Published online Nov 28, 2023. doi: 10.4329/wjr.v15.i11.304
Peer-review started: August 28, 2023
First decision: September 19, 2023
Revised: September 20, 2023
Accepted: October 23, 2023
Article in press: October 23, 2023
Published online: November 28, 2023
Processing time: 88 Days and 2.6 Hours
Radiomics can assess prognostic factors in several types of tumors, but consi
To evaluate the performance of two different radiomics software in assessing survival outcomes in pancreatic cancer patients.
We retrospectively reviewed pretreatment contrast-enhanced dual-energy com
3D analysis showed that higher mean tumor density [hazard ratio (HR) = 0.971, P = 0.041)] and higher median tumor density (HR = 0.970, P = 0.037) correlated with better OS. 2D analysis showed that higher mean tumor density (HR = 0.963, P = 0.014) and higher mean positive pixels (HR = 0.962, P = 0.014) correlated with better OS; higher skewness (HR = 3.067, P = 0.008) and higher kurtosis (HR = 1.176, P = 0.029) correlated with worse OS. Higher entropy correlated with better PFS (HR = 0.056, P = 0.036). Models determined that patients with increased tumor size greater than 1.35 cm were likely to have a higher percentage of residual tumors of over 10%.
Several radiomics features can be used as prognostic tools for pancreatic cancer. However, results vary between 2D and 3D analyses. Mean tumor density was the only variable that could reliably predict OS, irrespective of the ana
Core Tip: The use of radiomics to assess pancreatic cancer has been limited. This retrospective study evaluated the performance of 2-dimensional (2D) and 3D radiomic software in determining survival outcomes of pancreatic cancer patients. The mean tumor density was the only variable to reliably predict overall survival (OS) irrespective of the type of analysis. Mean tumor density may be able to differentiate survival and potentially may be help in treatment planning irrespective of the texture analysis software used. Higher skewness [hazard ratio (HR) = 3.067, P = 0.008] and higher kurtosis (HR = 1.176, P = 0.029) correlated with worse OS based on 2D analysis.
- Citation: Saleh M, Virarkar M, Mahmoud HS, Wong VK, Gonzalez Baerga CI, Parikh M, Elsherif SB, Bhosale PR. Radiomics analysis with three-dimensional and two-dimensional segmentation to predict survival outcomes in pancreatic cancer. World J Radiol 2023; 15(11): 304-314
- URL: https://www.wjgnet.com/1949-8470/full/v15/i11/304.htm
- DOI: https://dx.doi.org/10.4329/wjr.v15.i11.304
Pancreatic cancer is an aggressive malignancy causing 7%-8% of cancer-related deaths in the United States[1]. The 5-year overall survival (OS) rate is currently less than 5% despite aggressive multimodality treatment approaches, which mainly include neoadjuvant chemoradiation followed by surgery[2,3]. Up to 60% of patients experience recurrence following definitive therapy[3-5]. Pretreatment stratification of patients based on risk of recurrence and mortality may help de
Several risk factors may affect pancreatic cancer prognosis and OS, such as vascular invasion, lymph node metastasis, tumor stage, and tumor differentiation[6]. Although some prognostic factors may be evaluated by conventional imaging, several others require invasive histologic assessment, which is expensive, carries the risk of complications such as infections or bleeding, and may not provide a complete evaluation of the tumor owing to sampling variability.
There is increasing interest in radiomics because it converts qualitative and subjective imaging data into quantitative and objective data through complex algorithms to provide information that a radiologist cannot extract with the naked eye. Several studies have demonstrated that radiomics can noninvasively assess tumor grade, lymph node metastasis, and other prognostic factors for multiple types of tumors[7-9]. However, only a few studies have evaluated the use of radiomics for assessing prognosis for pancreatic tumors[10-13]. Additionally, texture analysis values extracted via artificial intelligence programs are software-dependent and may vary among software programs due to differing algorithms and processing. For example, MIM performs a 3-dimensional (3D) volumetric analysis, whereas TexRad performs a cross-sectional 2D analysis of only a single slice.
Given this discrepancy, the primary objective of the current study was to determine whether pancreatic tumor texture features can reliably be used as prognostic indicators to provide reproducible results independent of the artificial intelligence software or type of analysis used.
This retrospective single-center study was reviewed and approved by the Institutional Review Board in compliance with HIPAA guidelines. Institutional records between January 2012 and November 2020 were accessed. Our inclusion criteria included patients undergoing a baseline (pretreatment) contrast-enhanced dual-energy computed tomography (CT) study of the primary pancreatic tumor before chemoradiation and subsequent surgery. Patients who did not have: (1) Baseline pretreatment CT study data; (2) Histologic confirmation of a primary tumor of the pancreas; and/or (3) Visible tumors on the CT study were excluded from the analysis. Forty-eight patients met the inclusion criteria. An under
All patients underwent the pretreatment abdominal contrast-enhanced dual-energy CT study via a 64-detector row Discovery CT750 HD CT scanner (Gemstone Spectral Imaging, GE Healthcare, Milwaukee, WI) with a multiphasic pancreatic protocol with rapid switching. Images were acquired intravenously after injecting 125-150 mL of Omnipaque 350 (Mallinckrodt, St Louis, MO) at a rate of 4-5 mL/s. Bolus tracking was used. When a 100-Hounsfield unit (HU) increase was detected at the origin of the celiac axis, images were obtained with a diagnostic scan delay of 20 s, from the level of the hemidiaphragm to the iliac crest, using a rapid switching dual-energy technique (80 kVp and 140 kVp). The scan duration was 5 s for the abdomen. The late arterial/pancreatic parenchymal phase was obtained approximately 40-45 s after the start of contrast injection, and an additional 20-s delay scan using a 120-kVp conventional non-dual-energy imaging technique, resulting in a portal venous phase, was obtained approximately 65-70 s after the start of contrast injection. Images in the pancreatic parenchymal and portal venous phases were reconstructed at 2.5-mm slice thickness. The scan parameters were as follows: Tube current 125-600 ms, tube voltage 120 kVp, pitch 0.98:1, slice thickness 0.6-5 mm, revolution time 0.8 s, table feed speed 39.375 mm/rotation, and field of view 440 mm.
