Published online Nov 22, 2021. doi: 10.4291/wjgp.v12.i6.134
Peer-review started: January 27, 2021
First decision: March 1, 2021
Revised: March 13, 2021
Accepted: October 11, 2021
Article in press: October 11, 2021
Published online: November 22, 2021
Processing time: 293 Days and 8.4 Hours
Hereditary non-polyposis colon cancer is a dominantly inherited syndrome of colorectal cancer (CRC), with heightened risk for younger population. Previous studies link its susceptibility to the DNA sequence polymorphism along with Amsterdam and Bethesda criteria. However, those fail in term of applicability.
To determine a clear cut-off of MSH2 gene expression for CRC heredity grouping factor. Further, the study also aims to examine the association of risk factors to the CRC heredity.
The cross-sectional study observed 71 respondents from May 2018 to December 2019 in determining the CRC hereditary status through MSH2 mRNA expression using reverse transcription-polymerase chain reaction and the disease’s risk factors. Data were analyzed through Chi-Square, Fischer exact, t-test, Mann-Whitney, and multiple logistics.
There are significant differences of MSH2 within CRC group among tissue and blood; yet, negative for significance between groups. Through the blood gene expression fifth percentile, the hereditary CRC cut-off is 11059 fc, dividing the 40 CRC respondents to 32.5% with hereditary CRC. Significant risk factors include age, family history, and staging. Nonetheless, after multivariate control, age is just a confounder. Further, the study develops a probability equation with area under the curve 82.2%.
Numerous factors have significant relations to heredity of CRC patients. However, true important factors are staging and family history, while age and others are confounders. The study also established a definite cut-off point for heredity CRC based on mRNA MSH2 expression, 11059 fc. These findings shall act as concrete foundations on further risk factors and/or genetical CRC future studies.
Core Tip: The study has determined a definitive cut-off for grouping colorectal cancer (CRC) with its heredity using the MSH2 mRNA gene expression, which amounts to 11059 fc. The gene expressions may differ between blood and tissue sample of the CRC group, yet none between CRC and control group. Nevertheless, subsequent risk factors of family history and staging are found to be significant toward the heredity. The after-mentioned risk factors act as urgent reminder for highly risky people with family history of CRC and/or high CRC staging to have themselves and their immediate family members to undergo routine examinations as well as strict preventions.
- Citation: Tedjasaputra TR, Hatta M, Massi MN, Natzir R, Bukhari A, Masadah R, Parewangi ML, Prihantono P, Nariswati R, Tedjasaputra V. Prediction of hereditary nonpolyposis colorectal cancer using mRNA MSH2 quantitative and the correlation with nonmodifiable factor. World J Gastrointest Pathophysiol 2021; 12(6): 134-146
- URL: https://www.wjgnet.com/2150-5330/full/v12/i6/134.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v12.i6.134
Colorectal cancer (CRC) or also known as colorectal adenocarcinoma is a group of cancer that manifest in the colon and/or rectum. The cancer first arises as polyps that comprised of extraneous cells from the uncontrolled proliferation because of genetic mutations. These benign masses of flesh will then achieve even greater hyper-replication, survival, and angiogenesis, leading to a malignant carcinoma (CRC) which then can metastasize[1].
In fact, CRC holds the fourth place in the top ten of the most diagnosed global cancer with around 2000000 incidence cases, while taking third place for worldwide cancer mortality with approximately 1000000 deaths[2]. Within very high human development index countries, Hungary and Norway have the highest age-standardized rates of CRC over 100000 populations for male and female respectively (70.6 and 39.3)[3]. Meanwhile, the top age-standardized death rates are taken both for male and female by Hungary with 31.2 and 14.8 over 100000 population[3].
Specifically, CRC can be further divided according to its differentiation, epidemiology, and hereditary. Interesting potential lies dormant in CRC hereditary status, where as far as the author’s known, there has been no gold standard measure to classify one’s CRC into hereditary [i.e., hereditary non-polyposis colon cancer (HNPCC)] or sporadic.
HNPCC or sometimes known to the general citizens as lynch syndrome (LS) is a hereditary mutation of the MLH1, MSH2, MSH6, EpCAM, and PMS2 genes which contribute to the development of CRC yet also the passing of the autosomal dominant mutated genes and thus the heightened susceptibility to the offspring[4]. LS can be found in younger people compared to overall CRC as these mutations provide grounds for CRC rapid development. Within his or her lifetime, the risk of developing LS is around 4.1%-4.4%[5].
The incidence of LS can be said to mainly comes from the mutation of MSH2 gene on chromosome 2. The protein translated by that subsequent gene encode MutS Homolog 2 protein which functions as a DNA mismatch repair protein. When doing its intended functions, it bonds with MSH6 or MSH3 to procure MutSα or MutSb complex according to specifications of the DNA damage, namely: Transcription repair, base excision repair, homologous recombination, etc[6-8].
Identification of those who carry the mutated genes of MSH2 or the groups would benefit the patients as early detection and adequate prevention can reduce morbidity, mortality, and recurrence risk of LS[9,10]. Several studies have tried to implement the Bethesda and Amsterdam criteria to solve this dynamic screening of LS, yet the effort failed due to its complexity and arduousness especially in small family and late age of onset settings[11-15]. Consequently, when trying to address the problem from its roots of the mutated genes, establishment of the definite gene expression may act as a well cut-off point to categorize CRC into LS or non-LS with high potential of becoming a gold standard measure. The subsequent practices utilize specific enzymes that pinpoint the post-transcription mRNA strategically[16].
