Copyright
©2014 Baishideng Publishing Group Inc.
World J Cardiol. Aug 26, 2014; 6(8): 744-754
Published online Aug 26, 2014. doi: 10.4330/wjc.v6.i8.744
Published online Aug 26, 2014. doi: 10.4330/wjc.v6.i8.744
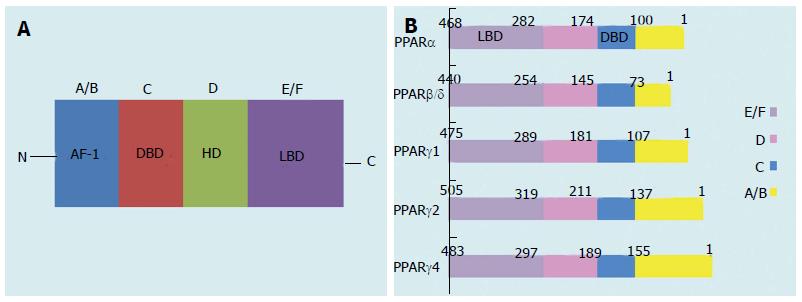
Figure 1 Schematic structure of peroxisome proliferator-activated receptor protein isoforms.
A/B, C, D, and E/F indicate the N-terminal A/B domain containing a ligand-independent AF-1, the DBD, the hinge region, and the C-terminal LBD containing AF-2, respectively. AF-1 is responsible for phosphorylation, while AF-2 promotes the recruitment of co-activators for gene transcription. PPAR: Peroxisome proliferator-activated receptor; AF-1: Activation function-1; DBD: DNA-binding domain; HD: Hinge domain; LBD: Ligand-binding domain. Figure adapted from reference[8].
Figure 2 Domain structure of the peroxisome proliferator-activated receptor γ isoforms.
PPAR: Peroxisome proliferator-activated receptor. Figure adapted from reference[8].
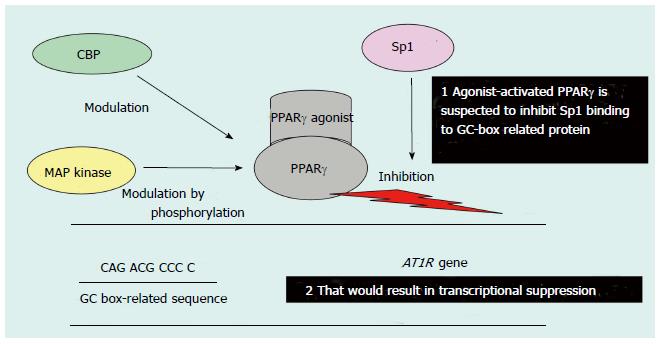
Figure 3 Possible mechanism of peroxisome proliferator-activated receptor γ-agonist-mediated transcriptional suppression of the Ang-II type 1 receptor gene promoter.
PPAR: Peroxisome proliferator-activated receptor; AT1R: Ang-II type 1 receptor; CBP: CERB-binding protein; MAP: Mitogen-activated protein. Figure adapted from reference[71].
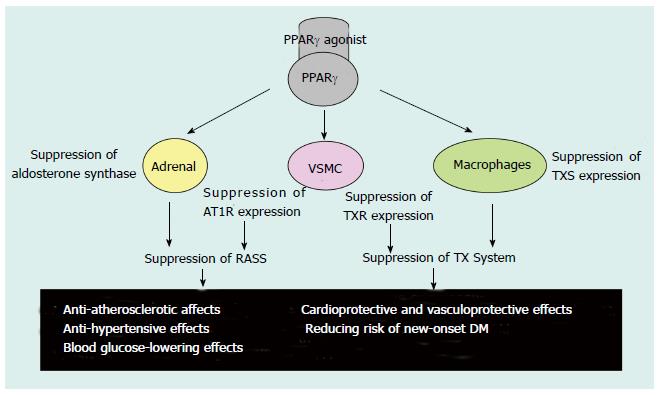
Figure 4 Possible effects of Peroxisome proliferator-activated receptor γ agonists.
PPAR: Peroxisome proliferator-activated receptor; AT1R: Ang-II type 1 receptor; RAAS: Renin-angiotensin-aldosterone system; TX: Thromboxane; TXS: TX synthase; TXR: TX receptor; VSMC: Vascular smooth muscle cells; DM: Diabetes mellitus. Table adapted from reference[71].
- Citation: Usuda D, Kanda T. Peroxisome proliferator-activated receptors for hypertension. World J Cardiol 2014; 6(8): 744-754
- URL: https://www.wjgnet.com/1949-8462/full/v6/i8/744.htm
- DOI: https://dx.doi.org/10.4330/wjc.v6.i8.744