Oberti F, Calès P, Diehl AM. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1050] [Article Influence: 116.7] [Reference Citation Analysis (0)] |
| 53. | Li W, Zhou Y, Pang N, Hu Q, Li Q, Sun Y, Ding Y, Gu Y, Xiao Y, Gao M, Ma S, Pan J, Fang EF, Zhang Z, Yang L. NAD Supplement Alleviates Intestinal Barrier Injury Induced by Ethanol Via Protecting Epithelial Mitochondrial Function. Nutrients. 2022;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 54. | Iogna Prat L, Tsochatzis EA. Pediatric NAFLD: lessons from the gut. Hepatobiliary Surg Nutr. 2020;9:534-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 55. | Fukui H, Brauner B, Bode JC, Bode C. Plasma endotoxin concentrations in patients with alcoholic and non-alcoholic liver disease: reevaluation with an improved chromogenic assay. J Hepatol. 1991;12:162-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 351] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 56. | Pérez-Paramo M, Muñoz J, Albillos A, Freile I, Portero F, Santos M, Ortiz-Berrocal J. Effect of propranolol on the factors promoting bacterial translocation in cirrhotic rats with ascites. Hepatology. 2000;31:43-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 195] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 57. | Reiberger T, Ferlitsch A, Payer BA, Mandorfer M, Heinisch BB, Hayden H, Lammert F, Trauner M, Peck-Radosavljevic M, Vogelsang H; Vienna Hepatic Hemodynamic Lab. Non-selective betablocker therapy decreases intestinal permeability and serum levels of LBP and IL-6 in patients with cirrhosis. J Hepatol. 2013;58:911-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 58. | Forslund SK, Chakaroun R, Zimmermann-Kogadeeva M, Markó L, Aron-Wisnewsky J, Nielsen T, Moitinho-Silva L, Schmidt TSB, Falony G, Vieira-Silva S, Adriouch S, Alves RJ, Assmann K, Bastard JP, Birkner T, Caesar R, Chilloux J, Coelho LP, Fezeu L, Galleron N, Helft G, Isnard R, Ji B, Kuhn M, Le Chatelier E, Myridakis A, Olsson L, Pons N, Prifti E, Quinquis B, Roume H, Salem JE, Sokolovska N, Tremaroli V, Valles-Colomer M, Lewinter C, Søndertoft NB, Pedersen HK, Hansen TH; MetaCardis Consortium*, Gøtze JP, Køber L, Vestergaard H, Hansen T, Zucker JD, Hercberg S, Oppert JM, Letunic I, Nielsen J, Bäckhed F, Ehrlich SD, Dumas ME, Raes J, Pedersen O, Clément K, Stumvoll M, Bork P. Combinatorial, additive and dose-dependent drug-microbiome associations. Nature. 2021;600:500-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 130] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 59. | Moore KP, Aithal GP. Guidelines on the management of ascites in cirrhosis. Gut. 2006;55 Suppl 6:vi1-v12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 208] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 60. | Mookerjee RP, Pavesi M, Thomsen KL, Mehta G, Macnaughtan J, Bendtsen F, Coenraad M, Sperl J, Gines P, Moreau R, Arroyo V, Jalan R; CANONIC Study Investigators of the EASL-CLIF Consortium. Treatment with non-selective beta blockers is associated with reduced severity of systemic inflammation and improved survival of patients with acute-on-chronic liver failure. J Hepatol. 2016;64:574-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 184] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 61. | Bajaj JS, Idilman R, Mabudian L, Hood M, Fagan A, Turan D, White MB, Karakaya F, Wang J, Atalay R, Hylemon PB, Gavis EA, Brown R, Thacker LR, Acharya C, Heuman DM, Sikaroodi M, Gillevet PM. Diet affects gut microbiota and modulates hospitalization risk differentially in an international cirrhosis cohort. Hepatology. 2018;68:234-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 62. | Gupta H, Youn GS, Shin MJ, Suk KT. Role of Gut Microbiota in Hepatocarcinogenesis. Microorganisms. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 63. | Yu LX, Yan HX, Liu Q, Yang W, Wu HP, Dong W, Tang L, Lin Y, He YQ, Zou SS, Wang C, Zhang HL, Cao GW, Wu MC, Wang HY. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52:1322-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 255] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 64. | Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 414] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 65. | Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, Gao B, Wisplinghoff H, Kasper P, Roderburg C, Tacke F, Steffen HM, Goeser T, Abraldes JG, Tu XM, Loomba R, Stärkel P, Pride D, Fouts DE, Schnabl B. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;159:1839-1852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 66. | Zheng R, Wang G, Pang Z, Ran N, Gu Y, Guan X, Yuan Y, Zuo X, Pan H, Zheng J, Wang F. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020;9:4232-4250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 67. | Floreani A, Restrepo-Jiménez P, Secchi MF, De Martin S, Leung PSC, Krawitt E, Bowlus CL, Gershwin ME, Anaya JM. Etiopathogenesis of autoimmune hepatitis. J Autoimmun. 2018;95:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 68. | Khalyfa AA, Punatar S, Yarbrough A. Hepatocellular Carcinoma: Understanding the Inflammatory Implications of the Microbiome. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 69. | Liu Q, Li F, Zhuang Y, Xu J, Wang J, Mao X, Zhang Y, Liu X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019;11:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 70. | Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol. 2015;8:5153-5160. [PubMed] |
| 71. | Hsu YC, Chen CC, Lee WH, Chang CY, Lee FJ, Tseng CH, Chen TH, Ho HJ, Lin JT, Wu CY. Compositions of gut microbiota before and shortly after hepatitis C viral eradication by direct antiviral agents. Sci Rep. 2022;12:5481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 72. | El-Mowafy M, Elgaml A, El-Mesery M, Sultan S, Ahmed TAE, Gomaa AI, Aly M, Mottawea W. Changes of Gut-Microbiota-Liver Axis in Hepatitis C Virus Infection. Biology (Basel). 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 73. | Marascio N, De Caro C, Quirino A, Mazzitelli M, Russo E, Torti C, Matera G. The Role of the Microbiota Gut-Liver Axis during HCV Chronic Infection: A Schematic Overview. J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, Drastich P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23:4548-4558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 215] [Cited by in RCA: 262] [Article Influence: 32.8] [Reference Citation Analysis (4)] |
| 75. | Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR, Li A, Shi HY, Jiang XW, Jiang HY, Xiao YH, Zheng SS, Li LJ. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. 2016;18:2272-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 76. | Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 2009;43:163-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 321] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 77. | Li J, Sung CY, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, El-Nezami H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A. 2016;113:E1306-E1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 425] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 78. | Grander C, Adolph TE, Wieser V, Lowe P, Wrzosek L, Gyongyosi B, Ward DV, Grabherr F, Gerner RR, Pfister A, Enrich B, Ciocan D, Macheiner S, Mayr L, Drach M, Moser P, Moschen AR, Perlemuter G, Szabo G, Cassard AM, Tilg H. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 457] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 79. | Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2493] [Cited by in RCA: 3799] [Article Influence: 474.9] [Reference Citation Analysis (0)] |
| 80. | Houron C, Ciocan D, Trainel N, Mercier-Nomé F, Hugot C, Spatz M, Perlemuter G, Cassard AM. Gut Microbiota Reshaped by Pectin Treatment Improves Liver Steatosis in Obese Mice. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 81. | Plamada D, Vodnar DC. Polyphenols-Gut Microbiota Interrelationship: A Transition to a New Generation of Prebiotics. Nutrients. 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 183] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 82. | Reshef N, Gophna U, Reshef L, Konikoff F, Gabay G, Zornitzki T, Knobler H, Maor Y. Prebiotic Treatment in Patients with Nonalcoholic Fatty Liver Disease (NAFLD)-A Randomized Pilot Trial. Nutrients. 2024;16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 83. | Hadi A, Mohammadi H, Miraghajani M, Ghaedi E. Efficacy of synbiotic supplementation in patients with nonalcoholic fatty liver disease: A systematic review and meta-analysis of clinical trials: Synbiotic supplementation and NAFLD. Crit Rev Food Sci Nutr. 2019;59:2494-2505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 84. | Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, Mastrojeni S, Malaguarnera G, Mistretta A, Li Volti G, Galvano F. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012;57:545-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 332] [Article Influence: 25.5] [Reference Citation Analysis (1)] |
| 85. | Ferrere G, Wrzosek L, Cailleux F, Turpin W, Puchois V, Spatz M, Ciocan D, Rainteau D, Humbert L, Hugot C, Gaudin F, Noordine ML, Robert V, Berrebi D, Thomas M, Naveau S, Perlemuter G, Cassard AM. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol. 2017;66:806-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 251] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 86. | Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727-1738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 453] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 87. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 797] [Article Influence: 56.9] [Reference Citation Analysis (3)] |
| 88. | Liu J, Wu D, Ahmed A, Li X, Ma Y, Tang L, Mo D, Ma Y, Xin Y. Comparison of the gut microbe profiles and numbers between patients with liver cirrhosis and healthy individuals. Curr Microbiol. 2012;65:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 89. | Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168-G175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 423] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 90. | Zhang Z, Zhai H, Geng J, Yu R, Ren H, Fan H, Shi P. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am J Gastroenterol. 2013;108:1601-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 91. | Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, Yeung DK, Law PT, Kwan HS, Yu J, Sung JJ, Chan HL. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One. 2013;8:e62885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 249] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 92. | Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 556] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 93. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 619] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 94. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1537] [Article Influence: 139.7] [Reference Citation Analysis (40)] |
| 95. | Bajaj JS. The role of microbiota in hepatic encephalopathy. Gut Microbes. 2014;5:397-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 96. | Bajaj JS. Review article: potential mechanisms of action of rifaximin in the management of hepatic encephalopathy and other complications of cirrhosis. Aliment Pharmacol Ther. 2016;43 Suppl 1:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 97. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 837] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 98. | Grąt M, Wronka KM, Krasnodębski M, Masior Ł, Lewandowski Z, Kosińska I, Grąt K, Stypułkowski J, Rejowski S, Wasilewicz M, Gałęcka M, Szachta P, Krawczyk M. Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc. 2016;48:1687-1691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 145] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 99. | Chen Y, Ji F, Guo J, Shi D, Fang D, Li L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep. 2016;6:34055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 100. | Ahluwalia V, Betrapally NS, Hylemon PB, White MB, Gillevet PM, Unser AB, Fagan A, Daita K, Heuman DM, Zhou H, Sikaroodi M, Bajaj JS. Impaired Gut-Liver-Brain Axis in Patients with Cirrhosis. Sci Rep. 2016;6:26800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 101. | Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, Kisseleva T, Torralba MG, Moncera K, Beeri K, Chen CS, Freese K, Hellerbrand C, Lee SM, Hoffman HM, Mehal WZ, Garcia-Tsao G, Mutlu EA, Keshavarzian A, Brown GD, Ho SB, Bataller R, Stärkel P, Fouts DE, Schnabl B. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829-2841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 361] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 102. | Dubinkina VB, Tyakht AV, Odintsova VY, Yarygin KS, Kovarsky BA, Pavlenko AV, Ischenko DS, Popenko AS, Alexeev DG, Taraskina AY, Nasyrova RF, Krupitsky EM, Shalikiani NV, Bakulin IG, Shcherbakov PL, Skorodumova LO, Larin AK, Kostryukova ES, Abdulkhakov RA, Abdulkhakov SR, Malanin SY, Ismagilova RK, Grigoryeva TV, Ilina EN, Govorun VM. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. 2017;5:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 338] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 103. | Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054-1062.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 752] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 104. | Liu Y, Jin Y, Li J, Zhao L, Li Z, Xu J, Zhao F, Feng J, Chen H, Fang C, Shilpakar R, Wei Y. Small Bowel Transit and Altered Gut Microbiota in Patients With Liver Cirrhosis. Front Physiol. 2018;9:470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 105. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Chen X, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 509] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 106. | Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, Sanguinetti M, Morelli D, Paroni Sterbini F, Petito V, Reddel S, Calvani R, Camisaschi C, Picca A, Tuccitto A, Gasbarrini A, Pompili M, Mazzaferro V. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology. 2019;69:107-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 474] [Article Influence: 79.0] [Reference Citation Analysis (1)] |
| 107. | Piñero F, Vazquez M, Baré P, Rohr C, Mendizabal M, Sciara M, Alonso C, Fay F, Silva M. A different gut microbiome linked to inflammation found in cirrhotic patients with and without hepatocellular carcinoma. Ann Hepatol. 2019;18:480-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 108. | Ni J, Huang R, Zhou H, Xu X, Li Y, Cao P, Zhong K, Ge M, Chen X, Hou B, Yu M, Peng B, Li Q, Zhang P, Gao Y. Analysis of the Relationship Between the Degree of Dysbiosis in Gut Microbiota and Prognosis at Different Stages of Primary Hepatocellular Carcinoma. Front Microbiol. 2019;10:1458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 109. | Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, Bross C, Durelle J, Goyal NP, Hamilton G, Holtz ML, Lavine JE, Mitreva M, Newton KP, Pan A, Simpson PM, Sirlin CB, Sodergren E, Tyagi R, Yates KP, Weinstock GM, Salzman NH. Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2019;157:1109-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 110. | Duarte SMB, Stefano JT, Oliveira CP. Microbiota and nonalcoholic fatty liver disease/nonalcoholic steatohepatitis (NAFLD/NASH). Ann Hepatol. 2019;18:416-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 111. | Monga Kravetz A, Testerman T, Galuppo B, Graf J, Pierpont B, Siebel S, Feinn R, Santoro N. Effect of Gut Microbiota and PNPLA3 rs738409 Variant on Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Youth. J Clin Endocrinol Metab. 2020;105:e3575-e3585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 112. | Lang S, Martin A, Zhang X, Farowski F, Wisplinghoff H, J G T Vehreschild M, Krawczyk M, Nowag A, Kretzschmar A, Scholz C, Kasper P, Roderburg C, Mohr R, Lammert F, Tacke F, Schnabl B, Goeser T, Steffen HM, Demir M. Combined analysis of gut microbiota, diet and PNPLA3 polymorphism in biopsy-proven non-alcoholic fatty liver disease. Liver Int. 2021;41:1576-1591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 113. | Behary J, Amorim N, Jiang XT, Raposo A, Gong L, McGovern E, Ibrahim R, Chu F, Stephens C, Jebeili H, Fragomeli V, Koay YC, Jackson M, O'Sullivan J, Weltman M, McCaughan G, El-Omar E, Zekry A. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat Commun. 2021;12:187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 290] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 114. | Trebicka J, Macnaughtan J, Schnabl B, Shawcross DL, Bajaj JS. The microbiota in cirrhosis and its role in hepatic decompensation. J Hepatol. 2021;75 Suppl 1:S67-S81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 115. | Solé C, Guilly S, Da Silva K, Llopis M, Le-Chatelier E, Huelin P, Carol M, Moreira R, Fabrellas N, De Prada G, Napoleone L, Graupera I, Pose E, Juanola A, Borruel N, Berland M, Toapanta D, Casellas F, Guarner F, Doré J, Solà E, Ehrlich SD, Ginès P. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship With Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology. 2021;160:206-218.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 116. | Matsumoto K, Ichimura M, Tsuneyama K, Moritoki Y, Tsunashima H, Omagari K, Hara M, Yasuda I, Miyakawa H, Kikuchi K. Fructo-oligosaccharides and intestinal barrier function in a methionine-choline-deficient mouse model of nonalcoholic steatohepatitis. PLoS One. 2017;12:e0175406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 117. | Lata J, Novotný I, Príbramská V, Juránková J, Fric P, Kroupa R, Stibůrek O. The effect of probiotics on gut flora, level of endotoxin and Child-Pugh score in cirrhotic patients: results of a double-blind randomized study. Eur J Gastroenterol Hepatol. 2007;19:1111-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 118. | Hsieh FC, Lee CL, Chai CY, Chen WT, Lu YC, Wu CS. Oral administration of Lactobacillus reuteri GMNL-263 improves insulin resistance and ameliorates hepatic steatosis in high fructose-fed rats. Nutr Metab (Lond). 2013;10:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 182] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 119. | Alisi A, Bedogni G, Baviera G, Giorgio V, Porro E, Paris C, Giammaria P, Reali L, Anania F, Nobili V. Randomised clinical trial: The beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2014;39:1276-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 342] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 120. | Dhiman RK, Rana B, Agrawal S, Garg A, Chopra M, Thumburu KK, Khattri A, Malhotra S, Duseja A, Chawla YK. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology. 2014;147:1327-37.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 255] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 121. | Mei L, Tang Y, Li M, Yang P, Liu Z, Yuan J, Zheng P. Co-Administration of Cholesterol-Lowering Probiotics and Anthraquinone from Cassia obtusifolia L. Ameliorate Non-Alcoholic Fatty Liver. PLoS One. 2015;10:e0138078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 122. | Liang Y, Liang S, Zhang Y, Deng Y, He Y, Chen Y, Liu C, Lin C, Yang Q. Oral Administration of Compound Probiotics Ameliorates HFD-Induced Gut Microbe Dysbiosis and Chronic Metabolic Inflammation via the G Protein-Coupled Receptor 43 in Non-alcoholic Fatty Liver Disease Rats. Probiotics Antimicrob Proteins. 2019;11:175-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 123. | Ahn SB, Jun DW, Kang BK, Lim JH, Lim S, Chung MJ. Randomized, Double-blind, Placebo-controlled Study of a Multispecies Probiotic Mixture in Nonalcoholic Fatty Liver Disease. Sci Rep. 2019;9:5688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 124. | Horvath A, Durdevic M, Leber B, di Vora K, Rainer F, Krones E, Douschan P, Spindelboeck W, Durchschein F, Zollner G, Stauber RE, Fickert P, Stiegler P, Stadlbauer V. Changes in the Intestinal Microbiome during a Multispecies Probiotic Intervention in Compensated Cirrhosis. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 125. | Zhang H, Liu M, Liu X, Zhong W, Li Y, Ran Y, Guo L, Chen X, Zhao J, Wang B, Zhou L. Bifidobacterium animalis ssp. Lactis 420 Mitigates Autoimmune Hepatitis Through Regulating Intestinal Barrier and Liver Immune Cells. Front Immunol. 2020;11:569104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 126. | Liu Q, Tian H, Kang Y, Tian Y, Li L, Kang X, Yang H, Wang Y, Tian J, Zhang F, Tong M, Cai H, Fan W. Probiotics alleviate autoimmune hepatitis in mice through modulation of gut microbiota and intestinal permeability. J Nutr Biochem. 2021;98:108863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 127. | Mohamad Nor MH, Ayob N, Mokhtar NM, Raja Ali RA, Tan GC, Wong Z, Shafiee NH, Wong YP, Mustangin M, Nawawi KNM. The Effect of Probiotics (MCP(®) BCMC(®) Strains) on Hepatic Steatosis, Small Intestinal Mucosal Immune Function, and Intestinal Barrier in Patients with Non-Alcoholic Fatty Liver Disease. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 128. | Bajaj JS, Salzman N, Acharya C, Takei H, Kakiyama G, Fagan A, White MB, Gavis EA, Holtz ML, Hayward M, Nittono H, Hylemon PB, Cox IJ, Williams R, Taylor-Robinson SD, Sterling RK, Matherly SC, Fuchs M, Lee H, Puri P, Stravitz RT, Sanyal AJ, Ajayi L, Le Guennec A, Atkinson RA, Siddiqui MS, Luketic V, Pandak WM, Sikaroodi M, Gillevet PM. Microbial functional change is linked with clinical outcomes after capsular fecal transplant in cirrhosis. JCI Insight. 2019;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 129. | Witjes JJ, Smits LP, Pekmez CT, Prodan A, Meijnikman AS, Troelstra MA, Bouter KEC, Herrema H, Levin E, Holleboom AG, Winkelmeijer M, Beuers UH, van Lienden K, Aron-Wisnewky J, Mannisto V, Bergman JJ, Runge JH, Nederveen AJ, Dragsted LO, Konstanti P, Zoetendal EG, de Vos W, Verheij J, Groen AK, Nieuwdorp M. Donor Fecal Microbiota Transplantation Alters Gut Microbiota and Metabolites in Obese Individuals With Steatohepatitis. Hepatol Commun. 2020;4:1578-1590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 130. | Craven L, Rahman A, Nair Parvathy S, Beaton M, Silverman J, Qumosani K, Hramiak I, Hegele R, Joy T, Meddings J, Urquhart B, Harvie R, McKenzie C, Summers K, Reid G, Burton JP, Silverman M. Allogenic Fecal Microbiota Transplantation in Patients With Nonalcoholic Fatty Liver Disease Improves Abnormal Small Intestinal Permeability: A Randomized Control Trial. Am J Gastroenterol. 2020;115:1055-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 131. | Liang M, Liwen Z, Jianguo S, Juan D, Fei D, Yin Z, Changping W, Jianping C. Fecal Microbiota Transplantation Controls Progression of Experimental Autoimmune Hepatitis in Mice by Modulating the TFR/TFH Immune Imbalance and Intestinal Microbiota Composition. Front Immunol. 2021;12:728723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 132. | Xue L, Deng Z, Luo W, He X, Chen Y. Effect of Fecal Microbiota Transplantation on Non-Alcoholic Fatty Liver Disease: A Randomized Clinical Trial. Front Cell Infect Microbiol. 2022;12:759306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 137] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 133. | Lambert JE, Parnell JA, Eksteen B, Raman M, Bomhof MR, Rioux KP, Madsen KL, Reimer RA. Gut microbiota manipulation with prebiotics in patients with non-alcoholic fatty liver disease: a randomized controlled trial protocol. BMC Gastroenterol. 2015;15:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 134. | Scorletti E, Afolabi PR, Miles EA, Smith DE, Almehmadi A, Alshathry A, Childs CE, Del Fabbro S, Bilson J, Moyses HE, Clough GF, Sethi JK, Patel J, Wright M, Breen DJ, Peebles C, Darekar A, Aspinall R, Fowell AJ, Dowman JK, Nobili V, Targher G, Delzenne NM, Bindels LB, Calder PC, Byrne CD. Synbiotics Alter Fecal Microbiomes, But Not Liver Fat or Fibrosis, in a Randomized Trial of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;158:1597-1610.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 155] [Article Influence: 31.0] [Reference Citation Analysis (0)] |