These techniques of separating the CRC intro groups, enable risk factor assessments toward hereditary and sporadic CRC types. Modifiable risk factors for LS and CRC are body mass index (BMI), physical activity, diet, lifestyle (i.e., smoking, alcohol, and narcotics), routine medications, and diabetes mellitus (DM). Insulin resistance and the hyperglycemic state of the body can predispose a person to CRC. The excess of blood sugar trigger Warburg effect (carcinogenic glycolysis) through modulation of glucose metabolism[17,18]. Studies on DM relations to CRC prove a 1.17-1.42 hazard ratio along with 11536/559375 DM patients have CRC[19,20]. On the other hand, non-modifiable risk factors for LS and CRC involve the race, age, gender, heredity, radiations, and some diseases (i.e., inflammatory bowel disease or cystic fibrosis).
Above all, the hazardous nature of CRC and LS, scarce information on CRC risk factors identifications, and the lack of gold standard for categorizing hereditary measurements, thus the present study urgently aimed to compute suitable MSH2 gene expression for appropriate cut-off and certify the associations from the risk factors.
The present study was conducted using cross-sectional design with 71 respondents divided into 31 respondents in the control group, those who have been sequentially matched (i.e., age, sex, and BMI) with the case group. The study involves tumor and malignancy sector which are a sensitive section of health as it rapidly deteriorates health while integrated to other bodily system. Henceforth, strict exclusion criteria were adapted in the current study, namely: (1) The presence or history of other cancer; (2) The presence or history of inflammatory bowel disease; (3) Chemotherapy or radiotherapy in progress or history; (4) Refusal of participation; and (5) Missing or incomplete data.
From May 2018 to December 2019, the respondents are taken from the internal medicine outpatient and inpatient department of Tarakan General Hospital and Siloam Hospitals Lippo Village through consecutive sampling. Sample size calculated using 5% alpha and power 80%.
Three major steps are contained within this study. Initially, the study collects respondents and their clinical data of demographics and malignancy characteristics. Then, the respective mRNA gene expression of MSH2 was quantified using polymerase chain reaction (PCR) and studied in assessing within groups and between groups differences as well as its hereditary significance. Whereas the last component of the study involves risk factor analysis toward hereditary of CRC and its probability model.
Biopsy tissues of suspected CRC tissues and venous blood are the key samples of this study. Then, the samples are placed in a L6 buffer and have their RNA extracted. The L6 buffer are concocted earlier according to the RNA Boom extraction method of the Hasanuddin laboratory. Next, RT-PCR targeting the MSH2 mRNA were done to measure the gene expression.
The PCR are conducted through the DNA multiplication, denaturation, primer attachment, and amplification stage. Specific Korean primers are supplied to specifically target the MSH2 gene: CAT-CCA-GGC-ATG-CTT-GTG-TTG-A (forward) and GCA-GTC-CAC-AAT-GGA-CAC-TTC (reverse). The mechanics and PCR analysis follow the Bio-Rad protocols from Unites States of America using the power SYBR green master mix kit[21-23].
Data tabulation was done through Microsoft Excel 365, while SPSS v26 is the software of choice for the statistical analysis. Missing data is excluded from the study. The respondents’ demographics are characterized using descriptive statistics; yet, Chi-Square or Fischer test are applied for categorical factors, while t-test or Mann-Whitney for numerical factors. Significance obtained if P value < 0.05.
The present study employs 71 respondents among the 19-mo study period, which comprised of 56.34% in the case group and 43.66% in the control group. Respondents within the control group are adequately matched according to the case group characteristics, proven with no significant difference (P > 0.05) in the demographic characteristics between the groups as depicted in Table 1.
| Characteristic | CRC group | Control group | P value |
| Age (yr) | 56.8 ± 8.4 | 51.6 ± 13.4 | > 0.05 |
| Sex | |||
| Male | 21 (52.5) | 13 (41.9) | |
| Female | 19 (47.5) | 18 (58.1) | |
| Body mass index (kg/m2) | 22.4 ± 3.3 | 23.6 ± 3.4 |
Among the respondents on the CRC group, histopathological samples are taken and observed. Specifically, 90.00% respondents have adenocarcinoma while 7.50% have adenocarcinoma with Signet ring cell and 2.50% have neuroendocrine carcinoma. Nevertheless, specifications on the histopathological profile can also be seen from the level of differentiation. Well-differentiated biopsies are found in 26 (66.7%) respondents, intermediately differentiated in 6 (15.4%) respondents, and poorly differentiated in 7 (17.9%).
Subsequently, the study utilizes PCR analysis to measure the MSH2 mRNA gene expression in blood and tissue between the groups. Significance is observed when comparing the gene expression within the CRC group between blood and tissue (12554.50 vs 7485.00). However, as pictured in Table 2, there is no significant difference of MSH2 mRNA expression between CRC and control groups (P = 0.116 and 0.465).
| MSH2 expression | CRC group | Control group | P value |
| Blood | |||
| Median (range) | 12554.50 (4230.00-14559.00) | 12146.00 (11029.00-13633.00) | 0.116 |
| mean ± SD | 11411.05 ± 2912.45 | 12219.87 ± 756.87 | 0.465 |
| Tissue | 7 485.00 (4174.00-14218.00) |
Moreover, the group with CRC were then subdivided based on each respondent hereditary status. One is considered having hereditary condition if his or her blood mRNA MSH2 gene expression less than the cut-off from the fifth percentile, 11059 fc. It was established that 67.50% of the CRC group respondents have non-hereditary status, even including one-third of those with positive family history of CRC. Likewise, Table 3 portrayed the relationship of risk factors to hereditary status.
| Factor | CRC hereditary, yes (n: 13) | CRC hereditary, no (n: 27) | OR (95%CI) | P value |
| Age | 50.69 ± 14.99 | 59.74 ± 11.68 | 0.043 | |
| < 50 | 8 (61.54) | 6 (22.22) | 5.60 (1.33-23.62) | 0.031 |
| > 50 | 5 (38.46) | 21 (77.78) | ||
| Gender | 0.69 (0.18-2.59) | 0.826 | ||
| Male | 6 (46.15) | 15 (55.56) | ||
| Female | 7 (53.85) | 12 (44.44) | ||
| Location | - | 0.595 | ||
| Caecum | 1 (7.69) | 1 (3.70) | ||
| Ascending colon | 2 (15.38) | 4 (14.81) | ||
| Transverse colon | 2 (15.38) | 2 (7.41) | ||
| Descending colon | 0 (0.00) | 1 (3.70) | ||
| Sigmoid | 3 (23.08) | 7 (25.93) | ||
| Rectum | 5 (38.46) | 12 (44.44) | ||
| Proximity | 1.79 (0.44-7.32) | 0.476 | ||
| Proximal colon | 5 (38.46) | 7 (25.93) | ||
| Distal colon | 8 (61.54) | 20 (74.07) | ||
| Staging | - | 0.020 | ||
| A | 0 (0.00) | 7 (25.93) | ||
| B | 4 (30.77) | 10 (37.04) | ||
| C | 7 (54.85) | 9 (33.33) | ||
| D | 2 (15.38) | 1 (3.70) | ||
| Group staging | 3.83 (0.93-15.72) | 0.116 | ||
| C-D | 9 (69.23) | 10 (37.04) | ||
| A-B | 4 (30.77) | 17 (62.96) | ||
| Family history | 9.20 (1.97-42.97) | 0.008 | ||
| Yes | 8 (61.54) | 4 (14.81) | ||
| No | 5 (38.46) | 23 (85.19) | ||
| Differentiation1 | ||||
| Poor | 4 (30.77) | 3 (11.54) | 2.52 (0.46-13.80) | 0.287 |
| Intermediate | 0 (0.00) | 6 (23.08) | - | 0.999 |
| Well | 9 (69.23) | 17 (65.38) |
CRC has numerous substantial risk factors in theory. However, within the 40 CRC group respondents, only three factors are deemed essential hereditarily: Age, tumor staging, and family history. Uniquely, among all locations and proximity potential for CRC, hereditary does not hold any significance (P = 0.595 and 0.476). There is also no difference on biopsies differentiation between the groups (P = 0.287 and 0.999).
The study found 5.60 times increase in risk of CRC between those < 50 years old and over, in which a 9.05-year difference is found between the subsequent groups. Similarly, respondents with hereditary CRC are mostly within the C stage (54.85%) while the non-group mostly in B (37.04%). This pattern holds true even when the stages are divided into C-D and A-B clusters, where the CRC group dominate the former cluster while the latter cluster for the rest. Yet, the clustering of stages is insignificant (P = 0.116).
Family history of CRC and its hereditary follow a significant linear relationship (P = 0.008). Those who has history of CRC in his/her family majorly belong to the hereditary group (61.54%) and vice versa. There is also a notable risk increase for those who has CRC history amounting to 9.20 times than those who don’t.
Bringing further to multivariate perspective, the current study applies multiple logistic analysis to find the truly significant risk factors toward CRC hereditary and its subsequent probability. Table 4 explain the regression where staging and family history are truly significant (P = 0.034 and 0.006), while age is just confounder. The unstandardized coefficients can be morphed to a LOGIT and probability functions of CRC hereditary as follows (Eq. 1 and 2).
| Factor | B | SE | P value | OR (95%CI) |
| Staging | 2.395 | 1.130 | 0.034 | 10.970 (1.199-100.382) |
| History | 3.126 | 1.143 | 0.006 | 22.784 (2.423-214.273) |
| Constant | -3.165 | 1.094 | 0.004 | 0.042 |
LOGIT = -3.165 + 2.395 × staging + 3.126 × history (Eq. 1)
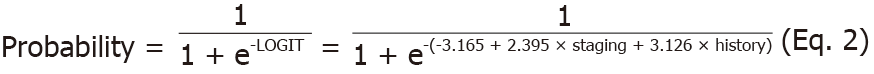
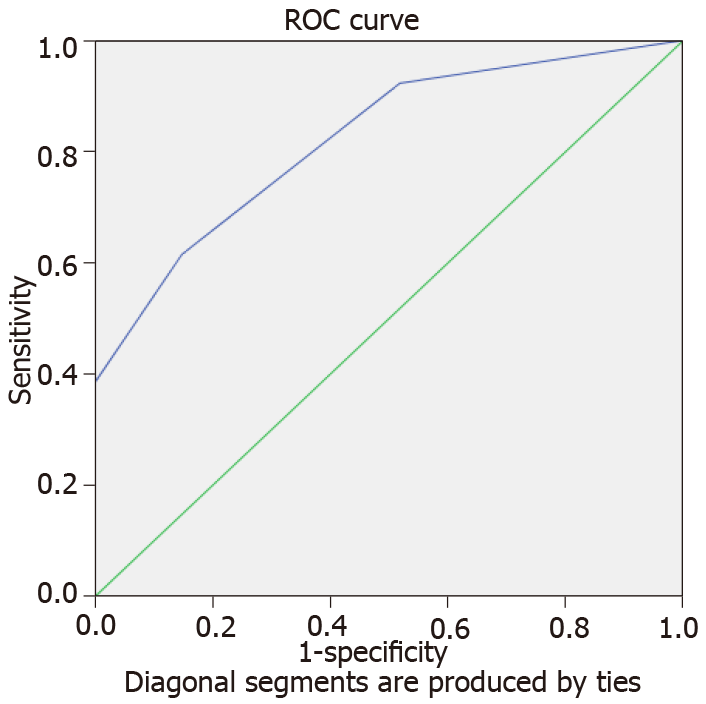
The variables of staging and history hold the value of either one or zero. Representatively, score of one amount to C or D in the staging component and presence of family history. Furthermore, the equations and probabilities are having good fit and not due to chance by having significant Hosmer and Lameshow statistics as well as 82.2% area under the curve (AUC) for receiving operator curve (ROC) (Figure 1).
From May 2018 to December 2019, 71 respondents were collected with 56.34% prevalence of CRC among the pre-elderly population (45-59 years old). The CRC group is predominantly male (52.5%) and classified with normal BMI. The discrepancy also found on similar studies in the Asia region (i.e., Japan, China, Korea, and Hong Kong) where CRC has 20.7-64.8 incidence rate over 100 thousand populations, in which differ by 6.3-28.1 compared to the female incidence rate[24-27]. The theory on hormonal difference between gender is suspected to be the leading cause of the CRC dominancy in male.
CRC higher prevalence in the male gender is the result of female protectiveness from the disease. The sex hormone of estradiol and progesterone acts as protective mechanism for CRC development in the body. The hormonal clinical trial in 2019 observe that introduction of estradiol and progesterone combination treatment provide increased apoptosis of tumor cells (P < 0.05) while lowering tumor cell proliferations (P < 0.01)[28]. Aside from its function as a sexual hormone, estrogen or estradiol play an important role in cell turnover. It acts as a bridge for ion transport that regulates cells’ pH, intracellular ions, and several protein activations[29]. Estrogen also exudes anti-inflammatory properties due to its ability to bond and modulate leukocytes including natural killer cell, neutrophils, dendritic cells, etc[30].
Moreover, associations are observed of body anthropometry to CRC within many global and Asian studies. For instance, a Japanese prospective study finds significant difference of BMI to colon cancer respondents (P = 0.004-0.007)[31]. Akin notion is discovered in a Korean study where every five cm increment in height increase the risk for CRC in men by 1.04-1.06 and in women by 1.00-1.08[32]. Both after multivariate control with other factors. Another Korean study also view significant association of BMI to CRC especially the distal colon and rectal cancer (P < 0.001 and 0.016)[33]. These trends are not restricted only to the Asian region. A global meta-analysis obtains a relative risk of 1.25 (1.18-1.32) for CRC with the highest vs lowest height[34]. Likewise, a prospective systematic review corroborates the idea where CRC’s risk multiply by 1.06 for every 5 kg/m2 increase in BMI[35].
BMI has indirect relations to CRC. Higher BMI equates to higher number of cells and tissues in the body, giving rise to higher chance of genetic mutations and malignancy. People with higher stature also found to have longer intestine length than others (r: 0.827), leading to more proliferation rate and chance of cancer[36]. Further, CRC can be influenced by other diseases. Acromegaly and insulin-like growth factor abnormality for example procure modulation in body height, BMI, and thus CRC[37-39].
With the advancement of technology in healthcare and information, genetic studies for diseases’ prevention, detection, and treatment have developed in a rapid pace. Specimens with DNA or RNA materials of the patients or family can be analyzed to account for the disease. This methodology has been implemented for several diseases[9]. Representatively, in assessing the hereditary status of a given CRC patient, his or her mRNA gene expression can be compared to a defined cut-off, where ≥ equates to positive status. Henceforth, the study uses 5th percentile cut-off points which are often used in M, C, D, and A statistic of circular data in a wrapped Cauchy distribution[40]. Although significant dissonance is present between blood and tissue samples in the CRC group, there are no significant difference between the first, third, and fifth percentile of MSH2 gene expression which leads to the acceptance of utilizing 5th percentile cut-off amounting to 11059 fc. Consequently, 32.5% of CRC patients are categorized to the hereditary group. The rate supports the discovery of 22% hereditary CRC by Chang et al[41].
Table 3 provide risk factors assessment between the hereditary and sporadic CRC groups. Significant risk factors fall on the age, staging, and family history. The sporadic CRC has older patients than the counterpart with ∆: 9.05 years and 77.78% proportion for those > 50 years old. Generally, age of 40 years old and over has significantly higher incidence of CRC[42]. A 2018 study paints that there is a sharp increase in CRC age-specific incidence as early as 35, then the pre-elderly age of 35-64 [∆: 60.2 (male) and ± 35 (female)], and over 65 years old [∆: 237.5 (male) and 131.4 (female)][24]. Nonetheless, Yurgelun et al[43] describe that LS mutation carriers have significantly younger age at CRC diagnosis with ∆: 11.1 years (P < 0.001)[43]. HNPCC specifically has only ± 20% probability to develop around the age of 50 and 50% for 70 years old or above[44].
Age is an unfortunate risk for malignancy. The older a person is getting, he or she accumulate a lot of endogenous factors (i.e., diet, chronic inflammation, metabolism, waning immune system, etc.) and exogenous factors (i.e., genotoxins, mutations, medications, environmental triggers, etc.) which stimulate oxidative stress and reactive oxygen species that initiate DNA damage, mutations, and uncontrolled cell growth[45]. The body proinflammatory state as time passes also become a progressive breeding ground for malignancy[46].
Insignificant relations are found in gender, tumor location, and histological differentiation. The hereditary CRC are predominated by female (53.85%) while the sporadic by male (55.56%) yet the difference is negligible (P = 0.826). This is consistent with earlier studies where Dominguez-Valentin et al[47] ascertain that in earlier years of elderly age, MSH2 carries similar risk of CRC in terms of gender[47]. Further, this ascertain that even though female is protected from overall CRC due to its hormonal effect, there is no importance to the heredity status of the CRC.
Both hereditary and sporadic CRCs tend to be in the distal colon (61.54% vs 74.07%), especially the rectum (38.46% vs 44.44%). Supremacy of CRCs in the rectum also seen in a 2020 general hospital study where rectal CRC amounts to 61.8% prevalence[48]. Yet the locations are inessential to the heredity status (P = 0.476). Theory upon this predicament include the intrinsic and extrinsic factors within everyone. Carethers[49] in his 2018 study disclose that the overall risk of CRC and the CRC risk of different regions of the colon are affiliated to one’s physical activity, gender, height, BMI, smoking status, alcohol intake, diabetes, medications, and hormonal therapy[49]. For example, physical activity reduces the overall CRC risk and the proximal colon CRC risk; while increase in height do not affect the rectal specific location while profoundly heightening risk of overall, proximal, and distal locations.
Likewise, no significance can be seen between histopathological differentiation and CRC heredity status. Current study observes higher poorly differentiated CRC in the hereditary compared to the sporadic group (30.77% vs 11.54%), which is akin to the study by Sun dictating that HNPCC features prominent lymphocyte infiltrations and RER+ status, which easily translates to poor differentiation and resulting in more within the HNPCC compared to the sporadic group[50]. On the contrary, the heredity group also has more well differentiated specimens (∆: 3.85%). The contrast may be due to amounts of proteins and cytokines within the tissue. The tissue staining with chromogranin A produce significant difference of 13.6% between hereditary and sporadic group[50]. The positive staining with after-mentioned stain has high correlation with tumor’s grade and stage[51,52]. Meanwhile, the dissonance may also happen due to defects on the sample when taken by colonoscopy biopsy as opposed to a surgery.
Independent staging of the CRC produces significant results between hereditary and sporadic groups (P = 0.020), with the former mostly in stage C (54.85%) and the latter in stage B (37.04%). The findings contradict data by Yurgelun et al[43], where most LS mutation carriers are in stage II (45.5%)[43]. Difference may occur due to the health system flaw in detecting cancer and the pathophysiology of the CRC.
HNPCC or LS is very hard to be detected as a cancer diagnosis must precede the genetic diagnosis. Even in the developed country of United States, only < 1% of the Americans with LS know about the disease presence[53,54]. The rate may lessen in Indonesia where technology is not as advanced in the United States, genetic testing is not a routine test and very expensive, as well as Indonesians’ tradition to not seek the healthcare center if there are no symptoms or still bearable.
Incidentally, family history has linear relationship to the hereditary vs sporadic type of CRC (P = 0.008). Hereditary CRC majorly has positive history patients (61.54%) while the sporadic group doesn’t (14.81%). The conditions amount to 9.20 (1.97-42.97) times increase of risk in developing hereditary CRC when one has family history of CRC. HNPCC is an autosomal dominant disease leading to its presence in every generation of the familial generation, as a dominant trait will always be expressed according to the mendelian law of inheritance. Simultaneously, similar relationship also observed in a 2017 LS study where LS mutation carriers have P < 0.001 for all first- and second-degree family history of CRC[43].
Multiple regression control of the factors demonstrates that staging and family history is truly significant (P = 0.034 and 0.006) while age just a confounder. The analysis then developed applicative equations (Eq. 1 and 2) to predict the heredity of CRC, where examples of their usage are listed in Table 5. The analysis has a satisfactory fit criteria with significant Lemeshow and adequate AUC.
Subsequently, the probability prediction model of the current study can be one of the prospective tools to overcome the weaknesses of the Amsterdam and Bethesda criteria. Personal family history and Mendelian family genogram are important for the diagnosis of Hereditary CRC, with the Amsterdam and Bethesda criteria being the standard diagnosis tools for LS. However, those tools often face difficulties, especially for smaller families and late age of disease onset[11-15,55,56]. In addition, individual specific genotype and environmental traits assessment may be utilized to overcome the hurdle of empirical recurrence risk removal because of its impracticality in incomplete penetrance and late onset[11-15]. Meanwhile, aside from the practical probability prediction model of the present study, mRNA MSH2 gene expression can be used through Bayesian theorem with prior pedigree risk modifications and conditional information.
Nevertheless, the limitation of the present study includes the un-generalization of the study sample. Participants are taken from the hospitals which indicates the possibility of selection bias and unrepresentativeness of the public. Future studies should determine whole genome sequencing to validate these findings and establish a gold standard for Hereditary CRC.
Numerous factors have significant relations to heredity of LS CRC patients. However, true important factors are staging and family history, while others (age) are confounders. The study also established a definite cut-off point for heredity LS CRC/HNPCC based on mRNA MSH2 expression, 11059 fc. These findings shall act as concrete foundations on further risk factors and/or genetical LS CRC future studies.
The lack of golden standard for categorizing hereditary status of colorectal cancer (CRC) poses diagnostic and management problems. Identifying proper techniques is urgent to procure the best care, prevention, risk factors management, and treatment of CRC be it hereditary or sporadic, along with judicious resource consumption.
The lack of golden standard leaves a gaping hole in the LS CRC healthcare system. Previous guideline of Bethesda and Amsterdam have tried yet fail in the applicability area especially with later age onset and smaller family. These coupled with the hazardous nature of CRC or lynch syndrome and scarce information on CRC risk factors identifications motivate the authors to commence the present study.
To determine the gold standard cut-off of MSH2 gene expression for hereditary cluster as well as to identify and examine the relationship of hereditary non-polyposis colon cancer (HNPCC) with its non-modifiable risk factors.
Consecutive sampling of the hospital internal medicine patients with CRC provides the case group. Then, the control group was concocted by matching the characteristics of the case group. MSH2 mRNA was then analyzed through blood and tissue collection and reverse transcription-polymerase chain reaction. Further, the gene expression cut-off determined using percentile technique akin to Cauchy distribution of M, C, A, and D circular data statistics. CRC groups then clustered into hereditary and sporadic according to the MSH2 gene expression against the cut-off. Lastly, risk factors are contrasted between each cluster and developed into a prediction model.
In a group of 40 CRCs differentiated into 13 hereditary and 27 sporadic through MSH2 mRNA cut-off in 11059 fc, significant risk factors for the hereditary CRC are family history and staging with (OR: 22.784, 95%CI: 2.423-214.273, P = 0.006; OR: 10.970, 95%CI: 1.199-100.382, P = 0.034). Moreover, a prediction model is concocted with area under the curve 82.2%.
The cut-off of MSH2 mRNA 5th percentile provided rough clustering of hereditary and sporadic CRC groups. Significant risk factors toward HNPCC are family history and staging, while age is just a confounder.
Future research directions include validation of the determined cut-off and reliability testing of the risk factors in a bigger sample size and/or with the general population. Further, a longitudinal study on the risk factors effects should be evaluated.
The authors highly appreciate Jonathan Salim, M.D. for his help and advice in the amendment of the manuscript.
Provenance and peer review: Invited article; Externally peer reviewed
Corresponding Author's Membership in Professional Societies: Indonesian Society of Gastroenterology.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Indonesia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gao F, Liu P S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Huck MB, Bohl JL. Colonic Polyps: Diagnosis and Surveillance. Clin Colon Rectal Surg. 2016;29:296-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Ewing I, Hurley JJ, Josephides E, Millar A. The molecular genetics of colorectal cancer. Frontline Gastroenterol. 2014;5:26-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 3. | World Health Organization. The Global Cancer Observatory. Very High HDI 2021. [cited 10 January 2021]. Available from: https://www.who.int/. |
| 4. | Sobocińska J, Kolenda T, Teresiak A, Badziąg-Leśniak N, Kopczyńska M, Guglas K, Przybyła A, Filas V, Bogajewska-Ryłko E, Lamperska K, Mackiewicz A. Diagnostics of Mutations in MMR/EPCAM Genes and Their Role in the Treatment and Care of Patients with Lynch Syndrome. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3270] [Article Influence: 654.0] [Reference Citation Analysis (2)] |
| 6. | Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 200] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 599] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 8. | Pitsikas P, Lee D, Rainbow AJ. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis. 2007;22:235-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Turnpenny PD, Elllard S. The Genetics of Cancer and Cancer Genetics. In: Emery’s Elements of Medical Genetics. 15th ed. United Kingdom: Elsevier, 2016: 177–199. |
| 10. | Wagner A, Tops C, Wijnen JT, Zwinderman K, van der Meer C, Kets M, Niermeijer MF, Klijn JG, Tibben A, Vasen HF, Meijers-Heijboer H. Genetic testing in hereditary non-polyposis colorectal cancer families with a MSH2, MLH1, or MSH6 mutation. J Med Genet. 2002;39:833-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Turnpenny PD, Elllard S. Pattern of Inheritance. In: Emery’s Elements of Medical Genetics. 15th ed. United Kingdom: Elsevier, 2016. |
| 12. | Nussbaum R, McInnes R, Willard H. Lynch Syndrome (DNA Mismatch Repair Gene Mutations, MM 120435). In: Thompson & Thompson Genetics in Medicine. 8th ed. Philadelphia: Elsevier, 2015. |
| 13. | Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1412] [Cited by in RCA: 1462] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 14. | Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418-2425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 391] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 15. | Kilpivaara O, Aaltonen LA. Diagnostic cancer genome sequencing and the contribution of germline variants. Science. 2013;339:1559-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 358] [Cited by in RCA: 333] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 17. | to: Personality and Problematic Internet Use Among Chinese College Students: The Mediating Role of Maladaptive Cognitions Over Internet Use by Zhou N, Geng X, Du H, Wu L, Xu J, Ma S, Zhang J, Yu C, Liang Y, Meng J, Yuan X, Cao H, and Fang X. Cyberpsychol Behav Soc Netw. 2018; 21(11):719-726. DOI: 10.1089/cyber.2018.0279. Cyberpsychol Behav Soc Netw. 2019;22:617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Kitazawa M, Hatta T, Sasaki Y, Fukui K, Ogawa K, Fukuda E, Goshima N, Okita N, Yamada Y, Nakagama H, Natsume T, Horimoto K. Promotion of the Warburg effect is associated with poor benefit from adjuvant chemotherapy in colorectal cancer. Cancer Sci. 2020;111:658-666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Ma Y, Yang W, Song M, Smith-Warner SA, Yang J, Li Y, Ma W, Hu Y, Ogino S, Hu FB, Wen D, Chan AT, Giovannucci EL, Zhang X. Type 2 diabetes and risk of colorectal cancer in two large U.S. prospective cohorts. Br J Cancer. 2018;119:1436-1442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 20. | Ali Khan U, Fallah M, Sundquist K, Sundquist J, Brenner H, Kharazmi E. Risk of colorectal cancer in patients with diabetes mellitus: A Swedish nationwide cohort study. PLoS Med. 2020;17:e1003431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (1)] |
| 21. | Zhou Z, Liu H, Wang C, Lu Q, Huang Q, Zheng C, Lei Y. Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep. 2015;5:15293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 22. | Hatta M, Surachmanto EE, Islam AA, Wahid S. Expression of mRNA IL-17F and sIL-17F in atopic asthma patients. BMC Res Notes. 2017;10:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Sirait RH, Hatta M, Ramli M, Islam AA, Arief SK. Systemic lidocaine inhibits high-mobility group box 1 messenger ribonucleic acid expression and protein in BALB/c mice after closed fracture musculoskeletal injury. Saudi J Anaesth. 2018;12:395-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Jung KW, Won YJ, Kong HJ, Lee ES; Community of Population-Based Regional Cancer Registries. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2015. Cancer Res Treat. 2018;50:303-316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 326] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 25. | Chen W, Sun K, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X, He J. Cancer incidence and mortality in China. Zhongguo Aizheng Yanjiu Zazhi. 2018;30:1-12. [RCA] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 705] [Article Influence: 100.7] [Reference Citation Analysis (2)] |
| 26. | Hong Kong Cancer Registry (HKCaR). Colorectal Cancer in 2016. [cited 10 January 2021]. Available from: https://www3.ha.org.hk/cancereg/. |
| 27. | National Cancer Registry (Ministry of Health L and W). Cancer Statistics in Japan. [cited 10 January 2021]. Available from: https://www.facs.org/quality-programs/cancer/ncdb/. |
| 28. | Sasso CV, Santiano FE, Campo Verde Arboccó F, Zyla LE, Semino SN, Guerrero-Gimenez ME, Pistone Creydt V, López Fontana CM, Carón RW. Estradiol and progesterone regulate proliferation and apoptosis in colon cancer. Endocr Connect. 2019;8:217-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Maingi JW, Tang S, Liu S, Ngenya W, Bao E. Targeting estrogen receptors in colorectal cancer. Mol Biol Rep. 2020;47:4087-4091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | McCarthy M, Raval AP. The peri-menopause in a woman's life: a systemic inflammatory phase that enables later neurodegenerative disease. J Neuroinflammation. 2020;17:317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 31. | Shimizu N, Nagata C, Shimizu H, Kametani M, Takeyama N, Ohnuma T, Matsushita S. Height, weight, and alcohol consumption in relation to the risk of colorectal cancer in Japan: a prospective study. Br J Cancer. 2003;88:1038-1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Sung J, Song YM, Lawlor DA, Smith GD, Ebrahim S. Height and site-specific cancer risk: A cohort study of a korean adult population. Am J Epidemiol. 2009;170:53-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Shin A, Joo J, Bak J, Yang HR, Kim J, Park S, Nam BH. Site-specific risk factors for colorectal cancer in a Korean population. PLoS One. 2011;6:e23196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 34. | Song X, Gong X, Zhang T, Jiang W. Height and risk of colorectal cancer: a meta-analysis. Eur J Cancer Prev. 2018;27:521-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Abar L, Vieira AR, Aune D, Sobiecki JG, Vingeliene S, Polemiti E, Stevens C, Greenwood DC, Chan DSM, Schlesinger S, Norat T. Height and body fatness and colorectal cancer risk: an update of the WCRF-AICR systematic review of published prospective studies. Eur J Nutr. 2018;57:1701-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 36. | Minko E, Pagano A, Caceres N, Adar T, Márquez S. Human intestinal tract length and relationship with body height. FASEB J. 2014;28. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 37. | Dworakowska D, Grossman AB. Colonic Cancer and Acromegaly. Front Endocrinol (Lausanne). 2019;10:390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 38. | Murphy N, Carreras-Torres R, Song M, Chan AT, Martin RM, Papadimitriou N, Dimou N, Tsilidis KK, Banbury B, Bradbury KE, Besevic J, Rinaldi S, Riboli E, Cross AJ, Travis RC, Agnoli C, Albanes D, Berndt SI, Bézieau S, Bishop DT, Brenner H, Buchanan DD, Onland-Moret NC, Burnett-Hartman A, Campbell PT, Casey G, Castellví-Bel S, Chang-Claude J, Chirlaque MD, de la Chapelle A, English D, Figueiredo JC, Gallinger SJ, Giles GG, Gruber SB, Gsur A, Hampe J, Hampel H, Harrison TA, Hoffmeister M, Hsu L, Huang WY, Huyghe JR, Jenkins MA, Keku TO, Kühn T, Kweon SS, Le Marchand L, Li CI, Li L, Lindblom A, Martín V, Milne RL, Moreno V, Newcomb PA, Offit K, Ogino S, Ose J, Perduca V, Phipps AI, Platz EA, Potter JD, Qu C, Rennert G, Sakoda LC, Schafmayer C, Schoen RE, Slattery ML, Tangen CM, Ulrich CM, van Duijnhoven FJB, Van Guelpen B, Visvanathan K, Vodicka P, Vodickova L, Vymetalkova V, Wang H, White E, Wolk A, Woods MO, Wu AH, Zheng W, Peters U, Gunter MJ. Circulating Levels of Insulin-like Growth Factor 1 and Insulin-like Growth Factor Binding Protein 3 Associate With Risk of Colorectal Cancer Based on Serologic and Mendelian Randomization Analyses. Gastroenterology. 2020;158:1300-1312.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 39. | Kasprzak A. Insulin-Like Growth Factor 1 (IGF-1) Signaling in Glucose Metabolism in Colorectal Cancer. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 40. | Abuzaid AH, El-hanjouri MM, Kulab MM. On Discordance Tests for the Wrapped Cau-chy Distribution. Open J Stat. 2015;5:245-253. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, Welton M, Shelton A, Ma L, Arber DA. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25:1128-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 260] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 42. | Noone A, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review 1975-2015. National Cancer Institute; 2018. [cited 10 January 2021]. Available from: https://www.researchgate.net/publication/285863025_SEER_Cancer_statistics_review_1975-2004. |
| 43. | Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, Ukaegbu CI, Brais LK, McNamara PG, Mayer RJ, Schrag D, Meyerhardt JA, Ng K, Kidd J, Singh N, Hartman AR, Wenstrup RJ, Syngal S. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. J Clin Oncol. 2017;35:1086-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 371] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 44. | Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, Guimbaud R, Buecher B, Bignon YJ, Caron O, Colas C, Noguès C, Lejeune-Dumoulin S, Olivier-Faivre L, Polycarpe-Osaer F, Nguyen TD, Desseigne F, Saurin JC, Berthet P, Leroux D, Duffour J, Manouvrier S, Frébourg T, Sobol H, Lasset C, Bonaïti-Pellié C; French Cancer Genetics Network. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304-2310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 794] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 45. | Ahmad S, Akhter F, Shahab U, Rafi Z, Khan MS, Nabi R, Ahmad K, Ashraf JM, Moinuddin. Do all roads lead to the Rome? Semin Cancer Biol. 2018;49:9-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Rea IM, Gibson DS, McGilligan V, McNerlan SE, Alexander HD, Ross OA. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front Immunol. 2018;9:586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 658] [Cited by in RCA: 837] [Article Influence: 119.6] [Reference Citation Analysis (0)] |
| 47. | Dominguez-Valentin M, Sampson JR, Seppälä TT, ten Broeke SW, Plazzer J-P, Nakken S, Engel C, Aretz S, Jenkins MA, Sunde L, Bernstein I, Capella G, Balaguer F, Thomas H, Evans DG, Burn J, Greenblatt M, Hovig E, Cappel WHDVTN, Sijmons RH, Bertario L, Tibiletti MG, Cavestro GM, Lindblom A, Valle AD, Lopez-Köstner F, Gluck N, Katz LH, Heinimann K, Vaccaro CA, Büttner R, Görgens H, Holinski-Feder E, Morak M, Holzapfel S, Hüneburg R, Doeberitz MVK, Loeffler M, Rahner N, Schackert HK, Steinke-Lange V, Schmiegel W, Vangala D, Pylvänäinen K, Renkonen-Sinisalo L, Hopper JL, Win AK, Haile RW, Lindor NM, Gallinger S, Marchand LL, Newcomb PA, Figueiredo JC, Thibodeau SN, Wadt K, Therkildsen C, Okkels H, Ketabi Z, Moreira L, Sánchez A, Serra-Burriel M, Pineda M, Navarro M, Blanco I, Green K, Lalloo F, Crosbie EJ, Hill J, Denton OG, Frayling IM, Rødland EA, Vasen H, Mints M, Neffa F, Esperon P, Alvarez K, Kariv R, Rosner G, Pinero TA, Gonzalez ML, Kalfayan P, Tjandra D, Winship IM, Macrae F, Möslein G, Mecklin J, Nielsen M, Møller P. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15-25. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 48. | Anthonysamy MA, Maker LPLI, Gotra IM, Saputra H. Prevalence of colorectal carcinoma based on microscopic type, sex, age and anatomical location in Sanglah General Hospital. Intisari Sains Medis. 2020;11:272-276. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 49. | Carethers JM. Risk factors for colon location of cancer. Transl Gastroenterol Hepatol. 2018;3:76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Sun MH. Neuroendocrine differentiation in sporadic CRC and hereditary nonpolyosis colorectal cancer. Dis Markers. 2004;20:283-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 51. | Jesinghaus M, Konukiewitz B, Keller G, Kloor M, Steiger K, Reiche M, Penzel R, Endris V, Arsenic R, Hermann G, Stenzinger A, Weichert W, Pfarr N, Klöppel G. Colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas are genetically closely related to colorectal adenocarcinomas. Mod Pathol. 2017;30:610-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 52. | Herold Z, Dank M, Herold M, Nagy P, Rosta K, Somogyi A. Histopathological Chromogranin A-Positivity Is Associated with Right-Sided Colorectal Cancers and Worse Prognosis. Cancers (Basel). 2020;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Butterworth AS, Higgins JP, Pharoah P. Relative and absolute risk of colorectal cancer for individuals with a family history: a meta-analysis. Eur J Cancer. 2006;42:216-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 289] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 54. | Hampel H, de la Chapelle A. The search for unaffected individuals with Lynch syndrome: do the ends justify the means? Cancer Prev Res (Phila). 2011;4:1-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 55. | Guttmacher AE, Collins FS, Carmona RH. The family history--more important than ever. N Engl J Med. 2004;351:2333-2336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 333] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 56. | Goecke T, Schulmann K, Engel C, Holinski-Feder E, Pagenstecher C, Schackert HK, Kloor M, Kunstmann E, Vogelsang H, Keller G, Dietmaier W, Mangold E, Friedrichs N, Propping P, Krüger S, Gebert J, Schmiegel W, Rueschoff J, Loeffler M, Moeslein G; German HNPCC Consortium. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol. 2006;24:4285-4292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |