Published online May 12, 2020. doi: 10.4291/wjgp.v11.i3.64
Peer-review started: December 30, 2019
First decision: January 13, 2020
Revised: February 27, 2020
Accepted: March 30, 2020
Article in press: March 30, 2020
Published online: May 12, 2020
Processing time: 133 Days and 12.3 Hours
The challenges for inflammatory bowel disease (IBD) diagnostics are to discriminate it from gut conditions with similar symptoms such as irritable bowel syndrome (IBS), to distinguish IBD subtypes, to predict disease progression, and to establish the risk to develop colorectal cancer (CRC). Alterations in gut microbiota have been proposed as a source of information to assist in IBD diagnostics. Faecalibacterium prausnitzii (F. prausnitzii), its phylogroups, and Escherichia coli (E. coli) have been reported as potential biomarkers, but their performance in challenging IBD diagnostic situations remains elusive. We hypothesize that bacterial biomarkers based in these species may help to discriminate these conditions of complex diagnostics.
To evaluate the usefulness of indices calculated from the quantification of these species as biomarkers to aid in IBD diagnostics.
A retrospective study of 131 subjects (31 controls (H); 45 Crohn’s disease (CD), 25 ulcerative colitis (UC), 10 IBS, and 20 CRC patients) was performed to assess the usefulness of bacterial biomarkers in biopsies. Further, the performance of biomarkers in faeces was studied in 29 stool samples (19 CD, 10 UC). Relative abundances of total F. prausnitzii (FP), its phylogroups (PHGI and PHGII), and E. coli (E) quantification were determined by qPCR. Loads were combined to calculate the FP-E index, the PHGI–E index and the PHGII-E index. Biomarkers accuracy to discriminate among conditions was measured by the area under the receiver operating characteristic curve (AUC).
In biopsies, FP-E index was good for discriminating IBS from CD (AUC = 0.752) while PHGII-E index was suitable for discriminating IBS from UC (AUC = 0.632). The FP-E index would be the choice to discriminate IBD from CRC, especially from all UC subtypes (AUC ≥ 0.875), regardless of the activity status of the patient. Discrimination between UC patients that had the longest disease duration and those with CRC featured slightly lower AUC values. Concerning differentiation in IBD with shared location, PHGI-E index can establish progression from proctitis and left-sided colitis to ulcerative pancolitis (AUC ≥ 0.800). PHG I-E index analysis in tissue would be the choice to discriminate within IBD subtypes of shared location (AUC ≥ 0.712), while in non-invasive faecal samples FP or PHGI could be good indicators (AUC ≥ 0.833).
F. prausnitzii phylogroups combined with E. coli offer potential to discriminate between IBD and CRC patients and can assist in IBD subtypes classification, which may help in solving IBD diagnostics challenges.
Core tip: This manuscript evaluates the usefulness of new indexes calculated from the quantification of Faecalibacterium prausnitzii, its phylogroups, and Escherichia coli as biomarkers to assist in challenges of inflammatory bowel disease diagnostics. Firstly, discrimination between inflammatory bowel disease and other intestinal disorders was tested. We present indices to distinguish colorectal cancer from inflammatory bowel disease, especially from subjects with ulcerative colitis. This is of significance given the association between chronic inflammation and the risk of colorectal cancer. In contrast, the proposed indices featured limited performance for discriminating inflammatory bowel disease from irritable bowel syndrome. Secondly, we approach if these biomarkers would be useful to discriminate within inflammatory bowel disease subtypes. We show here good biomarkers to differentiate inflammatory bowel disease subtypes of shared disease location, which may assist in monitoring the risk of progression of the inflamed area. Their application in non-invasive faecal samples is also demonstrated.
- Citation: Lopez-Siles M, Aldeguer X, Sabat-Mir M, Serra-Pagès M, Duncan SH, Flint HJ, Garcia-Gil LJ, Martinez-Medina M. Evaluation of bacterial biomarkers to aid in challenging inflammatory bowel diseases diagnostics and subtype classification. World J Gastrointest Pathophysiol 2020; 11(3): 64-77
- URL: https://www.wjgnet.com/2150-5330/full/v11/i3/64.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v11.i3.64
Inflammatory bowel diseases (IBD) are chronic inflammatory bowel disorders of unknown aetiology that follow a course with periods of activity or flare-ups and periods of remission[1-4]. Crohn’s disease (CD) and ulcerative colitis (UC) are the main idiopathic IBD[5-7]. Despite these disorders differing in location, histology, and distribution of inflamed areas, sometimes they feature overlapping clinical and pathological characteristics that hamper a distinct classification[8,9]. It is essential to discriminate both entities to establish an appropriate treatment strategy[10]. Besides, there are other intestinal disorders, such as irritable bowel syndrome (IBS), that share symptoms similar to those observed in the early stages of IBD thus increasing its likelihood of misdiagnosis[11,12]. In contrast, chronic inflammation can lead to tumour formation and promote colorectal cancer (CRC) development. It would, therefore, be interesting to have a biomarker for IBD-progression to CRC, but currently, there is a lack of tools to predict which cases may progress to CRC. Altogether, current IBD diagnostics challenges are to discriminate phenotype variations within IBD accurately, but also to differentiate IBD from other gut conditions with milder or worsening phenotypes.
Given the absence of pathognomonic features, IBD diagnosis currently involves a comprehensive examination of the patient that includes clinical, endoscopic, radiologic, and histological criteria. Besides, as clinical manifestations of IBD are unstable during the disease course, a long monitoring period is needed to classify the disease phenotype accurately[11,15]. As IBD patients feature an imbalanced gut microbial community in comparison to healthy subjects[16-25], in the last years the implementation of bacteria representative of this dysbiosis as biomarkers has been started to be explored as a novel strategy to support IBD diagnostics and/or prognostics[23,26-30].
We and others have pointed out that the abundance of faecal or mucosa-associated Faecalibacterium prausnitzii (F. prausnitzii) is a potential biomarker to discriminate between gut disorders[23,26-30] Moreover, F. prausnitzii in conjunction with Escherichia coli (E. coli) abundance (FP-E index) has been proven to be a better biomarker than total F. prausnitzii alone[26,29]. Besides, the quantification of F. prausnitzii phylogroups I (PHG I) and II (PHGII) has been proposed as a source of additional information to discriminate between IBD subtypes. However, the usefulness of an index using the quantification of the phylogroups in conjunction with E. coli remains to be explored. Also, there is a lack of comparative studies from a methodological aspect that would allow the establishment of the biomarker of choice.
It is against this background that we examined six options for biomarkers (F. prausnitzii, the two phylogroups or the combination of these three with E. coli) in a cohort of non-IBD controls (H), IBS, IBD and CRC subjects, to (1) establish which would be the best parameter to discriminate IBD patients from H and IBS subjects; (2) determine which would be the best parameter to discriminate IBD from CRC patients; and (3) identify which would be the most accurate parameter to discriminate within IBD subtypes by location. We hypothesize that bacterial biomarkers based in these species may be of help to discriminate these conditions of complex diagnostics.
In this study, data from two groups of subjects were included. Firstly, biomarker performance was tested in biopsy samples. We hypothesized that given the inflammatory nature of IBD, to look for biomarkers in the tissue would be strongly associated with disease course. Secondly, the usefulness of selected biomarkers was assessed in non-invasive samples (i.e., stools).
To test the performance of the mucosa-associated bacterial biomarkers, a re-analysis of the data from a Spanish cohort including IBD, IBS, CRC, and H was performed (Table 1). Subjects were consecutively recruited by the Department of Gastroenterology at the Hospital Universitari Dr. Josep Trueta (Girona, Spain) and the Gastroenterology Unit at the Hospital Santa Caterina (Institut d’Assistència Sanitària of Girona, Salt, Spain) between May 2009 and November 2010. Patients were gender- and age-matched, except CD patients who were significantly younger than those in the H and IBS groups (P < 0.001) (Table 1). During routine endoscopy, up to three biopsy samples per patient were taken from different locations along the gut (Table 2) following standard procedures.
| Healthy1 | Irritable bowel syndrome | IBD | Colorectal cancer | P value3 | |||
| Ulcerative colitis | Crohn’s disease | ||||||
| n (patients) | 31 | 10 | 25 | 45 | 20 | ||
| Cohort of subjects for biopsies samples collection | Age (mean ± SD, yr) | 48.1 ± 16.3 | 42.4 ± 11.4 | 40.1 ± 15.8 | 33.5 ± 11.1 | 58.6 ± 7.52 | < 0.0014 |
| Male, n (%) | 16 (51.6) | 2 (20.0) | 16 (64.0) | 26 (57.7) | 14 (70.0%) | 0.6056 | |
| Active, n (%) | NA | NA | 20 (80.0) | 28 (62.2) | NA | 0.1005 | |
| Treatment, n (%)2 | |||||||
| No treatment | NA | NA | 16 (64.0) | 17 (37.8) | NA | ||
| Moderate immunosuppressant | NA | NA | 3 (12.0) | 17 (37.8) | NA | ||
| Anti-TNFα | NA | NA | 4 (16.0) | 10 (22.2) | NA | ||
| UC location, n (%)2 | NA | ||||||
| Ulcerative proctitis (E1) | NA | NA | 6 (24.0) | NA | NA | ||
| Distal UC (E2) | NA | NA | 11 (44.0) | NA | NA | ||
| Extensive UC or ulcerative pancolitis (E3) | NA | NA | 6 (24.0) | NA | NA | ||
| CD location, n (%)2 | NA | ||||||
| Ileal-CD (L1) | NA | NA | NA | 19 (42.2) | NA | ||
| Colonic-CD (L2) | NA | NA | NA | 11 (24.4) | NA | ||
| Ileocolonic-CD (L3) | NA | NA | NA | 14 (31.1) | NA | ||
| Cohort of subjects for faecal samples collection | n (patients) | 10 | 19 | ||||
| Age (mean ± SD, yr) | 47.4 ± 18.3 | 43.5 ± 18.3 | 0.4294 | ||||
| Male, n (%) | 5 (50.0) | 10 (52.6) | 0.8936 | ||||
| Active, n (%)2 | 1 (10) | 7 (36.8) | 0.1855 | ||||
| Treatment, n (%)2 | NA | NA | 5 (50) | 5 (26.3) | NA | ||
| No treatment | NA | NA | 1 (10) | 2 (10.5) | NA | ||
| Moderate immunosuppres-sant | NA | NA | 3 (30) | 12 (63.2) | NA | ||
| Anti-TNFα | |||||||
| UC location, n (%)2 | NA | ||||||
| Distal UC (E2) | 3 (30.0) | NA | |||||
| Extensive UC or ulcerative pancolitis (E3) | 7 (70.0) | NA | |||||
| CD location, n (%) | |||||||
| Ileal-CD (L1) | NA | 10 (52.6) | NA | ||||
| Colonic-CD (L2) | NA | 3 (15.8) | |||||
| Ileocolonic-CD (L3) | NA | 6 (31.6) | |||||
| No. Patients | No. biopsies | |||||
| Terminal ileum | Transverse colon | Rectum | Unknown region | Total | ||
| H | 31 | 14 | 24 | 10 | 0 | 48 |
| IBS | 10 | 1 | 3 | 3 | 12 | 19 |
| CRC | 20 | 3 | 17 | 0 | 20 | |
| UC | 25 | 11 | 23 | 16 | 0 | 50 |
| Location | ||||||
| Ulcerative proctitis (E1) | 6 | 4 | 5 | 5 | 0 | 14 |
| Distal UC (E2) | 11 | 3 | 11 | 8 | 0 | 22 |
| Extensive UC or ulcerative pancolitis (E3) | 6 | 3 | 6 | 1 | 0 | 10 |
| CD | 45 | 16 | 31 | 16 | 0 | 63 |
| Location | ||||||
| Ileal-CD (L1) | 19 | 5 | 13 | 7 | 0 | 25 |
| Colonic-CD (L2) | 11 | 6 | 7 | 4 | 0 | 17 |
| Ileocolonic-CD (L3) | 14 | 4 | 10 | 4 | 0 | 18 |
To test the performance of bacterial biomarkers in faecal samples, a cohort consisting of 29 IBD (19 CD and 10 UC) patients was recruited by the Gastroenterology Services of the Hospital Universitari Dr. Josep Trueta (Girona, Spain) between March 2014 and May 2015. Subjects were age- and gender-matched for both the groups (Table 1). Participants were asked to collect a stool sample from one bowel movement in a sterile faecal collection container. Subjects brought samples to the hospital, where they were stored at −80ºC until DNA extraction was performed.
To control bias between centres, patients with IBD were diagnosed according to standard clinical, pathological, and endoscopic criteria and categorized according to the Montreal classification. Patients with IBS were diagnosed according to Rome III criteria (available at http://www.romecriteria.org/criteria/). CRC diagnosis was established by colonoscopy and biopsy examination, and none of the subjects underwent radiotherapy, chemotherapy or surgery. The control group consisted of subjects with normal colonoscopy who underwent this procedure for different reasons (Table 1). Clinically relevant data of all participants, such as age, gender, and disease activity at sampling, were collected (Table 1). Active CD were defined as those with CDAI > 150 whereas active UC patients had a Mayo score > 3.
Individuals included in this study were > 18 years old, did not have any other intestinal disease, and were not pregnant. Antibiotic treatment within the last month prior to sample collection was the only exclusion criterion. None of the subjects received probiotics before sample collection.
For biopsies, sample treatment and DNA extraction were performed as reported previously[26,27]. For faeces, 200-500 mg of faecal material were used for bacterial DNA extraction and purification with the NucleoSpin® Soil (Macherey-Nagel) and following the instructions from the manufacturer.
Previously designed and optimized 16S rRNA gene-targeted primers and probes were used for total F. prausnitzii[26], phylogroups, E. coli and total bacterial quantification using quantitative polymerase chain reaction (qPCR). Human cell numbers were determined with the control kit RT-CKFT-18S (Eurogentec, Belgium) according to the manufacturer’s instructions.
Amplification reactions were performed as described elsewhere[26,27,35,36]. In brief, quantifications were performed in a total volume of 20 μL reactions containing: 1× TaqMan Universal PCR Master Mix 2× (Applied Biosystems, Foster City, CA, United States), 900 nmol/L of each primer, 300 nmol/L of each probe, and up to 50 ng of genomic DNA template. Samples were run in duplicate in the same plate. For data analysis, the mean of the duplicate quantifications was used. Duplicates were considered valid if the standard deviation between quantification cycles (Cq) was <0.34 (i.e., a difference of < 10% of the quantity was tolerated). Quantification controls consisting of at least 5 reactions with a known number of target genes were performed to assess inter-run reproducibility. For samples with undetected values during quantification, the number of 16S rRNA gene copies equivalent to the detection limit of each reaction was used. Inhibition of total F. prausnitzii quantification was controlled by adding 103 copies of an internal amplification control (IAC) template to each reaction. It was considered that there was no inhibition if the obtained Cq was < 0.34 from those obtained when quantifying the IAC alone for any of the replicates. A non-template control (consisting of a reaction without F. prausnitzii DNA) and a non-amplification control (which did not contain any DNA template, either bacterial or IAC) were also included in each run. Negative controls resulted in undetectable Cq values in all cases.
All quantitative PCRs were performed using a 7500 Real Time PCR system (Applied Biosystems). The thermal profile was: a first step at 50 ºC during 2 min for amperase treatment, followed by a 95 ºC hold for 10 min to denature DNA and activate Ampli-Taq Gold polymerase, and a further 40 cycles consisting of a denaturation step at 95 ºC for 15 seconds followed by an annealing and extension step at 60 ºC (or at 64ºC for phylogroups quantification) for 1 min. Data were collected and analysed using the 7500 SDS system software version 1.4 (Applied Biosystems). All quantifications were performed under average PCR efficiencies of 89.51 ± 7.06%.
The sample size was defined after the number of patients analysed in similar studies of bacterial abundance in subjects suffering of these conditions[20,22,26,28].
Relative abundances of total F. prausnitzii, phylogroups, and E. coli copy numbers were calculated by normalizing each species load for the total bacterial 16S rRNA gene copies. Data are given as the log10 of the ratio between 16S rRNA gene copies of the target microorganism and millions of total bacterial 16S rRNA genes detected in the same sample.
For biopsies, species relative abundances were combined to calculate the FP-E index as previously reported[26]. Similarly, the PHGI–E index and the PHGII-E index were calculated as follows:
PHGI-E index = [log10 (PHGI/Hc)- log10 (E/Hc)]/ [log10 (TB/Hc)]
PHGII-E index = [log10 (PHGII/Hc)- log10 (E/Hc)]/ [log10 (TB/Hc)]
Being PHGI and PHGII the 16S rRNA gene copies of F. prausnitzii phylogroup I or II respectively; E the 16S rRNA gene copies of E. coli; Hc a million of human cells; and TB a million of 16S rRNA gene copies of total bacteria.
For faecal samples, as no Hc quantification was performed to normalize sample size, indexes were calculated as:
FP-E index = log10 (total F. prausnitzii/E)
PHGI-E index = log10 (PHGI/E)
PHGII-E index = log10 (PHGII/E)
Differences in categorical variables such as gender were assessed by the χ2 test. For continuous variables such as age or biomarkers load, data normality was assessed through the KolmogorovâSmirnov test. The non-parametric Kruskal-Wallis test was used to asses differences in variables with more than two categories, such as diagnostics, and CD or UC disease location. Pairwise comparisons of subcategories of these variables were analysed using a Mann-Whitney U test. This test was also used to compare, within a subgroup of patients, variables with two categories.
The receiver operating characteristic curve analysis, a plot of the true-positive rate (sensitivity) versus false-positive rate (1-specificity), was applied to establish the usefulness of F. prausnitzii, along with each phylogroup, alone or in conjunction to E. coli counts (FP-E index, PHGI-E index, and PHGII-E index) to distinguish different intestinal conditions. The accuracy of discrimination was measured by the area under the receiver operating characteristic curve (AUC). An AUC approaching 1 indicates that the test is highly sensitive and highly specific, whereas an AUC approaching 0.5 indicates that the test is neither sensitive nor specific. For the best cut-off value, specificity and sensitivity were established.
All the statistical analyses were performed using the SPSS 15.0 statistical package (LEAD Technologies, Inc.). Significance levels were established for P values ≤ 0.05.
The statistical methods of this study were reviewed by MSc. Oliver Valero Coppin from the Statistical Service at the Universitat Autònoma de Barcelona.
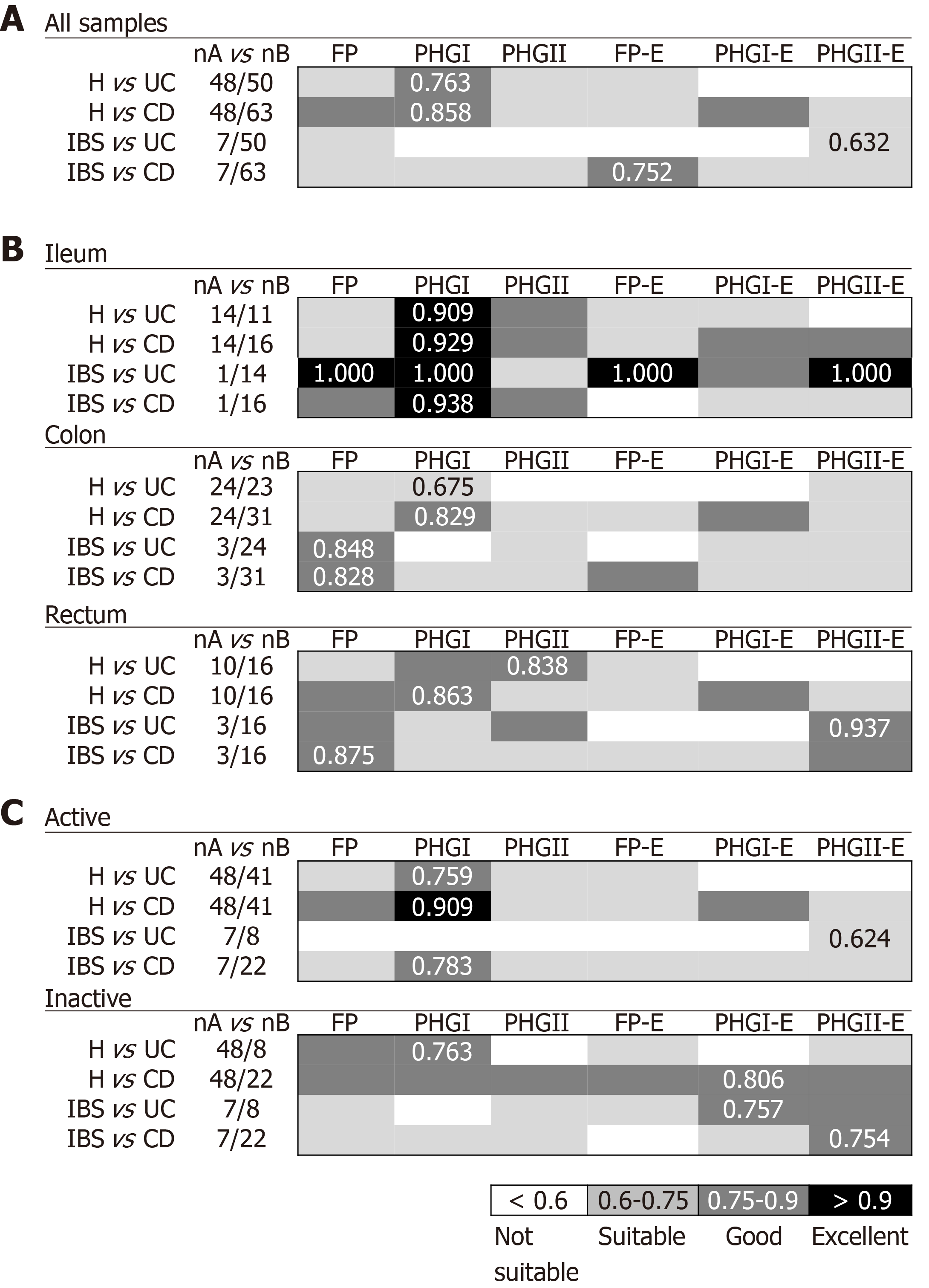
When considering all biopsy samples (Figure 1A), PHGI quantification was the most discriminative biomarker between H and IBD patients (AUC > 0.75). This can be attributed to higher load of PHGI in H in comparison to the other groups of subjects (Supplementary Table 1). Notably, discrimination was especially good between H and subjects with CD, which achieved 73% specificity and 91% sensitivity at the best cut-off value (Log10[16S rRNA phylogroup I/106 16S rRNA total bacteria] =2.3). This discrimination was particularly accurate when analysing ileal samples (AUC > 0.9) (Figure 1B). Besides, discrimination between H and IBD subjects achieved greater AUC values when considering only active IBD patients (Figure 1C). However, the discrimination was still good (AUC > 0.75) when taking into account only those with inactive disease. Therefore, our results support PHGI as an indicator of healthy gut status.
Regarding discrimination between IBD and IBS patients, different biomarkers performed best to distinguish IBS from UC or CD. When pooling all biopsy samples (Figure 1A), PHGII-E index was suitable to discriminate IBS from UC. This discrimination was excellent when considering ileal or rectal biopsies and suitable for colonic biopsies analyses (Figure 1B). It is of note that the PHGII-E index allowed good discrimination between these two conditions, even in the inactive cohort of patients (Figure 1C). In contrast, FP-E index was good for discriminating IBS from CD when pooling all samples, although this was not sustained for all sampled locations, probably due to the effect of the location of inflammation in CD. In contrast, FP was the best biomarker to discriminate IBS and CD in colonic and rectal samples, whereas PHGI counts discriminated best at the ileum (Figure 1B). This biomarker was good to discriminate IBS from active CD patients, whereas the PHGII-E index provided the best discrimination between IBS and inactive CD. Overall, to select a general biomarker to discriminate IBS from IBD, useful in all kinds of samples and conditions was challenging. However, FP could be an interesting candidate as performed in the suitable-excellent AUC range for all comparisons, regardless of the intestinal region selected for analysis.
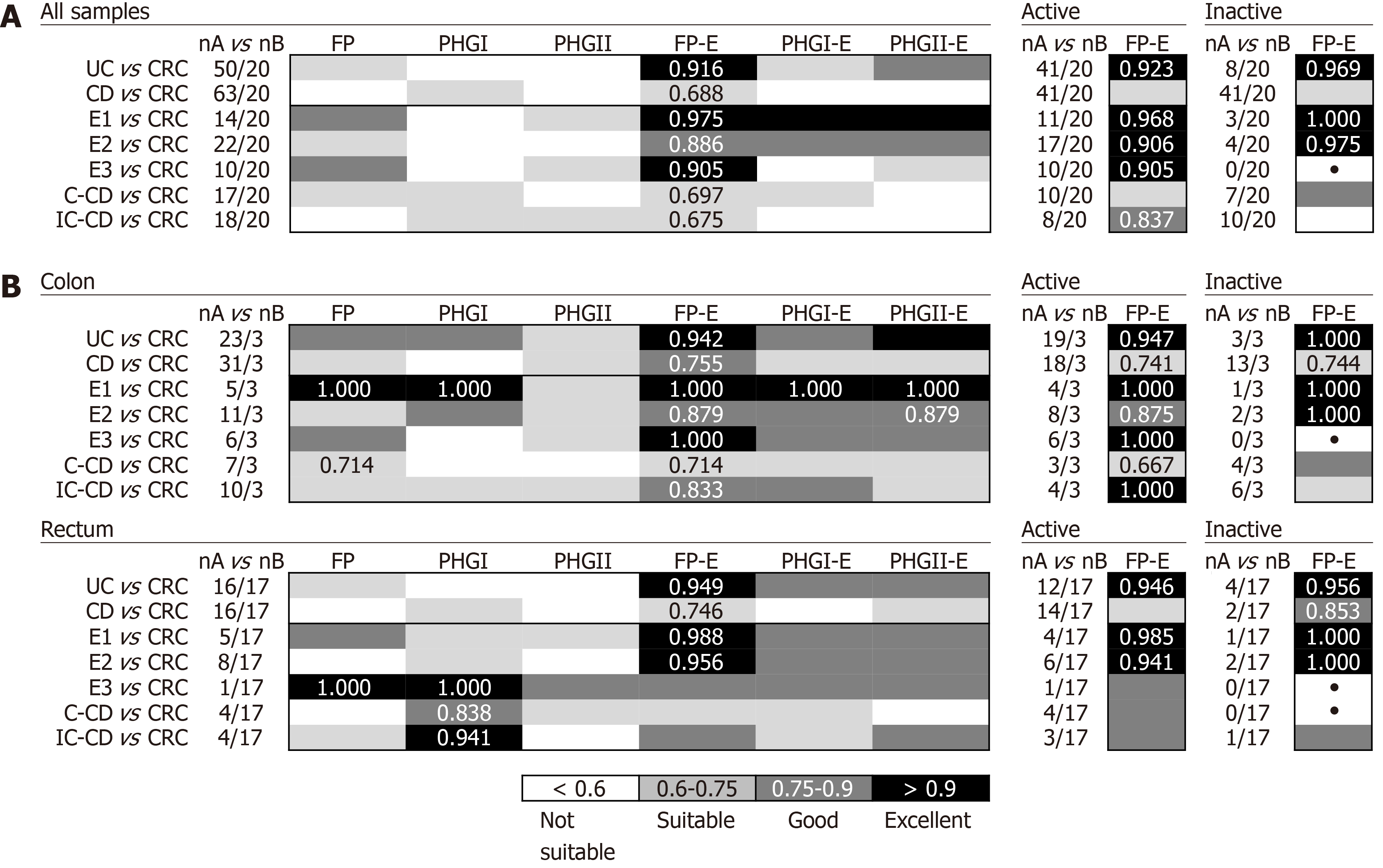
In general, the FP-E index was the most discriminatory between CRC and IBD patients when taking into account all biopsy samples together (Figure 2A), because lower FP-E values were associated with CRC subjects (Supplementary Table 1). Notably, discrimination was especially good between CRC and UC patients, which achieved 85% specificity and 94% of sensitivity at the best cut-off value (FP-E index= 0.009). This discrimination was excellent between CRC and patients with ulcerative proctitis (E1) and ulcerative pancolitis (E3) with 85% specificity and 100% sensitivity, while for patients with extensive UC (E2) sensitivity was reduced to 86% with the same specificity rate. Although good discrimination was achieved (AUC > 0.870) we observed that discern between E2 patients and those with CRC featured slightly lower AUC values, and in turn, these groups of patients had the longest disease duration (mean years of disease duration ± SD by UC subtype was: E1 = 0.93 ± 1.69; E2 = 7.10 ± 4.27; E3 = 2.63 ± 2.20.
In addition, this excellent discrimination was sustained regardless of the activity status of the patients. Regarding the location of sample (Figure 2B), for our particular cohort, colonic biopsies were the most discriminatory between CRC and those patients with E1 and E3, although good separation of groups was also achieved with rectal samples. In turn, rectal samples performed better to discriminate between E2 subjects and those with CRC.
The FP-E index was also suitable to classify CRC patients and those with CD of colonic location (i.e., C-CD and IC-CD). Interestingly, better AUC values were obtained for PHGI and when considering rectal samples alone, which needs further confirmation given the low number of samples analysed at this location for CD patients.
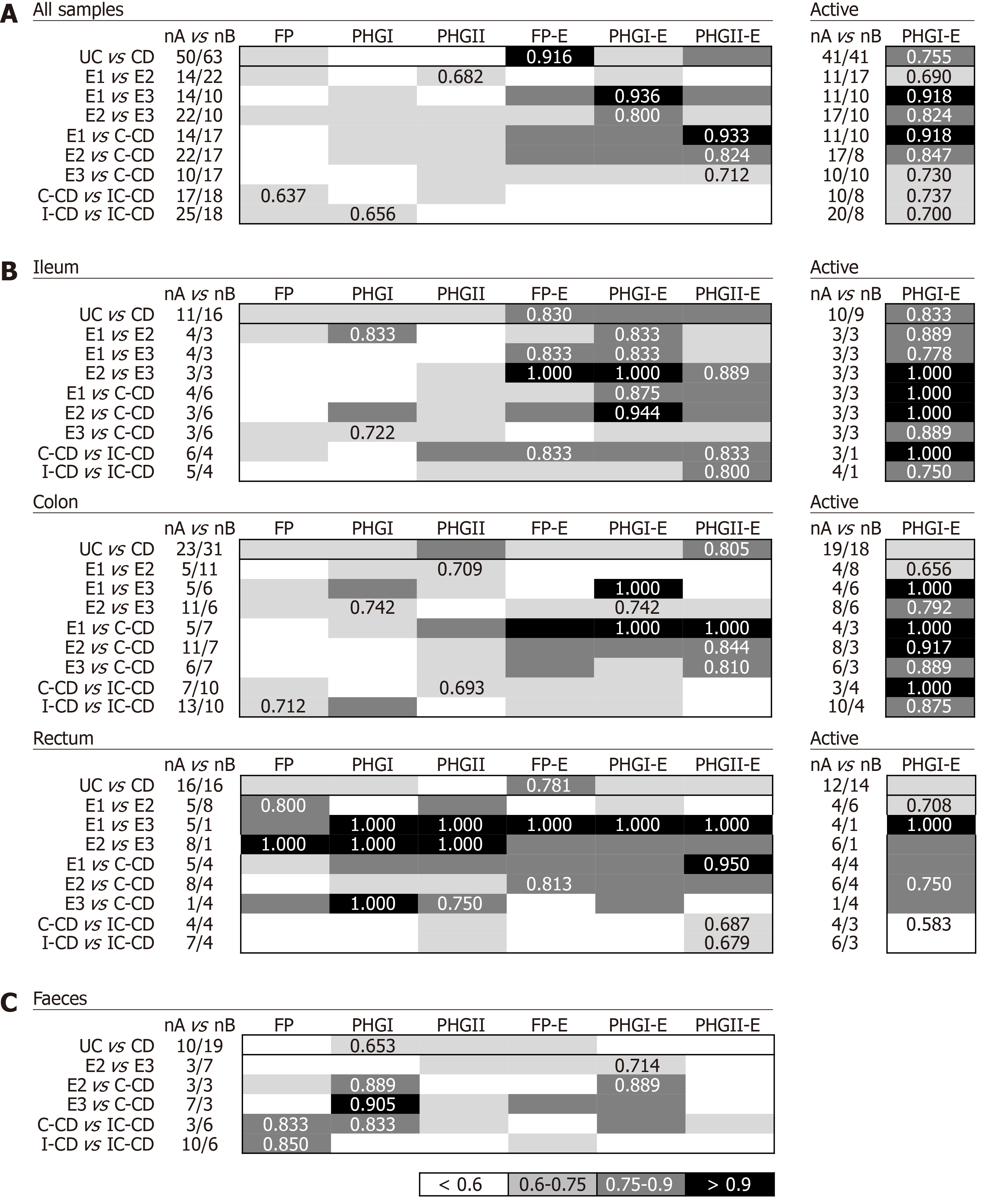
Biomarkers analyses in biopsy samples: The FP-E index was the best biomarker to differentiate UC from CD patients considering all locations (Figure 3A), given that UC patients had higher FP-E index values than CD patients (Supplementary Table 1). However, no consensus could be reached about the biomarker that performed best when comparing IBD subtypes with shared location of the inflammation. PHGI-E index was good to differentiate E1 and E2 from E3, particularly in ileal samples (AUC ≥ 0.875) although suitable discrimination was also obtained when analysing colonic biopsies. In contrast, the PHGII-E index was the most accurate to discriminate C-CD from all UC locations when considering all samples together, and this was sustained in colonic samples (Figure 3B).
As regards discrimination between CD locations, in general, all the biomarkers showed AUC ≤ 0.75 except for the PHGII-E index in ileal samples, which allowed for good discrimination between IC-CD from both C-CD and I-CD.
Interestingly, when considering only active patients, the PHGI-E index was the most discriminatory for all the comparisons when pooling samples, and when considering only those from the ileum and colon. Except for CD with ileal involvement, also suitable discrimination was obtained with the PHGI-E index from rectal samples. Analyses in inactive patients are not shown because, in most cases, they could not be conducted given the low number of samples with these characteristics when separating by IBD subtype.
Biomarkers analyses in faecal samples: Analyses of IBD faecal samples showed that the most suitable biomarker to discriminate between UC and CD conditions was PHGI (Figure 3C), whose load was higher in the former, regardless of the disease extent (Supplementary Table 2). This biomarker was different from that found in biopsies, and the AUC was 1.4 times lower than that obtained in tissue samples.
In contrast, better AUC values were achieved in faecal samples for PHGI and PHGI-E compared to those in biopsies to discriminate C-CD from E2, E3 and IC-CD, although corroboration by engaging more C-CD subjects is needed. It is of note that the results obtained for FP as a biomarker to distinguish IC-CD from I-CD, which substantially improved the biopsy results.
Quantification of bacterial biomarkers may be a valuable tool to assist in the diagnosis of intestinal disorders. In this work, we explored the usefulness of two species (E. coli and F. prausnitzii), extensively reported as dysbiosis representatives of IBD[16,18-20,22,23,26-30], to discriminate between different gastrointestinal disorders.
Firstly, we explored whether or not these bacterial biomarkers could assist in discriminating IBD from IBS, where symptoms can be similar at early stages of the disease. It was observed that a general biomarker to discriminate IBS from IBD could not be established, and therefore two biomarkers should be used. While FP-E index allowed discrimination between IBS and CD, PHGII-E index was the most appropriate to discriminate between IBS and UC. Our cohort of IBS patients was limited and not classified by IBS subtypes. As differences in gut microbiota composition have been found between patients with diarrhoea-predominant IBS and those with constipation-predominant IBS[37], we propose that in further studies aiming to define a biomarker between IBS and IBD, phenotype should be taken into account. Besides, the inclusion of newly diagnosed patients would be of interest to establish whether these biomarkers would be of assistance to discriminate between conditions at an early stage of the disease, particularly when symptoms are overlapping.
Secondly, as there is an association between IBD (especially those involving colonic inflammation) and risk of CRC[13,38], the usefulness of the six biomarkers to tell apart CRC and IBD patients with colonic inflammation was explored. Among the six options of biomarkers considered, the FP-E index was the most discriminatory between CRC and IBD patients, especially from UC, regardless of the activity status of the patient and irrespective of whether colonic or rectal samples were used. This observation is of particular relevance because it has been demonstrated that the extent and duration of the disease increase the risk of patients with UC developing CRC. Future follow-up studies to establish if this index would be useful to predict the risk of CRC development associated with IBD are needed. In contrast, discrimination for CD patients was somewhat limited. Therefore it would be of interest to determine if the combination of F. prausnitzii or its phylogroups with other representatives of CRC dysbiosis enriched in CRC patients[39,40] such as some phylotypes related to Bacteroides, P. stomatios or G. morbillorum could provide a clearer diagnostic test.
Finally, the usefulness of these biomarkers to discriminate IBD subtypes with shared location of inflammation was assessed. The PHGI-E index was a good parameter to discriminate UC subtypes, which is of interest for clinicians to monitor the risk of progression of the inflamed area. From our data, this index allowed the best discrimination within UC subtypes with active disease in ileal and colonic samples. However, a deeper analysis to decipher which sample is the best to analyse is required, as in our study, not all the subjects provided samples from all locations, and interindividual variability may be affecting our observations. Also, further confirmation is required concerning inactive patients since our cohort was limited.
In contrast, we have observed that PHGII load, in conjunction with E. coli counts, can distinguish with suitable accuracy between all UC patients regardless of their disease subtypes and patients with colonic CD (C-CD). The capacity to discriminate between patients with C-CD and E3 is noteworthy because inflammation in these two disorders affects a wide area of the colon and may present similar clinical manifestations, thus hampering a clear classification. Due to differences in treatment and management between UC and CD[10] it is extremely important to discriminate between these two entities accurately.
The best discrimination for CD vs UC was obtained for patients without shared inflamed area (data not shown), but the discrimination needs to be improved to differentiate IBDs with shared disease location, particularly within CD. The combination of F. prausnitzii or its phylogroups with other representatives of IBD dysbiosis may be a way to improve discrimination between IBD subtypes. For instance, depletion of Roseburia hominis has been reported as representative of UC dysbiosis, while the depletion of Ruminococcus gnavus and Ruminococcus torques, with a concomitant increase in Dialister invisus or Bifidobacterium adolescentis have been reported as signatures of CD dysbiosis[16,18]. In addition, the identification of novel species whose abundance differs between IBD subtypes sharing the inflamed location may be of assistance in this regard.
Overall, we observed that the FP-E index would be the selected biomarker to discriminate IBD from CRC while PHG I-E index would be the choice to discriminate within IBD subtypes, and yet no general biomarker of preference could be established to discriminate IBS from IBD. PHGII-E index would be suitable to tell apart IBS and UC patients, whereas the FP-E index could be of assistance to discriminate between IBS and CD although further confirmation on its usefulness for inactive patients is required. It has been reported that active CD and UC can be specifically diagnosed monitoring the faecal bacterial community in conjunction with leukocyte counts. Although in this previous study location of disease has not been considered, it demonstrates that serologic biomarkers may be a source of additional information. A recent study suggested that anti-E. coli, anti-Fusobacterium nucleatum, and anti-F. prausnitzii antibodies did not possess diagnostic value for CD or UC,. However, it would be worth testing if discrimination between gut conditions is enhanced when these bacterial indicators are combined with other previously reported serologic biomarkers of intestinal disease [such as calprotectin, lactoferrin, C-reactive protein, Perinuclear Anti-Neutrophil Cytoplasmic Antibodies (p-ANCA), and Anti-Saccharomyces cerevisiae antibodies (ASCA)].
In order to establish if the proposed indices are suitable for discriminating conditions in less invasive samples, data from the quantification of these species in faeces of IBD subjects was used. We restricted the proof of concept to IBD subjects as these are the conditions more similar in clinical traits and therefore more difficult to discriminate. Our results allowed to demonstrate that, despite our initial selection of biomarkers was based in tissue samples, they are also valuable in faeces. However, differences in which biomarkers performed the best were found. These differences could be because the cohorts used for faecal and biopsy analyses involved different subjects or may reflect the fact that gut microbiota composition is different between faeces and biopsies. Thus, in the future, if biomarkers are selected from tissue, it is crucial to test performance in faecal samples, ideally including the two kinds of samples from the same subject. We have observed that whereas quantification of E. coli in biopsies improved discrimination, the role in improving discrimination when faecal samples are used remained more limited.
On the one hand, this may be explained by the fact that this species may be directly involved in host-interaction during diseases. On the other hand, this information leads us to hypothesize that for future applications, other biomarkers selected from faecal samples analysis could also be included. Concerning F. prausnitzii, the observed differences on which subpopulation should be used as a biomarker, may be related to the distribution of phylogroups along with the gastrointestinal tract, each one with specific metabolic features[43,44].
To robustly validate our observations would require a larger cohort of completely independent patients, including volunteers from different ethnicities, to test these biomarkers as a tool for gut disease diagnostics. Moreover, it would be of interest to test whether F. prausnitzii or its phylogroups, in conjunction with E. coli as biomarkers could discriminate other intestinal disorders within IBD such as indeterminate colitis, unclassified IBD, pouchitis, microscopic colitis, and diverticulosis as these can also be possible confounding conditions.
Currently, inflammatory bowel diseases (IBD) diagnostics features several challenges mainly related to its accurate differentiation from other disease with similar symptoms. In the last years, some studies have shown that the abundance of Faecalibacterium prausnitzii (F. prausnitzii) is a potential biomarker to discriminate between gut disorders. This species load in conjunction with Escherichia coli (E. coli) abundance (F-E index) has been proven to be a better biomarker than total F. prausnitzii alone. Besides, the quantification of F. prausnitzii phylogroup I and phylogroup II has been proposed as a source of additional information to discriminate within IBD. However, the usefulness of an index including the quantification of the phylogroups in conjunction with E. coli remains to be explored, and also its applicability to tell apart these conditions from other gut disorders with milder or worsen phenotypes.
Currently, IBD diagnosis involves a comprehensive examination of the patient that includes clinical, endoscopic, radiologic, and histological criteria. In addition, as clinical manifestations of IBD are unstable during the disease course, a long monitoring period is needed to classify the disease phenotype accurately. As IBD patients feature an imbalanced gut microbial community in comparison to healthy subjects, in the last years the implementation of bacteria representative of this dysbiosis as biomarkers has been started to be explored as a novel strategy to support IBD diagnostics and/or prognostics.
The main objective of this study was to evaluate six options of bacterial biomarkers in terms of their capability to discriminate IBD from other gut disorders and within IBD subtypes.
Adult males and females undergoing routine colonoscopy at the Hospital Dr. Josep Trueta and Parc Hospitalari Martí i Julià in Girona (Spain) were asked to participate, providing either biopsy and/or faecal samples. Subjects included healthy controls as well as patients with IBD, CRC or irritable bowel syndrome (IBS). Genomic DNA extracts of samples were used to assess the load of bacterial markers candidates (total F. prausnitzii, phylogroup I and II of this species and E. coli) by qPCR using specific primers previously reported. Relative abundances to total Bacteria present in the sample, and indices combining F. prausnitzii and E. coli were calculated. Biomarkers accuracy to discriminate conditions was measured by the area under the receiver operating characteristic curve (AUC). To the best of our knowledge, this is the first study that tests combination of F. prausnitzii phylogroups and E. coli application to assist in discriminating challenging IBD diagnostic conditions, compares their performance with previously reported biomarkers and further corroborates results in non-invasive samples.
This study reveals that the F-E index would be the choice to discriminate IBD from colorectal cancer (CRC), especially from ulcerative colitis (UC), regardless of the activity status of the patient and irrespectively if a colonic or a rectal sample was used. This observation is of particular relevance because there is an association between IBD (especially those involving colonic inflammation) and the risk of CRC. Besides, we have observed that PHG I-E index is a good parameter to differentiate pancolitis from other UC subtypes, which is of interest for clinicians to monitor risk of progression of the inflamed area. The application of bacterial biomarkers in feces is also demonstrated, which is a non-invasive method and may represent a step forward to implement these biomarkers in clinical practice to support IBD diagnostics.
This study corroborates that F. prausnitzii combined with E. coli can help to discriminate within IBD subtypes both in tissue and fecal samples, as well as offer potential to differentiate IBD and CRC patients. Use of biopsy samples presented better performance, but we confirmed that suitable results in fecal samples were shown too. The comparison of the performance of new indices with those previously reported in the literature has allowed establishing the biomarker of choice to select depending on the conditions to discriminate. From these comparisons, we hypothesize that given the complexity of the disease in terms of multiple subtypes and phenotypes during the disease course, it would be complicated the establishment of a universal biomarker using only two species and total microbiota composition could be a more informative approximation in this regard. However, given the outcome obtained only with the biomarkers evaluated here, we envisage that implementation of bacterial load assessment in clinical routine may ease IBD diagnostics in the future, for example for initial screening.
This study contributes to providing evidence that bacterial biomarkers assessment may help in solving intestinal disorders diagnostic challenges. Because differences in performance were observed between tissue and faecal samples, attention should be paid to this issue in similar studies. Future directions of research could assess if discrimination between gut conditions is enhanced when these bacterial indicators are combined with other bacterial or serologic biomarkers of intestinal disease. Also, validation in a larger cohort of completely independent patients, including volunteers from different regions would be required to define a tool with worldwide application in clinical routine.
We are grateful for the generosity of the patients who freely gave their time and samples to make this study possible. We thank theatre staff of all centres for their dedication and careful sample collection, especially M.D David Busquets from the Hospital Dr. Josep Trueta (Girona, Spain) and M.D Carles López from the Hospital Santa Caterina (Salt, Spain). We appreciate Dr. Teresa Mas-de-Xaxars, Dr. Romà Surís-Valls and Dr. Anna Bahí for assistance in the analysis of samples.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Suzuki H, Yang BL S-Editor: Ma YJ L-Editor: A E-Editor: Liu MY
| 1. | Magro F, Rodrigues A, Vieira AI, Portela F, Cremers I, Cotter J, Correia L, Duarte MA, Tavares ML, Lago P, Ministro P, Peixe P, Lopes S, Garcia EB. Review of the disease course among adult ulcerative colitis population-based longitudinal cohorts. Inflamm Bowel Dis. 2012;18:573-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 2. | Moum B, Ekbom A, Vatn MH, Aadland E, Sauar J, Lygren I, Schulz T, Stray N, Fausa O. Clinical course during the 1st year after diagnosis in ulcerative colitis and Crohn's disease. Results of a large, prospective population-based study in southeastern Norway, 1990-93. Scand J Gastroenterol. 1997;32:1005-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 107] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Peyrin-Biroulet L, Loftus EV, Colombel JF, Sandborn WJ. Long-term complications, extraintestinal manifestations, and mortality in adult Crohn's disease in population-based cohorts. Inflamm Bowel Dis. 2011;17:471-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 4. | Solberg IC, Vatn MH, Høie O, Stray N, Sauar J, Jahnsen J, Moum B, Lygren I; IBSEN Study Group. Clinical course in Crohn's disease: results of a Norwegian population-based ten-year follow-up study. Clin Gastroenterol Hepatol. 2007;5:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 526] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 5. | Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1299] [Cited by in RCA: 1511] [Article Influence: 83.9] [Reference Citation Analysis (2)] |
| 6. | Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1247] [Cited by in RCA: 1358] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 7. | Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2894] [Cited by in RCA: 3353] [Article Influence: 186.3] [Reference Citation Analysis (11)] |
| 8. | Yantiss RK, Odze RD. Diagnostic difficulties in inflammatory bowel disease pathology. Histopathology. 2006;48:116-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2085] [Cited by in RCA: 2154] [Article Influence: 102.6] [Reference Citation Analysis (1)] |
| 10. | Mowat C, Cole A, Windsor A, Ahmad T, Arnott I, Driscoll R, Mitton S, Orchard T, Rutter M, Younge L, Lees C, Ho GT, Satsangi J, Bloom S; IBD Section of the British Society of Gastroenterology. Guidelines for the management of inflammatory bowel disease in adults. Gut. 2011;60:571-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 946] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 11. | Bernstein CN, Fried M, Krabshuis JH, Cohen H, Eliakim R, Fedail S, Gearry R, Goh KL, Hamid S, Khan AG, LeMair AW, Malfertheiner, Ouyang Q, Rey JF, Sood A, Steinwurz F, Thomsen OO, Thomson A, Watermeyer G. World Gastroenterology Organization Practice Guidelines for the diagnosis and management of IBD in 2010. Inflamm Bowel Dis. 2010;16:112-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 370] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 12. | Nikolaus S, Schreiber S. Diagnostics of inflammatory bowel disease. Gastroenterology. 2007;133:1670-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 292] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 13. | Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1985] [Cited by in RCA: 2080] [Article Influence: 86.7] [Reference Citation Analysis (1)] |
| 14. | Bernstein CN, Shanahan F. Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut. 2008;57:1185-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Louis E, Collard A, Oger AF, Degroote E, Aboul Nasr El Yafi FA, Belaiche J. Behaviour of Crohn's disease according to the Vienna classification: changing pattern over the course of the disease. Gut. 2001;49:777-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 705] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 16. | Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 797] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 17. | Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1677] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 18. | Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 198] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 19. | Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, Marteau P, Doré J, Leclerc M. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis. 2011;17:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 215] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 20. | Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 362] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 21. | Andoh A, Sakata S, Koizumi Y, Mitsuyama K, Fujiyama Y, Benno Y. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3433] [Article Influence: 190.7] [Reference Citation Analysis (1)] |
| 23. | Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1036] [Cited by in RCA: 1337] [Article Influence: 121.5] [Reference Citation Analysis (3)] |
| 24. | Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, Leleiko N, Kenche H, Stolfi A, Wine E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 214] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 25. | Noor SO, Ridgway K, Scovell L, Kemsley EK, Lund EK, Jamieson C, Johnson IT, Narbad A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 26. | Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan SH, Flint HJ, Aldeguer X, Garcia-Gil LJ. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes. Int J Med Microbiol. 2014;304:464-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 27. | Lopez-Siles M, Martinez-Medina M, Surís-Valls R, Aldeguer X, Sabat-Mir M, Duncan SH, Flint HJ, Garcia-Gil LJ. Changes in the Abundance of Faecalibacterium prausnitzii Phylogroups I and II in the Intestinal Mucosa of Inflammatory Bowel Disease and Patients with Colorectal Cancer. Inflamm Bowel Dis. 2016;22:28-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 29. | Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, Vermeire S, Sokol H, Guarner F, Manichanh C. A microbial signature for Crohn's disease. Gut. 2017;66:813-822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 454] [Cited by in RCA: 597] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 30. | Guo S, Lu Y, Xu B, Wang W, Xu J, Zhang G. A Simple Fecal Bacterial Marker Panel for the Diagnosis of Crohn's Disease. Front Microbiol. 2019;10:1306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, Caprilli R, Colombel JF, Gasche C, Geboes K, Jewell DP, Karban A, Loftus EV, Peña AS, Riddell RH, Sachar DB, Schreiber S, Steinhart AH, Targan SR, Vermeire S, Warren BF. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19 Suppl A:5A-36A. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2148] [Cited by in RCA: 2369] [Article Influence: 215.4] [Reference Citation Analysis (0)] |
| 32. | Best WR, Becktel JM, Singleton JW, Kern F. Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study. Gastroenterology. 1976;70:439-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2405] [Cited by in RCA: 2338] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 33. | Pineton de Chambrun G, Peyrin-Biroulet L, Lémann M, Colombel JF. Clinical implications of mucosal healing for the management of IBD. Nat Rev Gastroenterol Hepatol. 2010;7:15-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 34. | Huijsdens XW, Linskens RK, Mak M, Meuwissen SG, Vandenbroucke-Grauls CM, Savelkoul PH. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J Clin Microbiol. 2002;40:4423-4427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 262] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 35. | Furet JP, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, Doré J, Corthier G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009;68:351-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 321] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 36. | Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 305] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 37. | Maukonen J, Satokari R, Mättö J, Söderlund H, Mattila-Sandholm T, Saarela M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 113] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 38. | Castaño-Milla C, Chaparro M, Gisbert JP. Systematic review with meta-analysis: the declining risk of colorectal cancer in ulcerative colitis. Aliment Pharmacol Ther. 2014;39:645-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 39. | Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, Xiang C, Wang Y, Zhao F, Gao GF, Wang S, Li L, Zhang H, Zhu B. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 343] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 40. | Malagón M, Ramió-Pujol S, Serrano M, Serra-Pagès M, Amoedo J, Oliver L, Bahí A, Mas-de-Xaxars T, Torrealba L, Gilabert P, Miquel-Cusachs JO, García-Nimo L, Saló J, Guardiola J, Piñol V, Cubiella J, Castells A, Aldeguer X, Garcia-Gil J. Reduction of faecal immunochemical test false-positive results using a signature based on faecal bacterial markers. Aliment Pharmacol Ther. 2019;49:1410-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 42. | Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol. 2014;5:213-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 127] [Cited by in RCA: 156] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 43. | Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 553] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 44. | Fitzgerald CB, Shkoporov AN, Sutton TDS, Chaplin AV, Velayudhan V, Ross RP, Hill C. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genomics. 2018;19:931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |