Published online Dec 28, 2023. doi: 10.4329/wjr.v15.i12.359
Peer-review started: October 3, 2023
First decision: October 9, 2023
Revised: November 13, 2023
Accepted: December 5, 2023
Article in press: December 5, 2023
Published online: December 28, 2023
Processing time: 83 Days and 5.1 Hours
Missing occult cancer lesions accounts for the most diagnostic errors in retro
To achieve appropriate AI model training, a large annotated dataset is necessary to train the AI models. Our goal in this research is to compare two methods for decreasing the annotation time to establish ground truth: Skip-slice annotation and AI-initiated annotation.
We developed a 2D U-Net as an AI second observer for detecting colorectal cancer (CRC) and an ensemble of 5 differently initiated 2D U-Net for ensemble technique. Each model was trained with 51 cases of annotated CRC computed tomography of the abdomen and pelvis, tested with 7 cases, and validated with 20 cases from The Cancer Imaging Archive cases. The sensitivity, false positives per case, and estimated Dice coefficient were obtained for each method of training. We compared the two methods of annotations and the time reduction associated with the technique. The time differences were tested using Friedman’s two-way analysis of variance.
Sparse annotation significantly reduces the time for annotation particularly skipping 2 slices at a time (P < 0.001). Reduction of up to 2/3 of the annotation does not reduce AI model sensitivity or false positives per case. Although initializing human annotation with AI reduces the annotation time, the reduction is minimal, even when using an ensemble AI to decrease false positives.
Our data support the sparse annotation technique as an efficient technique for reducing the time needed to establish the ground truth.
Core Tip: Minimizing diagnostic errors for colorectal cancer may be most effectively performed with artificial intelligence (AI) second observer. Supervised training of AI-observer will require high volume of annotated training cases. Comparing skip-slice annotation and AI-initiated annotation shows that skipping slices does not affect the training outcome while AI-initiated annotation does not significantly improve annotation time.
- Citation: Grudza M, Salinel B, Zeien S, Murphy M, Adkins J, Jensen CT, Bay C, Kodibagkar V, Koo P, Dragovich T, Choti MA, Kundranda M, Syeda-Mahmood T, Wang HZ, Chang J. Methods for improving colorectal cancer annotation efficiency for artificial intelligence-observer training. World J Radiol 2023; 15(12): 359-369
- URL: https://www.wjgnet.com/1949-8470/full/v15/i12/359.htm
- DOI: https://dx.doi.org/10.4329/wjr.v15.i12.359
Colorectal cancer (CRC) is the third most common cancer in the United States, developing in about 4.3% of men and 4.0% of women. It is the second highest cause of cancer-related deaths in the United States, responsible for about 53200 deaths per year[1]. Likewise, CRC has also become the third most common cancer in China and is increasing in incidence in major countries such as Brazil and Russia[2]. Although early detection of CRC through established screening can greatly increase survival probability, resistance to the various invasive and noninvasive forms of screening persists[3-5]. This is reflected in the fact that up to 40% of CRC is diagnosed in the emergency department[6]. The rise in CRC incidence in adults younger than 55 also indicates a need for improved detection through non-screening methods[7-9]. Therefore, cross-sectional imaging remains important in early incidental diagnosis of CRC. However, up to 40% of the features of early CRC can be missed by radiologists when analyzing these scans[10-13]. This indicates a need for a “second-observer” to assist the busy radiologist in order to minimize false negatives which can result in reduced survival due to a delay in diagnosis[10].
Artificial intelligence (AI) has the potential to improve early disease detection, as shown by the recently approved algorithm for detecting intracranial hemorrhage on computed tomography (CT) and has been proposed for similar application in gastric cancer[14,15]. This model can be trained with the relatively low 39000 cases because of the low variation in brain anatomy and the simpler disease pattern on CT. CRC varies significantly in location and appearance because of the heterogeneity in anatomy and disease. The model training could be accomplished by using supervised training (requires ground truth with potentially fewer training cases) rather than unsupervised training (uses more training cases without ground truth).
In supervised training, inputs with established ground-truth is important for training model. Although supervised training requires lower volume of data, the necessary volume of training data is quite large and would require significant amount of time to label the ground-truth by trained personnel[16-18]. Several potential methods for decreasing the time to establish the ground truth for supervised training have been evaluated. These methods include image level labeling, bounding boxes to localize the site of cancer, sparse labeling, and deploying an incompletely trained AI-model for first pass segmentation followed by human adjustment (AI-Init)[19,20]. Image level labeling and bounding box techniques consider different level of image information for localizing cancer and are expected to require significantly less human intervention. However, these may require more cases for training. For true supervised training, the level of annotation contains significantly more detail, which requires human interaction. This human interaction can be minimized with sparse annotation that skips slices during segmentation or having the rudimentarily trained AI algorithm perform the initial segmentation, which is subsequently modified by humans. In this report, we compared the improvement in amount of time spent on annotating the CRC between the last two techniques (sparse annotation by skip-slicing and AI-Init).
The CRC cases were obtained from our respective cancer centers, which are tertiary and quaternary referral centers, between the years of 2012 through 2018 as well as cases from The Cancer Imaging Archive (TCGA-COAD) public domain images which were used to compare the outcomes of the techniques[21]. Our training and validating cases included 58 CRC cancer cases (27 males and 31 females) and 59 normal cases and consisted of CT scans of the abdomen and pelvis cases with a mixture of intravenous (IV) contrast enhanced and unenhanced studies. 51 of the cancer cases were used for training while 7 were retained for validation. The cases were retrieved from the picture archiving and communications system of the respective institutions and de-identified. The de-identified cases and their annotations were transferred between the research sites and the medical centers through the HIPPA compliant cloud server from Box.com. 20 of the 25 cases (8 males and 12 female) from TCGA-COAD were used to test the outcome of the training using the two different training techniques. The 5 excluded cases did not have clearly identifiable CRC on CT scans. The imaging stage of the 58 CRC cases and the 20 The Cancer Imaging Archive (TCIA) cases are listed in Table 1.
| Training cases (n = 58) | Test cases (n = 20) | |
| AJCC stage | ||
| Stage 1 | 0 | 5 |
| Stage 2 | 15 | 5 |
| Stage 3 | 14 | 7 |
| Stage 4 | 29 | 3 |
| T stage | ||
| T1 | 0 | 2 |
| T2 | 0 | 4 |
| T3 | 21 | 13 |
| T4 | 37 | 1 |
| Location | ||
| Right | 39 | 17 |
| Transverse | 3 | 2 |
| Left | 16 | 1 |
| CT slice thickness (mm) | ||
| 7 | 0 | 1 |
| 5 | 29 | 17 |
| 3-4 | 25 | 0 |
| 2 or less | 4 | 2 |
| Contrast | ||
| IV+PO | 27 | 18 |
| IV | 22 | 1 |
| PO | 4 | 1 |
| None | 5 | 0 |
The location and slices of the cancer were identified on the CT images using ITK-SNAP (versions 3.6.0 and 3.8.0; www.itksnap.org)[22]. For annotation, the CT axial slices containing the CRC from our cancer center and from TCGA-COAD were identified and a contour outlining the edges of the cancer was drawn using the drawing tool. All the CT slices containing the tumor were segmented. At the time of training, to simulate sparse annotation, the skip-slice training would evenly skip one or two annotated slices among those containing the tumor from being used in training AI methods for every annotated tumor slice used for training. For AI-Init technique, the TCGA-COAD cases were initially segmented by the trained AI-model after training with the 51 CRC and 59 normal cases described in the previous section. This segmented model was then viewed with ITK-SNAP and adjusted to the ground truth established by human segmentation. The time required to fully adjust the contour and to eliminate false positives and to correct the false negatives was recorded for each TCGA-COAD case.
The AI algorithm used in the project is a 2D U-Net, which is a convolutional neural network (CNN). The U-Net is a popular image segmentation algorithm for medical image segmentation tasks because it requires less training inputs than other techniques and is more robust with small training dataset[23]. In addition, recent research findings show that 2D U-Net has equivalent performance as that of 3D U-Net, but with lower computational requirement[24,25]. Inputs to the CNN consisted of 2D images with 512 × 512 pixels. The training dataset was augmented using the standard affine transformation with up to 30° rotation and up to 30% scale variation applied to the training patches. The CNN uses a 3 × 3 kernel and has 5 encoding layers containing 32, 64, 128, 256, and 512 filters and 5 decoding layers with each layer containing 512, 256, 128, 64, and 32 filters, respectively. Adam optimizer was used for training the model, and the model with the best validation accuracy was chosen. The network was trained using all the training cases, treating each image of a subject’s study as a case, with 1 case per batch and trained with 200 epochs. The 7 validation cases were used to choose the optimal model parameters while the 20 TCGA-COAD cases were used to validate the final accuracy of the different techniques. For evaluating the effects of sparse-annotation, the AI model was trained with either all of the slices of annotation or evenly skipping either 1 or 2 slices of annotation for every slice used for training.
In order to determine how to improve the AI-Init technique in establishing the ground-truth, we also developed a simple ensemble model with each individual component of the ensemble being an independently trained 2D U-Net model with a random initiation. This was obtained to improve the specificity of the AI segmentation. To this end, we trained five randomly initialized U-Net models for voting-based model ensemble. Each of these five model is trained as described in the previous paragraph. The difference is that the final decision is based on the voter ensemble inference technique. For voting-based model ensemble, a voxel is labeled as tumor by the algorithm if and only if at least certain number of automatic segmentation models label the voxel as tumor. The U-Net models were also trained with the skip annotation technique (with skip 0, 1, and 2 slices). We then compared the accuracy and number of false positives per case to see how the ensemble method would influence the time required to adjust the segmentation results. For this approach, the model ensemble approach generated 3 unique segmentation algorithms (for each annotation technique) based on whether at least one, two, or three member(s) of the ensemble identified the same lesion. The segmentation was reviewed using ITK-SNAP.
To analyze the algorithm’s accuracy, the AI-generated segmentations were compared to ground-truth annotations, which were established as described previously. The AI model generated a DICOM image series for each case with the number of slices for each case ranging from 60-400. The cancer segmented by the AI model was compared to the annotated ground truth in 3D to determine the false positives, false negatives, and the Dice coefficient (DSC). A false positive was considered any segmentation created by the algorithm that did not overlap any part of the ground-truth segmentation. A false negative was considered as any image series with human annotated tumor that was not identified by the algorithm’s segmentations. For true positive segmentation, DSC was visually estimated and categorized to be 0%-25%, 26%-50%, and > 50%. We obtained visual estimate as we do not have readily available software for full DSC calculation.
In order to measure the amount of time saved by initiating annotation by the rudimentarily-trained AI model (as described earlier), we recorded the time required for annotating the CRC. The time required for initial, full annotation of CRC as well as the time required to adjust the AI produced model were acquired. For adjusting the AI-model, the obtained time included the time to adjust the boundary of the CRC and for erasing the false positives. We randomly selected 3 large, 3 medium, and 3 small CRC from the TCIA dataset and analyzed these times. The sizes of the CRC were considered small, medium, or large if the lesion spanned ≤ 5 CT slices, 5-15 CT slices, and ≥ 15 CT slices, respectively. The median and average times were calculated. These same 9 cases were used for measuring the time needed to complete skip-slice annotation by annotating every other or every third slices of the tumor mass.
The details of the location of the tumor and the scanning protocols from the training and TCIA testing datasets are listed in Table 1. The training dataset contains more higher stage cancers with 74 % of cases at stage 3 or 4 while the TCIA dataset has 50% of the cases at stage 3 or 4. All of the training cases is T3 or T4 in T-stage, while the TCIA dataset has 70% of the cases being T3 or T4. 67% of the training dataset has right-sided tumor (being defined as ascending colon) while the TCIA test dataset has 85% of the cases being right-sided. In terms of scanning protocol, 50% of the training dataset has 5 mm slice thickness while 90% of the TCIA testing dataset has slice thickness being 5 mm or more. In terms of contrast administration, the training dataset has 47% of the cases with IV and positive oral (PO) contrast while the TCIA dataset has 90% of the cases with both IV and PO contrast. 38% of the training dataset has just IV contrast whereas TCIA data has 5% of cases with IV contrast only.
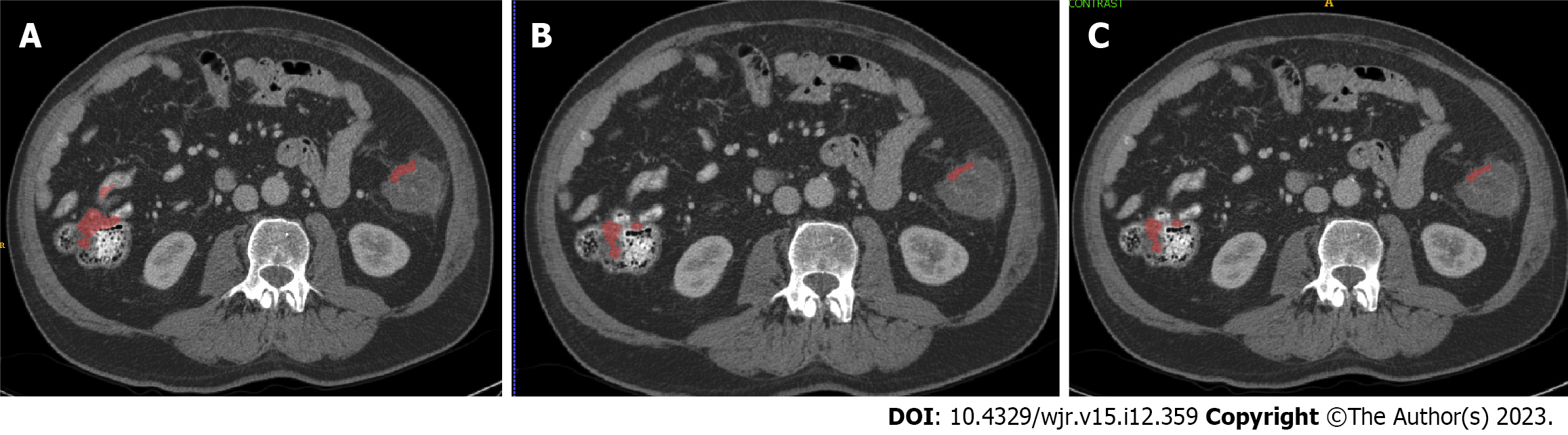
The AI-models generated by skip-slice annotation did not significantly alter the segmentation outcome of the AI-model. Figure 1 shows two separate cases segmented by the AI-models; although there is very subtle difference in the segmentations, the difference could not be detected by the measure that we chose (false positives per case, sensitivity, and DSC). For all three models, the sensitivity was 80% and the false positive lesions identified per case was 22. The DSC category distribution was 25% for 0-0.25, 60% for 0.26-0.5, and 15% for > 0.5.
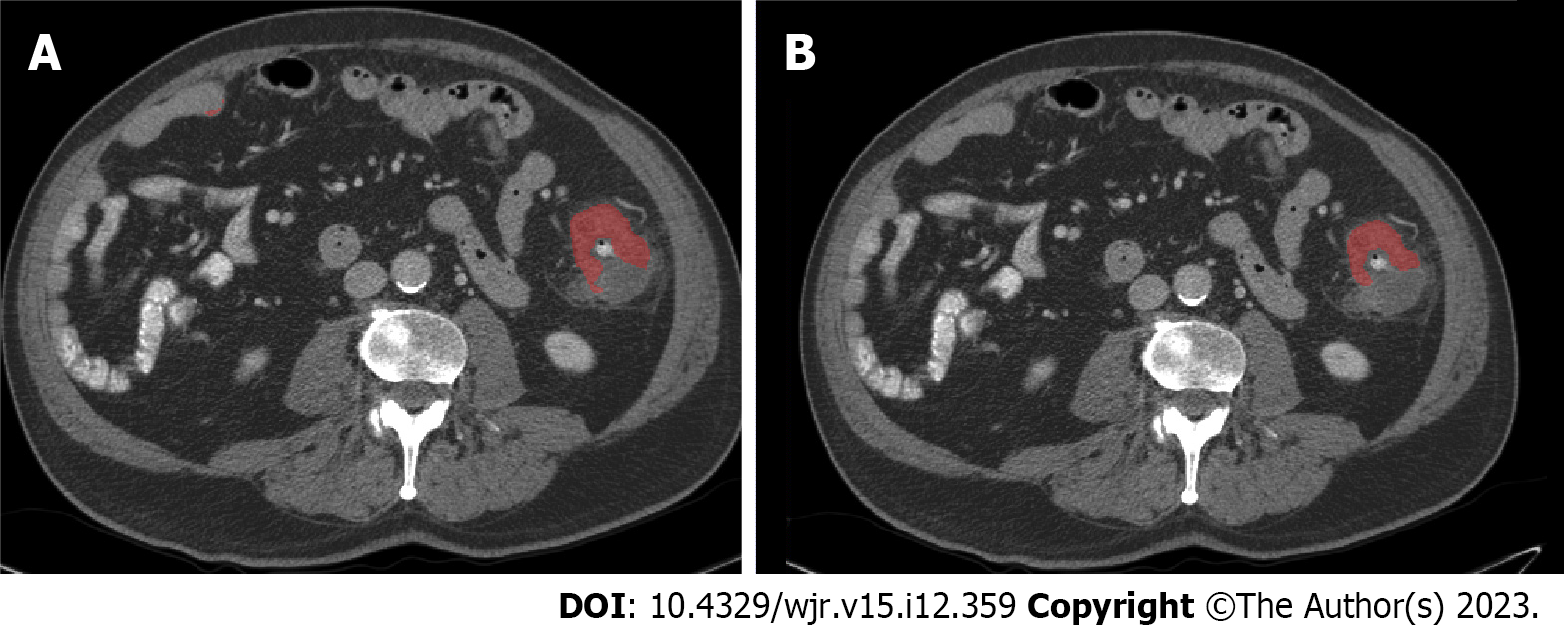
Prior to obtaining the AI-initiated annotation, we aimed to minimize the number of false positives per case as the false positives could decrease the efficiency of this technique in establishing the ground-truth. To do this, we chose a simple voting-based ensemble method to reduce the number of false positives per case. When the number of votes required by the ensemble technique for determining tumor segmentation is increased, there is a corresponding drop in false positives per case while there is also a decrease in sensitivity, although the drop in false positives was much greater than the drop in sensitivity. The DSC distribution also shifts toward more cases being in 0 to 0.25 category. These data are shown in Tables 2 and 3. Figures 2 and 3 show an example of both agreement and disagreement between 1- and 2-voter models.
| Single voter | 2 voter | 3 voter | |
| Sensitivity | 0.8 | 0.6 | 0.3 |
| False positives/case | 21.95 | 7.55 | 3.7 |
| Percentage of cases | Estimated dice coefficient | |||
| 0 | 0-0.25 | 0.25-0.5 | > 0.5 | |
| Single voter | 20 | 5 | 60 | 15 |
| 2 voter | 40 | 35 | 20 | 5 |
| 3 voter | 70 | 15 | 10 | 5 |
The models from the section above were used to generate the initial annotation of CRC which was then adjusted manually to fit the established ground truth. The amount of time required to modify these annotations to the ground truth was then recorded for 3 randomly selected cases from each of the large, medium, and small tumors. The complexity of these cases was determined as described in the methods section. The amount of time required to adjust these cases is listed in Table 4, along with the median and average. The measured time includes the time needed to remove the false positives as well as contouring the false negative lesions. The data show that AI-Init does decrease the time required to annotate the cases, although a statistical test of the distributions among the measured annotation time from the original, 1-voter, and 2-voter model using the Friedman’s two-way analysis of variance by ranks did not yield statistical significance (P = 0.121). Some improvement is seen, primarily, with medium sized tumors.
| Lesion size | Annotation time based on technique (Min:Sec ± min) | ||||
| Manual (n = 3 each) | AI-single voter (n = 3 each) | AI-2-voter (n = 3 each) | Skip-1 (n = 3 each) | Skip-2 (n = 3 each) | |
| Large | 22:09 ± 0.18 | 21:00 ± 0.23 | 20:29 ± 0.22 | 8:58 ± 1.22 | 5:34 ± 1.19 |
| Medium | 15:06 ± 0.4 | 10:37 ± 0.25 | 9:13 ± 0.15 | 4:58 ± 2.57 | 1:14 ± 1.38 |
| Small | 5:54 ± 0.07 | 6:26 ± 0.03 | 5:44 ± 0.02 | 2:23 ± 0.14 | 1:24 ± 0.28 |
For skip-slice annotation, the actual timed annotation revealed significant reduction in time needed to complete the annotation (Table 4). Although the reduction is not proportional, the differences are significant between full annotation and either skip-1 or skip-2 slice methods, using the Friedman’s two-way analysis of variance by ranks. The P-values for univariate analysis between fully-annotated and skip-1, fully-annotated and skip 2, and skip-1 and skip-2 annotation style are 0.034, < 0.001, and 0.034. When using multivariate analysis, the same P-values are 0.102, < 0.001, and 0.102. This suggests that skipping slices can reduce the labor necessary for establishing the ground-truth for supervised or semi-supervised training of AI models, and in multivariate analysis, the time different is statistically significant when higher number of slices are skipped.
Our results provide a direct comparison of annotation techniques for supervised training of AI models as second observer for detecting CRC. Supervised AI-model training by skipping-slices of CRC did not appreciably influence the outcome of segmentation. There were very subtle visual differences, but these were not detectable with the measures used. No significant segmentation difference could be detected when skipping up to 2 slices. For AI-initiated annotation, the model does not improve the time spent annotating large and small cancers, but does show some improvement for medium sized tumors. These methods allow for time-reduction in annotating the ground truth so supervised training of AI-models could be more efficient and allow greater participation by busy radiologists.
Skip-slice annotation is similar to a sparse-annotation technique, which has been explored in the literature. This technique was tested in confocal images of Xenopus kidney segmentation[19]. Cicek et al[19] showed that the DSC equivalent (intersection of union) improved with increasing slices in each axis, starting with one slice in each direction. Increasing the annotated slices was equivalent to increasing the number of ground-truth pixels based on their study. The authors achieved their annotation goal using up to 9% of all the available pixels as ground truth for training. With this training, the network achieved segmentation with 85% overlap with the ground truth. Our data also support this finding that not all ground truth needs to be presented to an AI-algorithm to train the algorithm properly. The difference between Cicek et al[19] and our study is that they began with the minimal number of slices while we evaluated from the maximum number of slices. Increasing from the minimum showed that minimalist approach may underfit the algorithm, while maximal approach may over-fit the network. The minimum necessary amount of established ground truth pixel for optimal network training is yet to be identified so that the amount of human effort in establishing ground truth can be minimized while maintaining optimal AI model training.
The literature reports multiple techniques for minimizing false positives, particularly regarding pulmonary nodule reduction which can be categorized into ones that use single modes of information (imaging only) or multimodal technique (combines imaging with clinical information). Jin et al[26] constructed a false-positive reduction algorithm for pulmonary nodule detection. This is constructed as a separate algorithm that could serve as add on to a nodule detector. They combined several methods to avoid the traps that cause false-positives. First, they deployed a 3D residual CNN which minimizes the effect of diminishing gradient in the stochastic gradient descent algorithm during training, so as to avoid local minima that may trap the algorithm. They also combined spatial pooling and cropping which provides multi-scale contextual information to improve the learning process. A similar technique uses multiscale contextual information where variable amount of the pulmonary nodule and surrounding normal lung is included for training[27,28]. The multiple levels of information are integrated and significantly lower the false positives[27,28]. Lastly, online hard sample selection training was chosen to maximize training of hard-to-discern examples so that the network can learn from its own failures. This technique replaces a portion of correctly detected training cases with ones that were previously missed so that the network can learn the features of the missed cases to improve its outcome[29].
The multimodal technique is a broad category of AI technique where different aspects of a patient’s clinical information are integrated to improve the classification and prediction algorithm. The additional information restricts the bias and variance of the model to improve the accuracy of the outcome[30]. For our algorithm, we employed the simulated multimodal technique with ensemble voting by integrating information from different instances of the AI model. This is similar to selecting 5 different models from the same model space to restrict bias and variance[30]. By generating 5 models from the base CNN technique, we chose a lesion to be cancer only if 2 or 3 of the 5 models agreed on a pixel being cancerous. This allowed us to dramatically decrease the number of false positives per case. This, however, also decreased sensitivity of the model. This trade-off is also seen with ensemble techniques trained with clinical data in predicting diabetic retinopathy[31]. Other studies have shown that the information contains different degree of relevance[32,33]. In the study by Boehm et al[32], applying all available information regarding a patient (clinical, genetic, histo
The findings that skip-slice annotation may reduce the time required for establishing the ground-truth for AI model training can significantly impact the development of AI models in imaging research. Annotating the ground truth requires trained personnel capable of identifying normal and abnormal structures on CT images, who are typically physicians or physicians in training. As we have shown in Table 4, full annotation of a case will require anywhere from 5-20 min, which can be reduced significantly with skip-slice annotation. This will significantly reduce the time of the highly trained personnel, who are involved in busy clinical work. Minimizing the time spent in establishing the ground-truth should theoretically improve participation of these highly trained personnel in assisting AI research in medical imaging.
There are several limitations to the present study. The first is the small size of the training and testing dataset. These training and testing dataset is also unbalanced and unmatched in both stage of the disease and the scanning protocol. Table 1 showed that the training dataset consists of higher stage disease than the testing dataset which may limit detection of the earlier stage disease in the testing dataset. In addition, the training dataset also has lower proportion of the cases containing both IV and PO contrast and has thinner slice images. It is uncertain if the blur from the thicker slices may influence the decision of the model, but the trained model have been exposed to thicker slices with IV and PO contrast. It will be interesting to evaluate the dilutive effect on model performance when the model is trained with a broader range of protocols and stages of the disease. The current model with limited training dataset is not generalizable, but it does show the potential of the second observer with better trained model[34]. Another limitation of the study is the lack of full software for calculating the precision, F1 score, and DSC of the model outcome. The sensitivity provided in the present study is equivalent to the recall measure of the model.
In comparing the different techniques for reducing annotation time to establish the ground truth, we developed a U-NET model in detecting CRC. This pilot model has the potential to serve as a second observer with further research. In order to accelerate AI second observer training, we compared different techniques of annotation in minimizing this data preparation work. Our results showed that skip slice annotation may lead to the most time reduction as there was minimal effect on model outcome when slices are skipped, leading to proportional decrease in time needed to annotate. Although AI-initiated segmentation may lead to reduced annotation time, it tends to reduce time for medium sized lesion while large complex and small lesions do not benefit. At this time, skipping slices may result in the most time efficient method for annotating cancer on training images.
Up to 40% of colorectal cancer (CRC) goes undetected on initial computed tomography (CT) scan performed in either the emergency department or outpatient imaging setting. This delay in diagnosis significantly impacts the overall survival of the patients. The ultimate goal is to develop an artificial intelligence (AI)-based second observer for clinical integration so as to improve the clinical diagnosis of CRC on CT studies.
The development of deep learning has shown that AI can potentially serve as a second observer to assist busy radiologist at a reasonable cost, as second reader has been shown in past research to improve imaging diagnosis. However, to develop an AI second observer, large number of training cases with annotated ground truth is required necessitating significant time commitment on the part of the research radiologists.
Our main objective in this research is to compare skip-slice annotation with AI-initiated annotation in time savings for annotating the ground truth for training dataset preparation. Saving annotation time will help improve the efficiency in dataset preparation. Our secondary objective was to evaluate whether ensemble technique could help improve false positive rate for AI-initiated annotation technique. Decreasing false positives per case will make the model more acceptable by clinical radiologist.
The dataset was manually annotated for the entire tumor as well as skipping annotation by one or two slices was measured; 9 total cases were randomly selected to measure the time required to annotate these tumors. These datasets were used to train 2D U-Net model with 5 encoding and 5 decoding layers, using the Adam optimizer. The model accuracy consisting of sensitivity, Dice coefficient estimate, and false positive per case were used to evaluate the model accuracy. The rudimentary AI model was also used to annotate the ground truth; the times required to adjust the annotation for the 9 cases from manually annotation were also measured.
We found that the model trained on skip-slice annotation did not have significant difference in tumor segmentation as a fully annotated dataset and which is statistically significant, thus showing that skip slice annotation can reduce the data preparation time. Although AI-initiated annotation also reduces time, the difference was not statistically significant. Ensemble technique is shown to reduce false positive per case, but at decreased sensitivity.
This study proposes that skip-slice annotation can improve the efficiency in data preparation for AI model training. The significance is that it will reduce the time commitment of highly trained medical personnel in participating in AI medical imaging research.
The future direction of the present research is that this should improve the efficiency in training dataset development given the decreased annotation time.
The authors would like to thank Gene A Hoyer and Judith C Hoyer for their generous support in funding the publication of this manuscript.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: RSNA.
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fu L, China; Zhu L, China S-Editor: Wang JJ L-Editor: A P-Editor: Zhao S
| 1. | American Cancer Society. Key Statistics for Colorectal Cancer. [cited 15 July 2023]. Available from: https://www.cancer.org/cancer/types/colon-rectal-cancer/about/key-statistics.html. |
| 2. | Xie Y, Shi L, He X, Luo Y. Gastrointestinal cancers in China, the USA, and Europe. Gastroenterol Rep (Oxf). 2021;9:91-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 128] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 3. | Akram A, Juang D, Bustamante R, Liu L, Earles A, Ho SB, Wang-Rodriguez J, Allison JE, Gupta S. Replacing the Guaiac Fecal Occult Blood Test With the Fecal Immunochemical Test Increases Proportion of Individuals Screened in a Large Healthcare Setting. Clin Gastroenterol Hepatol. 2017;15:1265-1270.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Weiser E, Parks PD, Swartz RK, Thomme JV, Lavin PT, Limburg P, Berger BM. Cross-sectional adherence with the multi-target stool DNA test for colorectal cancer screening: Real-world data from a large cohort of older adults. J Med Screen. 2021;28:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Gupta S. Screening for Colorectal Cancer. Hematol Oncol Clin North Am. 2022;36:393-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 97] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 6. | Esteva M, Ruidíaz M, Sánchez MA, Pértega S, Pita-Fernández S, Macià F, Posso M, González-Luján L, Boscá-Wats MM, Leiva A, Ripoll J; DECCIRE GROUP. Emergency presentation of colorectal patients in Spain. PLoS One. 2018;13:e0203556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Dharwadkar P, Zaki TA, Murphy CC. Colorectal Cancer in Younger Adults. Hematol Oncol Clin North Am. 2022;36:449-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 8. | Fisher DA, Saoud L, Finney Rutten LJ, Ozbay AB, Brooks D, Limburg PJ. Lowering the colorectal cancer screening age improves predicted outcomes in a microsimulation model. Curr Med Res Opin. 2021;37:1005-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang AJ, Fang S, Tao L, Li Y, Cheng S, He X, Lan P, Tian C, Liu NN, Zhu L. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun. 2021;12:3063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 137] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 10. | Rodriguez R, Perkins B, Park PY, Koo PJ, Kundranda M, Chang JC. Detecting early colorectal cancer on routine CT scan of the abdomen and pelvis can improve patient's 5-year survival. Arch Biomed Clin Res. 2019;1:1-6. [DOI] [Full Text] |
| 11. | Mangat S, Kozoriz MG, Bicknell S, Spielmann A. The Accuracy of Colorectal Cancer Detection by Computed Tomography in the Unprepared Large Bowel in a Community-Based Hospital. Can Assoc Radiol J. 2018;69:92-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Balthazar EJ, Megibow AJ, Hulnick D, Naidich DP. Carcinoma of the colon: detection and preoperative staging by CT. AJR Am J Roentgenol. 1988;150:301-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 168] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 13. | Johnson CD, Flicek KT, Mead-Harvey C, Quillen JK. Strategies for improving colorectal cancer detection with routine computed tomography. Abdom Radiol (NY). 2023;48:1891-1899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 14. | Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, Suever JD, Geise BD, Patel AA, Moore GJ. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit Med. 2018;1:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 242] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 15. | Cao R, Tang L, Fang M, Zhong L, Wang S, Gong L, Li J, Dong D, Tian J. Artificial intelligence in gastric cancer: applications and challenges. Gastroenterol Rep (Oxf). 2022;10:goac064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Bangert P, Moon H, Woo JO, Didari S, Hao H. Active Learning Performance in Labeling Radiology Images Is 90% Effective. Front Radiol. 2021;1:748968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Sermesant M, Delingette H, Cochet H, Jaïs P, Ayache N. Applications of artificial intelligence in cardiovascular imaging. Nat Rev Cardiol. 2021;18:600-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 18. | Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, Folio LR, Summers RM, Rubin DL, Lungren MP. Preparing Medical Imaging Data for Machine Learning. Radiology. 2020;295:4-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 424] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 19. | Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. 2016 Preprint. Available from: arXiv:1606.06650. [DOI] [Full Text] |
| 20. | Kervadec H, Dolz J, Wang S, Granger E, Ayed IB. Bounding boxes for weakly supervised segmentation: Global constraints get close to full supervision. 2004 Preprint. Available from: arXiv:2004.06816v1. [DOI] [Full Text] |
| 21. | The Cancer Imaging Archive. The Cancer Genome Atlas Colon Adenocarcinoma Collection (TCGA-COAD). [cited 15 July 2023]. Available from: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=16712033. |
| 22. | Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4899] [Cited by in RCA: 6001] [Article Influence: 315.8] [Reference Citation Analysis (0)] |
| 23. | Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. 2015 Preprint. Available from: arXiv:1505.04597v1. [DOI] [Full Text] |
| 24. | Srikrishna M, Heckemann RA, Pereira JB, Volpe G, Zettergren A, Kern S, Westman E, Skoog I, Schöll M. Comparison of Two-Dimensional- and Three-Dimensional-Based U-Net Architectures for Brain Tissue Classification in One-Dimensional Brain CT. Front Comput Neurosci. 2021;15:785244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Zettler N, Mastmeyer A. Comparison of 2D vs. 3D U-Net Organ Segmentation in abdominal 3D CT images. 2021 Preprint. Available from: arXiv:2107.04062v1. [DOI] [Full Text] |
| 26. | Jin H, Li Z, Tong R, Lin L. A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Med Phys. 2018;45:2097-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 27. | Dou Q, Chen H, Yu L, Qin J, Heng PA. Multilevel Contextual 3-D CNNs for False Positive Reduction in Pulmonary Nodule Detection. IEEE Trans Biomed Eng. 2017;64:1558-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 28. | Kim BC, Yoon JS, Choi JS, Suk HI. Multi-scale gradual integration CNN for false positive reduction in pulmonary nodule detection. Neural Netw. 2019;115:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 29. | Shrivastava A, Gupta A, Girschick R. Training region-based object detectors with online hard example mining. 2016 Preprint. Available from: arXiv:1604.03540v1. [DOI] [Full Text] |
| 30. | Seni G, Elder JF. Ensemble Methods in Data Mining: Improving Accuracy Through Combining Predictions. In: Synthesis Lectures on Data Mining and Knowledge Discovery. Germany: Springer, 2010. |
| 31. | Yang T, Zhang L, Yi L, Feng H, Li S, Chen H, Zhu J, Zhao J, Zeng Y, Liu H. Ensemble Learning Models Based on Noninvasive Features for Type 2 Diabetes Screening: Model Development and Validation. JMIR Med Inform. 2020;8:e15431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Boehm KM, Aherne EA, Ellenson L, Nikolovski I, Alghamdi M, Vázquez-García I, Zamarin D, Long Roche K, Liu Y, Patel D, Aukerman A, Pasha A, Rose D, Selenica P, Causa Andrieu PI, Fong C, Capanu M, Reis-Filho JS, Vanguri R, Veeraraghavan H, Gangai N, Sosa R, Leung S, McPherson A, Gao J; MSK MIND Consortium, Lakhman Y, Shah SP. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat Cancer. 2022;3:723-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 155] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 33. | Iseke S, Zeevi T, Kucukkaya AS, Raju R, Gross M, Haider SP, Petukhova-Greenstein A, Kuhn TN, Lin M, Nowak M, Cooper K, Thomas E, Weber MA, Madoff DC, Staib L, Batra R, Chapiro J. Machine Learning Models for Prediction of Posttreatment Recurrence in Early-Stage Hepatocellular Carcinoma Using Pretreatment Clinical and MRI Features: A Proof-of-Concept Study. AJR Am J Roentgenol. 2023;220:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 34. | Korfiatis P, Suman G, Patnam NG, Trivedi KH, Karbhari A, Mukherjee S, Cook C, Klug JR, Patra A, Khasawneh H, Rajamohan N, Fletcher JG, Truty MJ, Majumder S, Bolan CW, Sandrasegaran K, Chari ST, Goenka AH. Automated Artificial Intelligence Model Trained on a Large Data Set Can Detect Pancreas Cancer on Diagnostic Computed Tomography Scans As Well As Visually Occult Preinvasive Cancer on Prediagnostic Computed Tomography Scans. Gastroenterology. 2023;165:1533-1546.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 15.5] [Reference Citation Analysis (0)] |