Our study included two separate segmentation programs: MIM software version 6.9.4 (MIM Software Inc) and TexRad Research version 3.9 (Cambridge Computed Imaging LTD). Both radiomics workflows started with tumor segmentation on the treatment-naïve imaging studies, followed by feature extraction. The tumors were segmented by a research fellow in the abdominal radiology department under the supervision of an oncologic radiologist with ten years of experience.
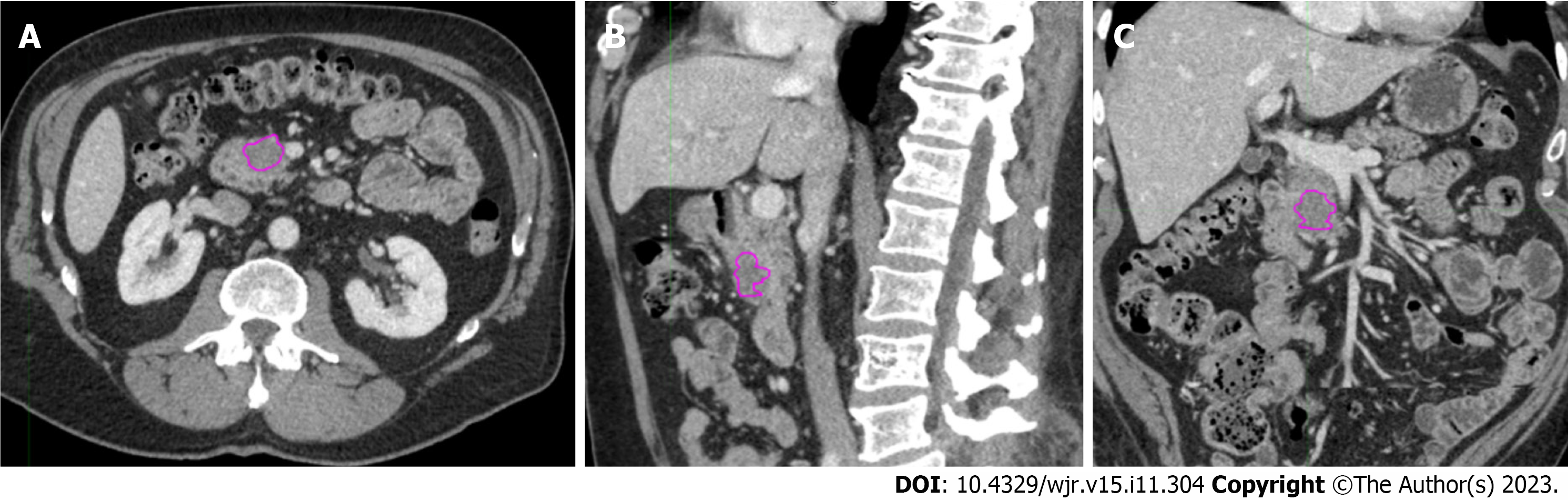
3D segmentation: The portal venous phase was used for segmentation. The tumors were contoured on MIM software using the 3D brush on the axial, coronal, and sagittal planes (Figure 1) by an abdominal radiologist with ten years of clinical experience. A research fellow assisted in extracting the texture features on MIM and saved them on an encrypted server. Seventeen texture features [integral total value (HU × Ml), kurtosis, maximum HU, mean HU, maximum mean HU ratio, median HU, median minimum HU ratio, minimum HU, minimum mean HU ratio, skewness, sphere value (cm), standard deviation, standard deviation mean HU ratio, total HU, volume, voxel count, and entropy], all belonging to first-order statistics, were extracted via an algorithm developed by MIM Software Inc for the texture analysis of CT scans (Table 1).
| MIM Software Inc (3D segmentation) | Cambridge Computed Imaging LTD (2D segmentation) |
| Integral total value | Entropy |
| Kurtosis | Kurtosis |
| Maximum HU | Mean HU |
| Mean HU | Mean positive pixels |
| Maximum mean HU ratio | Skewness |
| Median HU | Standard deviation |
| Median minimum HU ratio | |
| Minimum HU | |
| Minimum mean HU ratio | |
| Skewness | |
| Sphere value | |
| Standard deviation | |
| Standard deviation mean HU ratio | |
| Total HU | |
| Volume | |
| Voxel count | |
| Entropy |
2D segmentation: The images evaluated in 2D segmentation were also derived from the portal venous phase uploaded to the commercially available TexRad research software. 2D segmentation was performed using the polygon region of interest tool. The slice with the greatest tumor diameter was used, and textural radiomic features were extracted automatically from the images within the region of interest. A total of 6 texture features (entropy, kurtosis, mean HU, mean positive pixels, skewness, and standard deviation) were extracted. Because TexRad software applies spatial scale filters and MIM’s algorithm does not, only values without using a spatial scale filter were considered to compare the two soft
For 2D and 3D analysis, the correlation between tumor size and percentage of residual tumor based on histological evaluation was assessed using simple linear regression. The Wilcoxon test was used to show the relationship between tumor volume and the percent of residual tumor. The Youden index was used to determine the optimal cutoff for predicting residual tumors based on tumor size. Also, a receiver operating characteristic curve was generated to predict the performance of tumor size in estimating residual tumors.
Simple logistic regression was used to correlate the texture features with post-treatment adenopathy. A Cox proportional hazards model was used to fit univariate models identifying associations between texture features and OS and progression-free survival (PFS). Cox regression was used to detect any significant association between OS and the per
For 2D and 3D analysis, recursive partitioning analysis was carried out using the R package “rpart” to identify a cutoff that can predict OS by mean HU value. All tests were two-sided, and P < 0.05 was considered statistically significant. Statistical analysis was done using SAS version 9.4 (SAS Institute, Cary, NC). The Kaplan-Meier analysis was used to compare survival in patients with different tumor densities in HU for both 2D and 3D analysis.
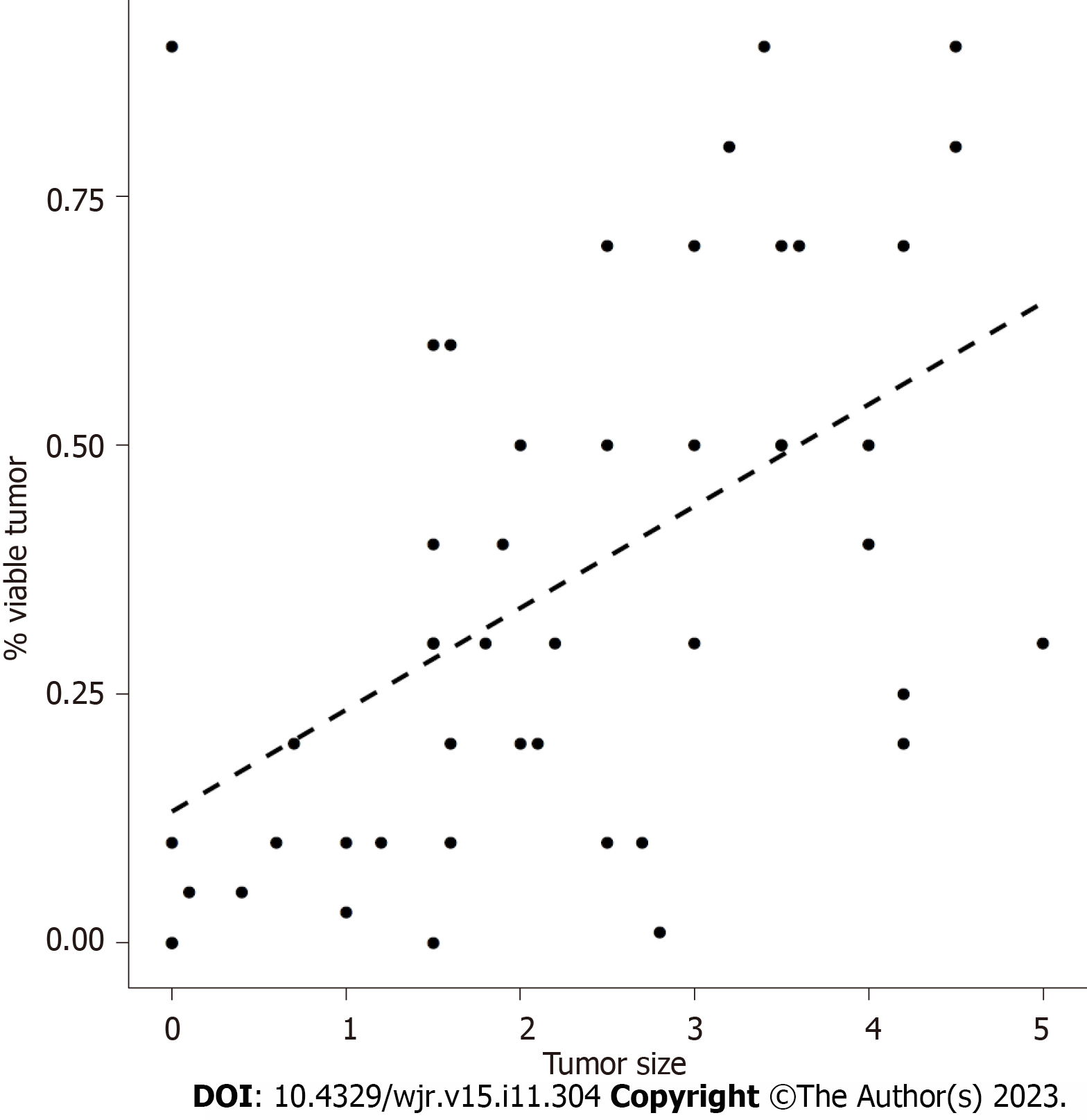
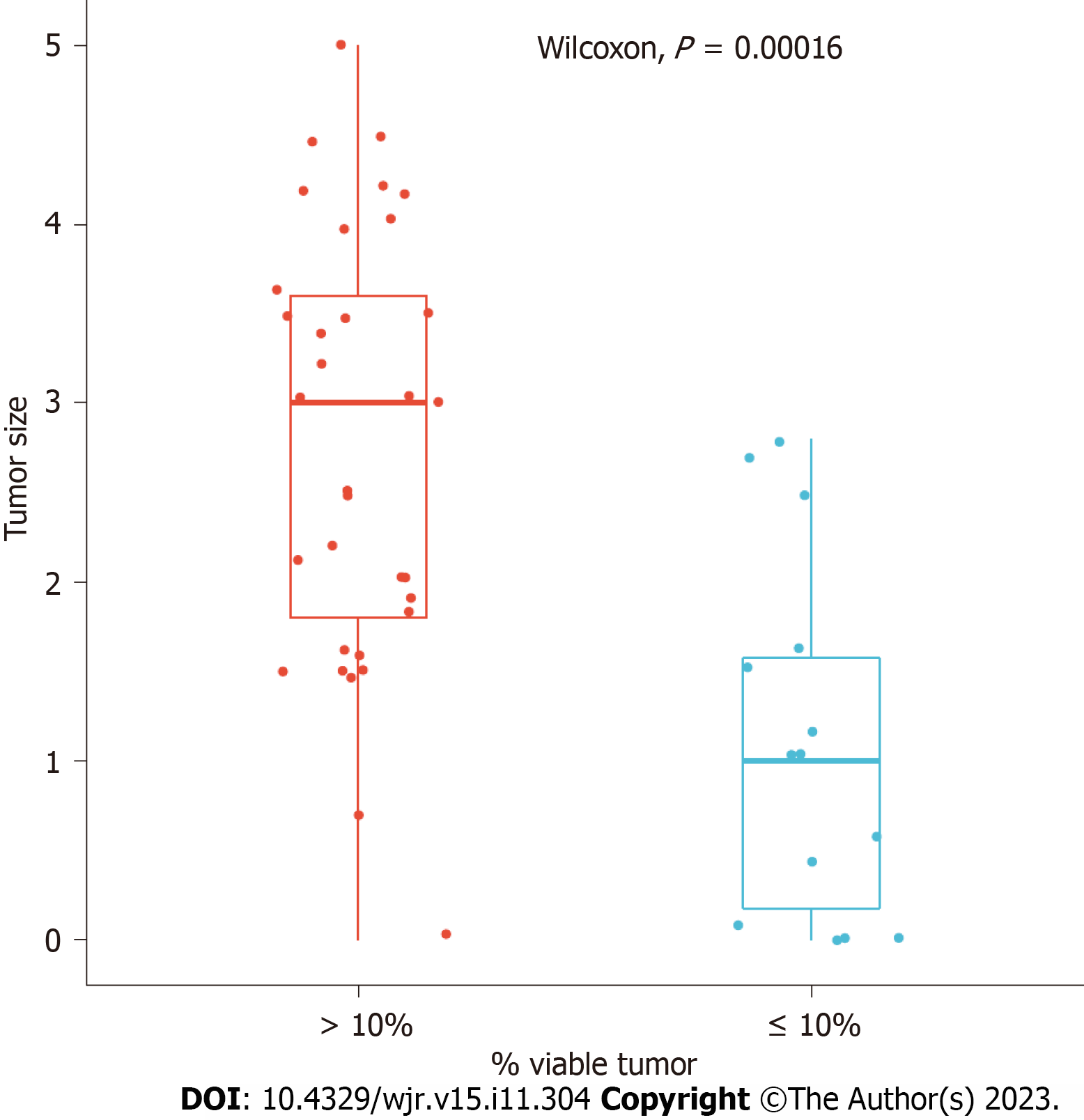
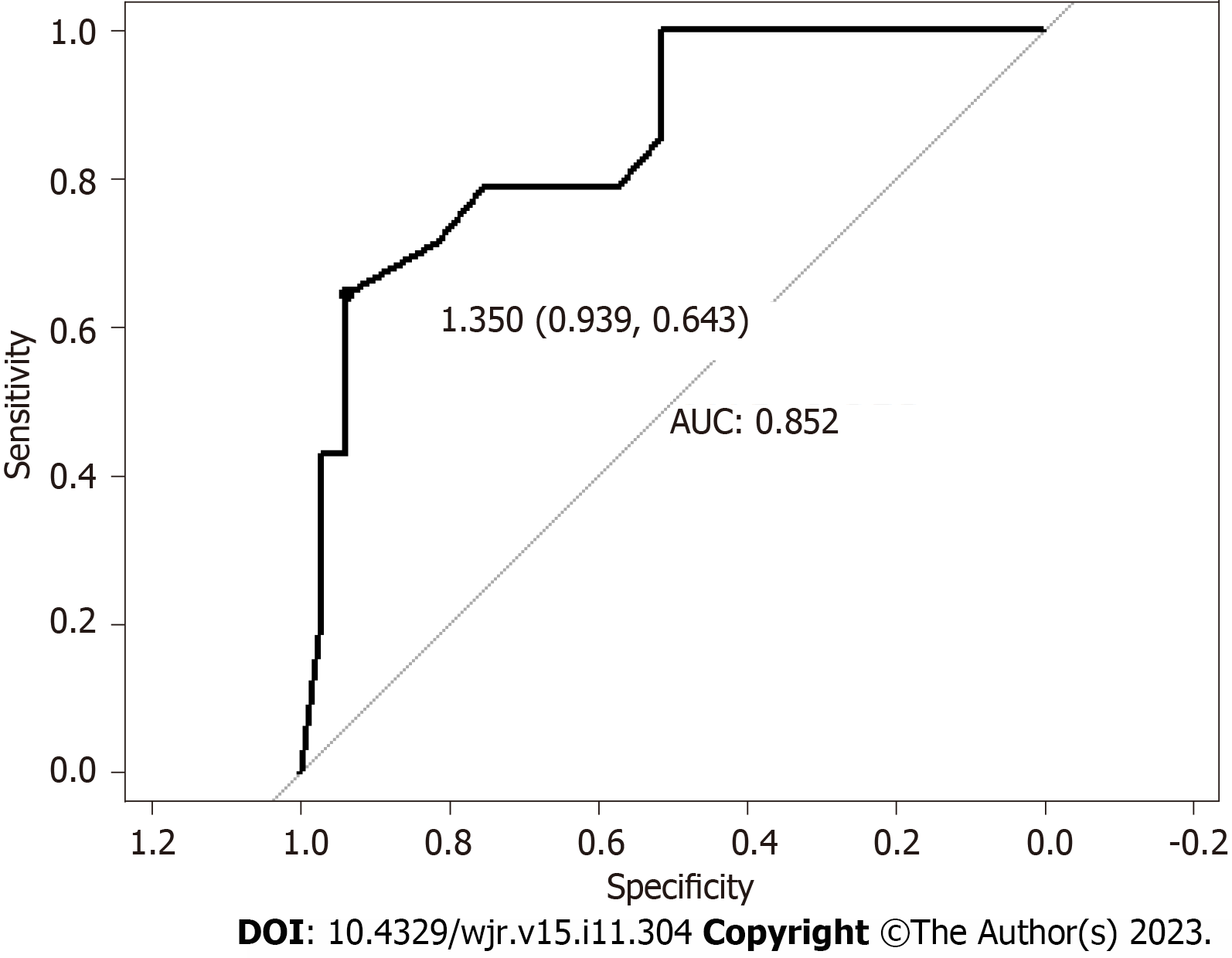
The patients’ mean ± SD age was 61.2 ± 12.8 years, with a median age of 61.9. Patient ages ranged from 18.6 years to 88.9 years. Linear regression showed that histologic tumor size was correlated with residual tumor [correlation coefficient = 0.51, 95% confidence interval (CI): 0.26-0.70, Figure 2]. Using a cutoff of 1.35 cm, based on histologic tumor size, our models showed that patients with a tumor size greater than 1.35 cm are at risk of having more than 10% residual tumor (sensitivity = 0.64, specificity = 0.94, accuracy = 0.85, P = 0.015; Figures 3 and 4).
Linear regression showed that histologic tumor size was correlated with residual tumor (correlation coefficient = 0.51, 95%CI: 0.26-0.70, Figure 2). Using a cutoff of 1.35 cm, based on histologic tumor size, our models showed that patients with a tumor size greater than 1.35 cm are at risk of having more than 10% residual tumor (sensitivity = 0.64, specificity = 0.94, accuracy = 0.85, P = 0.015; Figures 3 and 4).
Linear regression analysis showed that mean HU [correlation coefficient = -0.0040, standard error (SE) = 0.0018, P = 0.0326], median HU (correlation coefficient = -0.0039, SE = 0.0019, P = 0.0373), and minimum mean HU ratio (correlation coefficient = -0.1038, SE = 0.0499, P = 0.0406) were inversely correlated with the percentage of residual tumor following treatment.
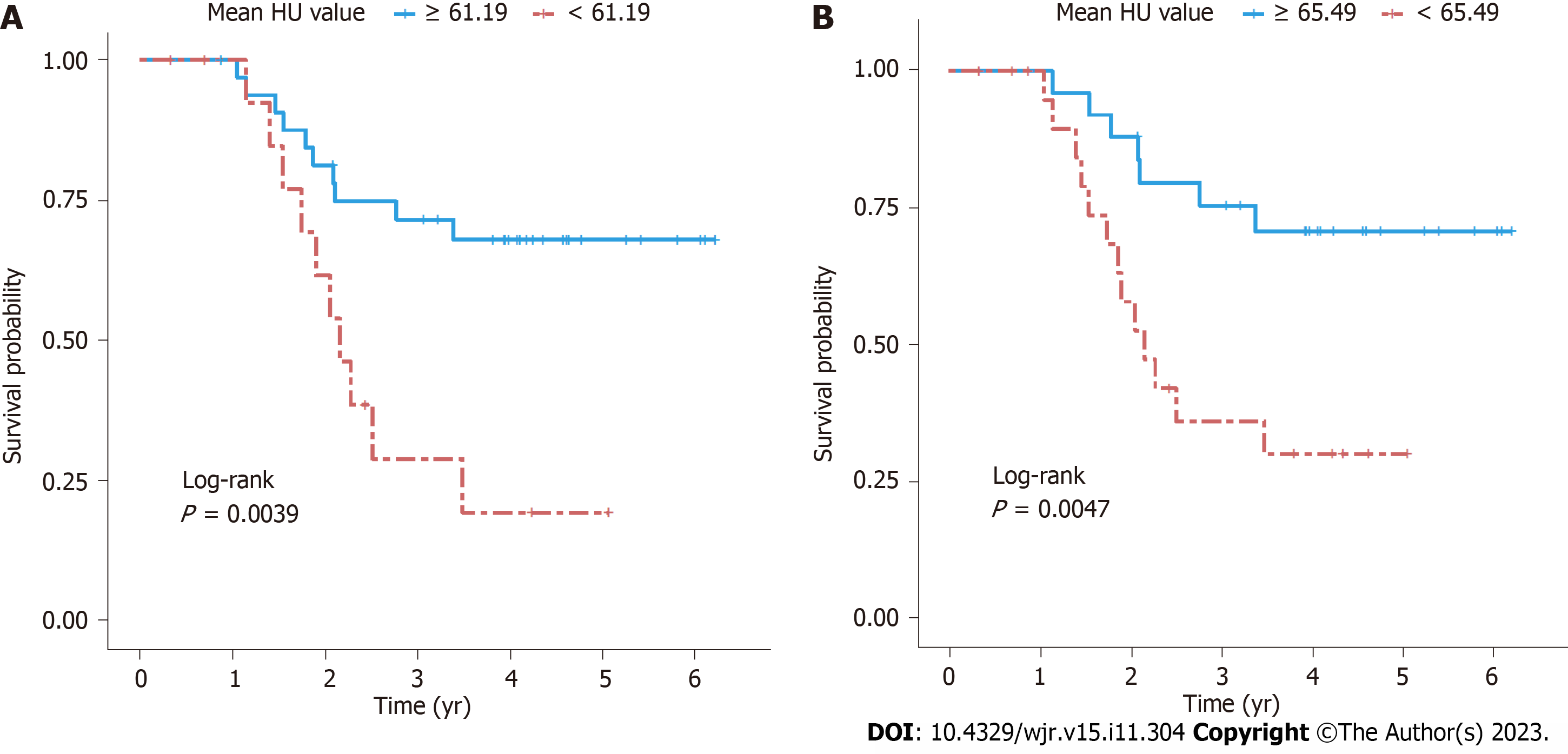
The univariate Cox proportional hazards model showed that mean HU and median HU were significantly correlated with OS [mean HU: hazard ratio (HR) = 0.971, 95%CI: 0.945-0.999, P = 0.041; median HU: HR = 0.970, 95%CI: 0.942-0.998, P = 0.037]. A cutoff value of mean HU ≥ 61.185 significantly predicted better OS (P = 0.0039; Figure 5A). None of the texture features significantly correlated with post-treatment adenopathy or PFS risk.
Without a spatial scale filter, high tumor entropy (correlation coefficient = -0.641, SE = 0.301, P = 0.039), increased mean HU (correlation coefficient = -0.004, SE = 0.002, P = 0.032), and high mean positive pixels (correlation coefficient = -0.005, SE = 0.002, P = 0.032) correlated with less than 10% residual tumor following treatment. Entropy is also positively associated with PFS (HR = 0.056, 95%CI: 0.004-0.831, P = 0.036).
For 2D values, a cutoff of 65.485 for mean HU was appropriate for differentiating mortality risk; patients with equal or higher values than the threshold had significantly better OS (P = 0.0047; Figure 5B). High mean positive pixels were associated with better OS (P = 0.014), whereas high kurtosis (P = 0.029) and skewness (P = 0.007) were associated with worse OS (Table 2). No significant correlations existed between texture features at 0 spatial scale filter and post-treatment adenopathy.
| SSF | Variable | Hazard ratio | 95% CI | P value |
| 0 | Kurtosis | 1.176 | 1.017-1.361 | 0.029 |
| 0 | Mean HU | 0.963 | 0.935-0.993 | 0.014 |
| 0 | MPP | 0.962 | 0.933-0.992 | 0.014 |
| 0 | Skewness | 3.067 | 1.345-6.993 | 0.008 |
| 2 | Entropy | 0.042 | 0.003-0.636 | 0.022 |
| 2 | Mean HU | 0.975 | 0.961-0.989 | 0.001 |
| 3 | Mean HU | 0.980 | 0.968-0.991 | < 0.001 |
| 4 | Kurtosis | 1.071 | 1.000-1.146 | 0.048 |
| 4 | Mean HU | 0.985 | 0.974-0.995 | 0.003 |
| 4 | MPP | 1.012 | 1.000-1.023 | 0.046 |
| 4 | SD | 1.006 | 1.001-1.011 | 0.016 |
| 5 | MPP | 1.009 | 1.002-1.017 | 0.012 |
| 5 | SD | 1.006 | 1.001-1.011 | 0.010 |
| 6 | MPP | 1.007 | 1.002-1.013 | 0.008 |
| 6 | SD | 1.006 | 1.001-1.011 | 0.011 |
Our study suggests that baseline CT-based texture features are noninvasive prognostic indicators that can help predict residual tumors, response to therapy, and prognosis in patients with pancreatic cancer. We also found that some of these textural features are reproducible irrespective of the software or analysis used. 3D and 2D analyses showed that higher tumor density correlated with better OS and lower residual tumors. However, unlike 3D analysis, the 2D analysis also showed that higher skewness and higher kurtosis were correlated with worse OS and higher entropy was correlated with better PFS.
Our study showed that tumor density was a predictor of OS, irrespective of the texture analysis software used. The findings of Cassinotto et al[14] corroborate our findings. In that study, hypoattenuating pancreatic adenocarcinomas on preoperative scans were associated with worse disease-free survival. In addition, hypoattenuating tumors were asso
Higher tumor density (mean HU) and smaller tumor size correlated with a lower percentage of residual tumors following treatment. Our findings suggest that smaller and/or hyperattenuating tumors on the portal venous phase of contrast-enhanced CT will likely respond to chemotherapy because they showed a lower percentage of residual tumors following treatment. To our knowledge, no previous studies have evaluated this correlation. Several studies have shown that patients with pancreatic cancer showing greater than 10% residual tumor following treatment have a worse pro
2D analysis showed that skewness and kurtosis were inversely correlated with OS. Attiyeh et al[22] developed two separate models with 255 radiomics features that measure pixel spatial variation, including kurtosis and skewness; their models demonstrated that tumors with greater heterogeneity were associated with poor OS. Their datasets predicted OS with a concordance index of 0.69 to 0.74. Data from Cozzi et al[23] corroborated the finding that higher heterogeneity correlates with worse prognosis; they found that tumors with lower homogeneity and higher dissimilarity textural features were associated with worse OS. However, skewness and kurtosis were not associated with survival outcomes in their study.
Increasing heterogeneity has also been associated with poor prognosis, irrespective of the imaging modality. For example, Hyun et al[24] reported that intratumoral heterogeneity measured by positron emission tomography textural features in pancreatic cancer patients predicted 2-year OS with an area under the curve of up to 0.72 using entropy fea
Although our study and other studies assessing pancreatic and colorectal cancers[27-29] have shown that higher entropy correlates with better prognosis, several others have shown that higher entropy correlates with worse prognosis in various cancers. This variability in the current literature suggests that entropy should not be used to predict prognosis in pancreatic cancer without further exploration. In our study, 2D analysis showed that higher entropy correlated with better PFS and less residual tumor following treatment. Sandrasegaran et al[29] reported similar findings; they observed better median OS times in patients with tumors with high entropy. However, this did not reach statistical significance. Cassinotto et al[14] showed that higher entropy correlated with less perineural invasion in pancreatic cancer, with an odds ratio of 0.018, which can help explain the positive prognostic implications of high entropy.
Although entropy reflects tumor heterogeneity similarly to skewness and kurtosis, entropy correlated differently with OS and PFS in our study; higher entropy was correlated with better prognosis. Other studies have shown that high entropy is associated with prognosis, poor treatment response, and aggressiveness in colorectal, pulmonary, and central nervous system tumors[27,30-32]. Several factors might explain these results. Unlike skewness and kurtosis, entropy analyzes randomness in the gray levels rather than in the distribution of gray levels in a region of interest[33]. Fur
Additionally, entropy is prone to alterations in processing and image acquisition because entropy is area-dependent, whereby any region of interest covering less than 200 pixels can lead to the inaccurate estimation of entropy’s relationship with any variable[34,35]. TexRad estimates entropy based on Shannon’s model, the most straightforward and earliest model for estimating entropy. Still, TexRad might overestimate entropy by assuming that the pixels within a region of interest have an identical distribution and are entirely independent of neighboring pixels[34,36]. Given the insufficient data about the prognostic implications of entropy for pancreatic cancers, further studies are required to accurately assess the prognostic impact of entropy.
Our study has several strengths. Because all images were obtained on the same scanner, any heterogeneity in the results that may arise from using different scanners has been ruled out. Additionally, most studies using radiomics to evaluate response to treatment usually use delta radiomics to compare pretreatment and post-treatment scans. For example, Nasief et al[37] demonstrated that delta-radiomic features obtained during treatment periods could distinguish poor responders from good responders with an area under the curve of 0.94. Our pretreatment findings allow identifying patients who are more likely to respond before any treatment using baseline imaging, allowing for treatment selection that minimizes morbidity and thus limits expenses. This contrasts delta radiomics, which evaluates response after treatment has begun. Another strength of our study is comparing 2D and 3D analysis using TexRad and MIM software, respectively. It is well known that radiomics depend on how they are processed, and the software used[38,39]. However, our comparison showed that tumor density (mean HU) is a consistent and valid predictor of OS and PFS, irrespective of the type of analysis used.
Our study has some limitations. Our sample size was small, and it was a retrospective study. Therefore, more extensive prospective studies are needed to validate our findings. Additionally, CT acquisition factors might affect texture analysis variables; however, the effect is minimal. Lastly, our results were not externally validated and can only be directly app
In conclusion, tumor density (mean HU) was the only variable in our study that could reliably predict OS and PFS, irrespective of the type of analysis used. This variable may be used as a prognostic indicator to differentiate high-risk patients from low-risk patients and could be used for treatment planning. However, prospective studies will be beneficial to validate our findings externally.
Radiomics can determine prognostic factors of several types of tumors.
Lack of evidence supporting radiomic studies on pancreatic cancer.
Compare two different radiomic softwares in assessing survival outcomes in pancreatic cancer patients.
Retrospective review of pretreatment dual energy computed tomography (CT) images of 48 patients with biopsy confirmed lesions. Tumors were segmented using TexRad [2-dimensional (2D)] analysis software and MIM (3D) analysis software and radiomic features were extracted to compare with overall surgical (OS) and progression free survival (PFS).
3D analysis demonstrates that higher mean tumor density and median tumor density correlated with better OS, while 2D analysis showed that higher mean tumor density and mean positive pixels correlated with better OS. 2D analysis also showed higher skewness and kurtosis correlated with worse OS. Higher entropy correlated with better PFS. Patients with increased tumor size greater than 1.35 cm were likely to have a higher percentage of residual tumor above 10%.
Radiomic features can serve as prognosis tools for pancreatic cancer and determine OS.
This study serves as a guide for future research that can be verified through a prospective approach, while also con
We thank Erica Goodoff, Senior Scientific Editor in the Research Medical Library at The University of Texas MD Ander
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lin W, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15469] [Article Influence: 2578.2] [Reference Citation Analysis (2)] |
| 2. | Malla M, Fekrmandi F, Malik N, Hatoum H, George S, Goldberg RM, Mukherjee S. The evolving role of radiation in pancreatic cancer. Front Oncol. 2022;12:1060885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 3. | Krishnan S, Chadha AS, Suh Y, Chen HC, Rao A, Das P, Minsky BD, Mahmood U, Delclos ME, Sawakuchi GO, Beddar S, Katz MH, Fleming JB, Javle MM, Varadhachary GR, Wolff RA, Crane CH. Focal Radiation Therapy Dose Escalation Improves Overall Survival in Locally Advanced Pancreatic Cancer Patients Receiving Induction Chemotherapy and Consolidative Chemoradiation. Int J Radiat Oncol Biol Phys. 2016;94:755-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 274] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 4. | Griffin JF, Smalley SR, Jewell W, Paradelo JC, Reymond RD, Hassanein RE, Evans RG. Patterns of failure after curative resection of pancreatic carcinoma. Cancer. 1990;66:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 5. | Brody JR, Costantino CL, Potoczek M, Cozzitorto J, McCue P, Yeo CJ, Hruban RH, Witkiewicz AK. Adenosquamous carcinoma of the pancreas harbors KRAS2, DPC4 and TP53 molecular alterations similar to pancreatic ductal adenocarcinoma. Mod Pathol. 2009;22:651-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Dell'Aquila E, Fulgenzi CAM, Minelli A, Citarella F, Stellato M, Pantano F, Russano M, Cursano MC, Napolitano A, Zeppola T, Vincenzi B, Tonini G, Santini D. Prognostic and predictive factors in pancreatic cancer. Oncotarget. 2020;11:924-941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 7. | Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 357] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 8. | Wu S, Zheng J, Li Y, Yu H, Shi S, Xie W, Liu H, Su Y, Huang J, Lin T. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin Cancer Res. 2017;23:6904-6911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 300] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 9. | Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology. 2018;287:833-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 260] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 10. | Tang TY, Li X, Zhang Q, Guo CX, Zhang XZ, Lao MY, Shen YN, Xiao WB, Ying SH, Sun K, Yu RS, Gao SL, Que RS, Chen W, Huang DB, Pang PP, Bai XL, Liang TB. Development of a Novel Multiparametric MRI Radiomic Nomogram for Preoperative Evaluation of Early Recurrence in Resectable Pancreatic Cancer. J Magn Reson Imaging. 2020;52:231-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 11. | Nasief H, Zheng C, Schott D, Hall W, Tsai S, Erickson B, Allen Li X. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis Oncol. 2019;3:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 12. | Zhang Y, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA, Khalvati F. Prognostic Value of Transfer Learning Based Features in Resectable Pancreatic Ductal Adenocarcinoma. Front Artif Intell. 2020;3:550890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Chakraborty J, Midya A, Gazit L, Attiyeh M, Langdon-Embry L, Allen PJ, Do RKG, Simpson AL. CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas. Med Phys. 2018;45:5019-5029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 14. | Cassinotto C, Chong J, Zogopoulos G, Reinhold C, Chiche L, Lafourcade JP, Cuggia A, Terrebonne E, Dohan A, Gallix B. Resectable pancreatic adenocarcinoma: Role of CT quantitative imaging biomarkers for predicting pathology and patient outcomes. Eur J Radiol. 2017;90:152-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 15. | Fukukura Y, Takumi K, Higashi M, Shinchi H, Kamimura K, Yoneyama T, Tateyama A. Contrast-enhanced CT and diffusion-weighted MR imaging: performance as a prognostic factor in patients with pancreatic ductal adenocarcinoma. Eur J Radiol. 2014;83:612-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Zhu L, Shi X, Xue H, Wu H, Chen G, Sun H, He Y, Jin Z, Liang Z, Zhang Z. CT Imaging Biomarkers Predict Clinical Outcomes After Pancreatic Cancer Surgery. Medicine (Baltimore). 2016;95:e2664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 17. | Kong B, Cheng T, Wu W, Regel I, Raulefs S, Friess H, Erkan M, Esposito I, Kleeff J, Michalski CW. Hypoxia-induced endoplasmic reticulum stress characterizes a necrotic phenotype of pancreatic cancer. Oncotarget. 2015;6:32154-32160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Hattori Y, Gabata T, Zen Y, Mochizuki K, Kitagawa H, Matsui O. Poorly enhanced areas of pancreatic adenocarcinomas on late-phase dynamic computed tomography: comparison with pathological findings. Pancreas. 2010;39:1263-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 19. | White RR, Xie HB, Gottfried MR, Czito BG, Hurwitz HI, Morse MA, Blobe GC, Paulson EK, Baillie J, Branch MS, Jowell PS, Clary BM, Pappas TN, Tyler DS. Significance of histological response to preoperative chemoradiotherapy for pancreatic cancer. Ann Surg Oncol. 2005;12:214-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 126] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Chatterjee D, Katz MH, Rashid A, Varadhachary GR, Wolff RA, Wang H, Lee JE, Pisters PW, Vauthey JN, Crane C, Gomez HF, Abbruzzese JL, Fleming JB. Histologic grading of the extent of residual carcinoma following neoadjuvant chemoradiation in pancreatic ductal adenocarcinoma: a predictor for patient outcome. Cancer. 2012;118:3182-3190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 21. | Okubo S, Kojima M, Matsuda Y, Hioki M, Shimizu Y, Toyama H, Morinaga S, Gotohda N, Uesaka K, Ishii G, Mino-Kenudson M, Takahashi S. Area of residual tumor (ART) can predict prognosis after post neoadjuvant therapy resection for pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:17145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Attiyeh MA, Chakraborty J, Doussot A, Langdon-Embry L, Mainarich S, Gönen M, Balachandran VP, D'Angelica MI, DeMatteo RP, Jarnagin WR, Kingham TP, Allen PJ, Simpson AL, Do RK. Survival Prediction in Pancreatic Ductal Adenocarcinoma by Quantitative Computed Tomography Image Analysis. Ann Surg Oncol. 2018;25:1034-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 23. | Cozzi L, Comito T, Fogliata A, Franzese C, Franceschini D, Bonifacio C, Tozzi A, Di Brina L, Clerici E, Tomatis S, Reggiori G, Lobefalo F, Stravato A, Mancosu P, Zerbi A, Sollini M, Kirienko M, Chiti A, Scorsetti M. Computed tomography based radiomic signature as predictive of survival and local control after stereotactic body radiation therapy in pancreatic carcinoma. PLoS One. 2019;14:e0210758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 24. | Hyun SH, Kim HS, Choi SH, Choi DW, Lee JK, Lee KH, Park JO, Kim BT, Choi JY. Intratumoral heterogeneity of (18)F-FDG uptake predicts survival in patients with pancreatic ductal adenocarcinoma. Eur J Nucl Med Mol Imaging. 2016;43:1461-1468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 25. | Verbeke C. Morphological heterogeneity in ductal adenocarcinoma of the pancreas - Does it matter? Pancreatology. 2016;16:295-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 26. | Bandyopadhyay S, Basturk O, Coban I, Thirabanjasak D, Liang H, Altinel D, Adsay NV. Isolated solitary ducts (naked ducts) in adipose tissue: a specific but underappreciated finding of pancreatic adenocarcinoma and one of the potential reasons of understaging and high recurrence rate. Am J Surg Pathol. 2009;33:425-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology. 2013;266:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 342] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 28. | Lubner MG, Stabo N, Lubner SJ, del Rio AM, Song C, Halberg RB, Pickhardt PJ. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331-2337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 224] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 29. | Sandrasegaran K, Lin Y, Asare-Sawiri M, Taiyini T, Tann M. CT texture analysis of pancreatic cancer. Eur Radiol. 2019;29:1067-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 30. | Chee CG, Kim YH, Lee KH, Lee YJ, Park JH, Lee HS, Ahn S, Kim B. CT texture analysis in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy: A potential imaging biomarker for treatment response and prognosis. PLoS One. 2017;12:e0182883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 31. | Zhao Q, Shi CZ, Luo LP. Role of the texture features of images in the diagnosis of solitary pulmonary nodules in different sizes. Chin J Cancer Res. 2014;26:451-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 32. | Ryu YJ, Choi SH, Park SJ, Yun TJ, Kim JH, Sohn CH. Glioma: application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS One. 2014;9:e108335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 33. | Guan Y, Li W, Jiang Z, Chen Y, Liu S, He J, Zhou Z, Ge Y. Whole-Lesion Apparent Diffusion Coefficient-Based Entropy-Related Parameters for Characterizing Cervical Cancers: Initial Findings. Acad Radiol. 2016;23:1559-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 34. | Dercle L, Ammari S, Bateson M, Durand PB, Haspinger E, Massard C, Jaudet C, Varga A, Deutsch E, Soria JC, Ferté C. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci Rep. 2017;7:7952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 35. | Shannon CE. A Mathematical Theory of Communication. New York: Nokia Bell Labs, 1948; 27: 379-423. |
| 36. | Razlighi QR, Kehtarnavaz N, Nosratinia A. Computation of image spatial entropy using quadrilateral Markov random field. IEEE Trans Image Process. 2009;18:2629-2639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 37. | Nasief H, Hall W, Zheng C, Tsai S, Wang L, Erickson B, Li XA. Improving Treatment Response Prediction for Chemoradiation Therapy of Pancreatic Cancer Using a Combination of Delta-Radiomics and the Clinical Biomarker CA19-9. Front Oncol. 2019;9:1464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Defeudis A, De Mattia C, Rizzetto F, Calderoni F, Mazzetti S, Torresin A, Vanzulli A, Regge D, Giannini V. Standardization of CT radiomics features for multi-center analysis: impact of software settings and parameters. Phys Med Biol. 2020;65:195012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG 3rd. Variation in algorithm implementation across radiomics software. J Med Imaging (Bellingham). 2018;5:044505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |