Copyright
©The Author(s) 2025.
World J Cardiol. Jan 26, 2025; 17(1): 102147
Published online Jan 26, 2025. doi: 10.4330/wjc.v17.i1.102147
Published online Jan 26, 2025. doi: 10.4330/wjc.v17.i1.102147
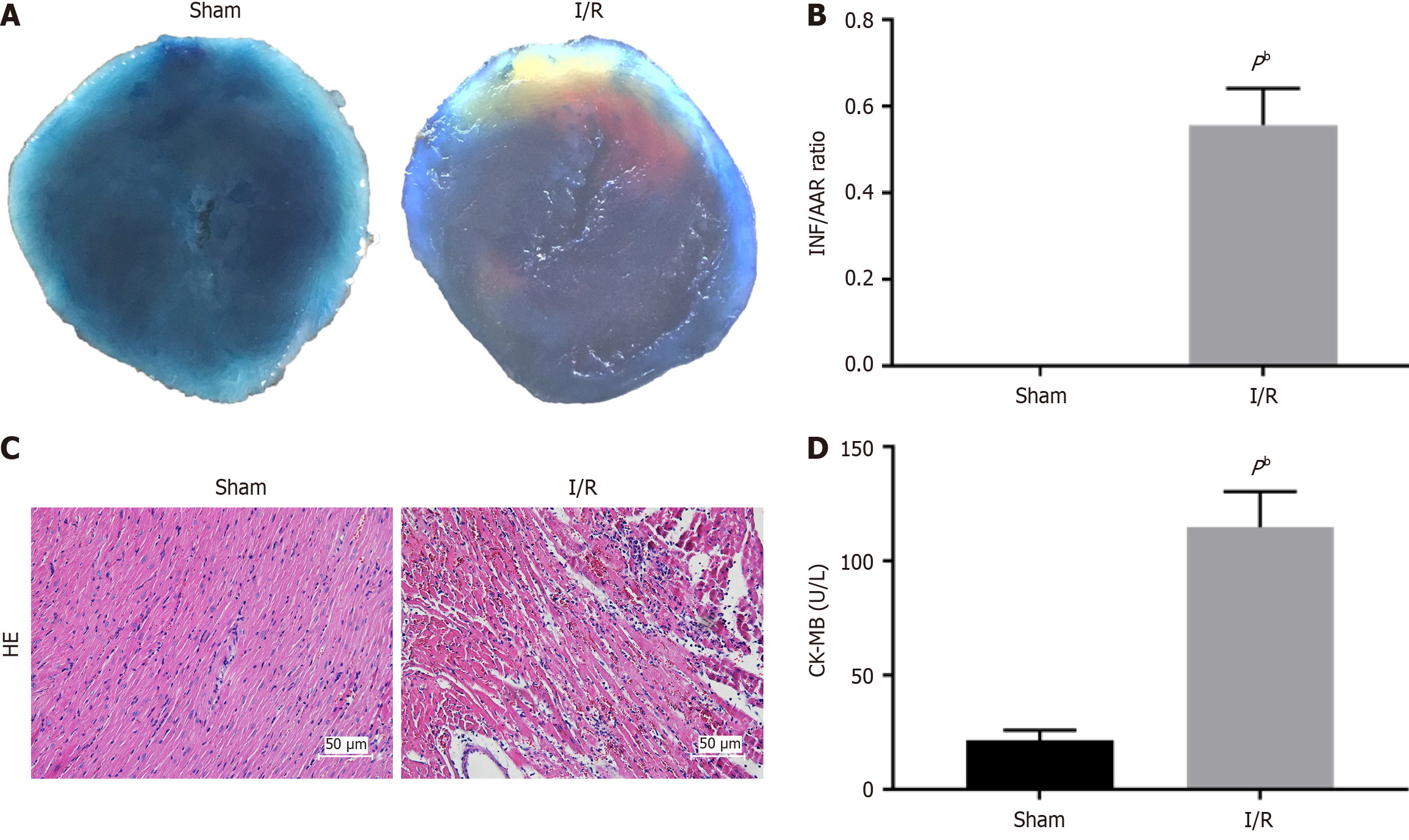
Figure 1 Ischemia/reperfusion induced myocardial injury in mice.
A: Representative images of the infract area international normalized ratio (INR) (white), area at risk (AAR) (red and white), and normal area (blue); B: Quantitative analysis of infarct size and the ratio of INF/AAR; C: Representative images of myocardial tissues from the myocardial ischemia/reperfusion group and sham group; D: Quantitative analysis of creatine kinase-myocardial bound level. All Data are shown as means ± SD. aP < 0.05, bP < 0.01 vs sham group, n = 6 per group. AAR: Area at risk; CK-MB: Creatine kinase-myocardial bound; HE: Hematoxylin-eosin; INR: International normalized ratio; I/R: Ischemia/reperfusion.
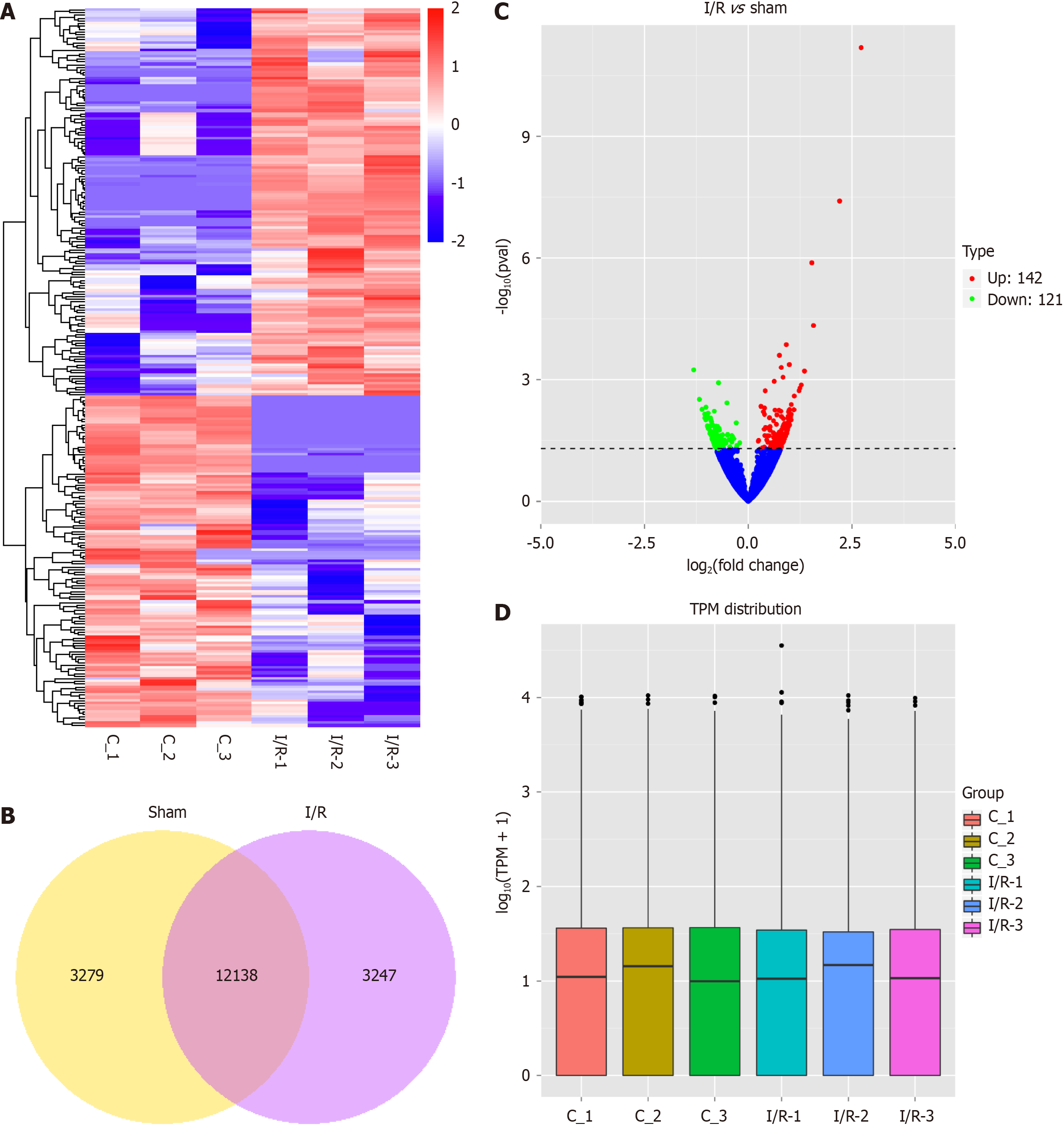
Figure 2 Circular RNA expression profile comparison between the ischemia/reperfusion group (ischemia/reperfusion 1-3) and sham group (C 1-3).
A: Hierarchical clustering demonstrates the differential circular RNA (circRNA) expression profiling between the ischemia/reperfusion group and sham group; B: The number of circRNAs identified in the two groups; C: Volcano plot shows a difference of circRNA expression between the two groups. In the volcano plots, the red and green points represent the circRNAs with log 20-fold changes (up and down regulated) with statistical significance (P < 0.05); D: Box plot shows the distribution of circRNA expression patterns of two group of samples was not different. I/R: Ischemia/reperfusion.
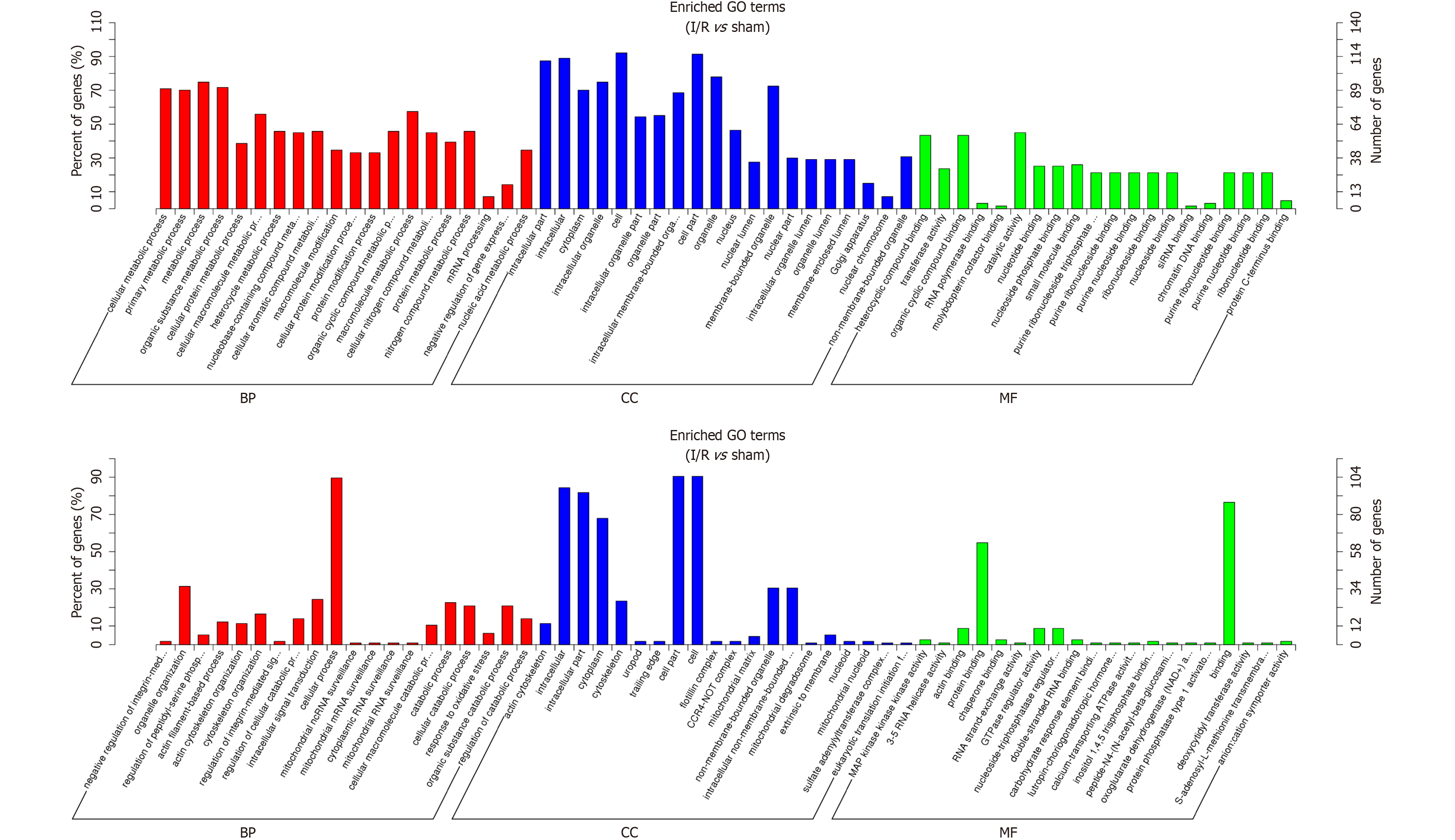
Figure 3 Top 10 classes of Gene Ontology enrichment terms.
A: Upregulated Gene Ontology (GO) terms; B: Downregulated GO terms. The GO terms were classified into three parts including biological processes, cellular components, and molecular functions. BP: Biological processes; CC: Cellular components; GO: Gene Ontology; MF: Molecular functions; I/R: Ischemia/reperfusion.
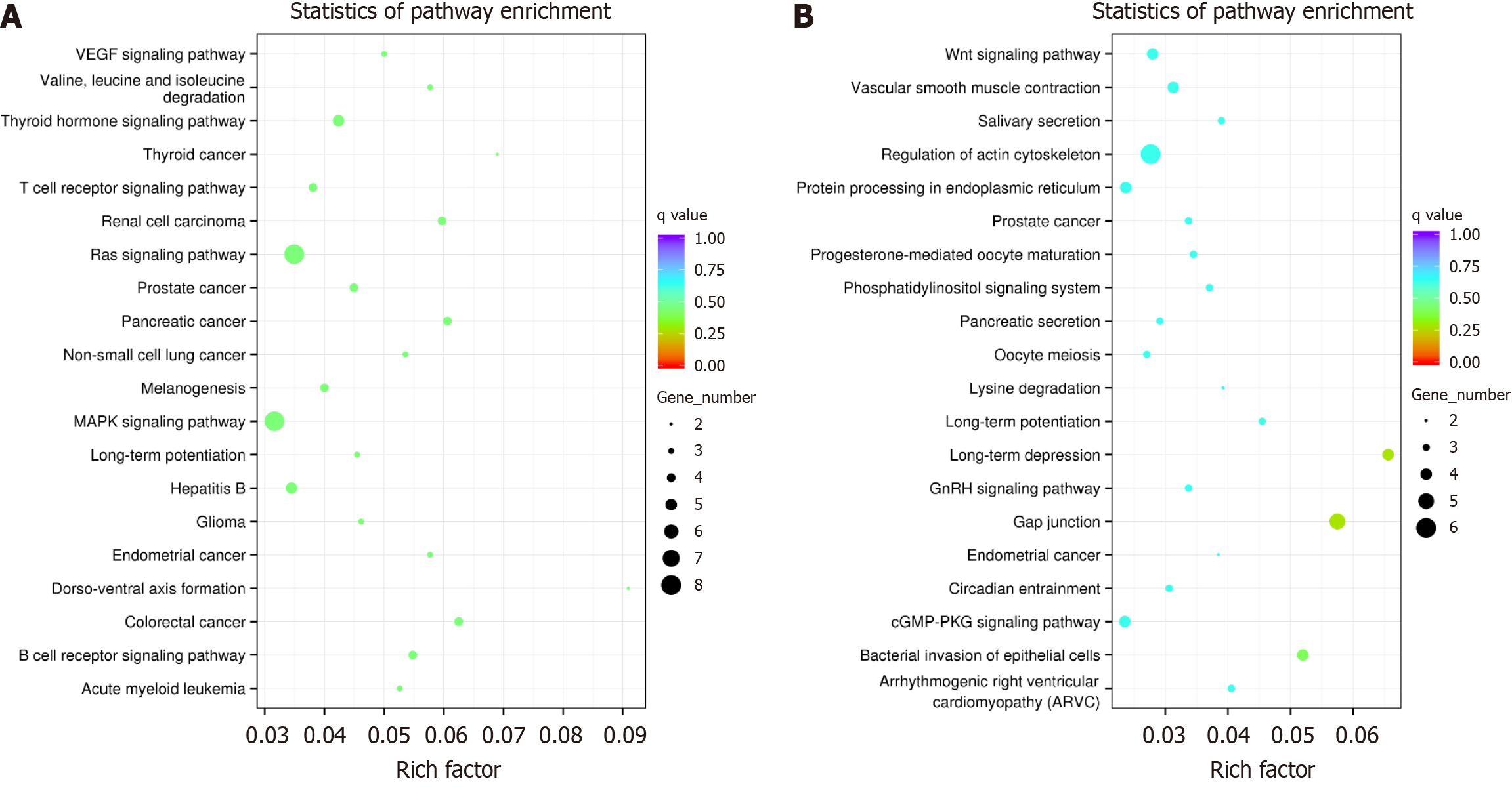
Figure 4 Top 20 classes of Kyoto Encyclopedia of Genes and Genomes pathway enrichment terms.
A: Upregulated gene pathways; B: Downregulated gene pathways. The size of the points represents the number of genes mapped, and the lower q value is, the more significant the pathway was enriched. VEGF: Vascular endothelial growth factors.
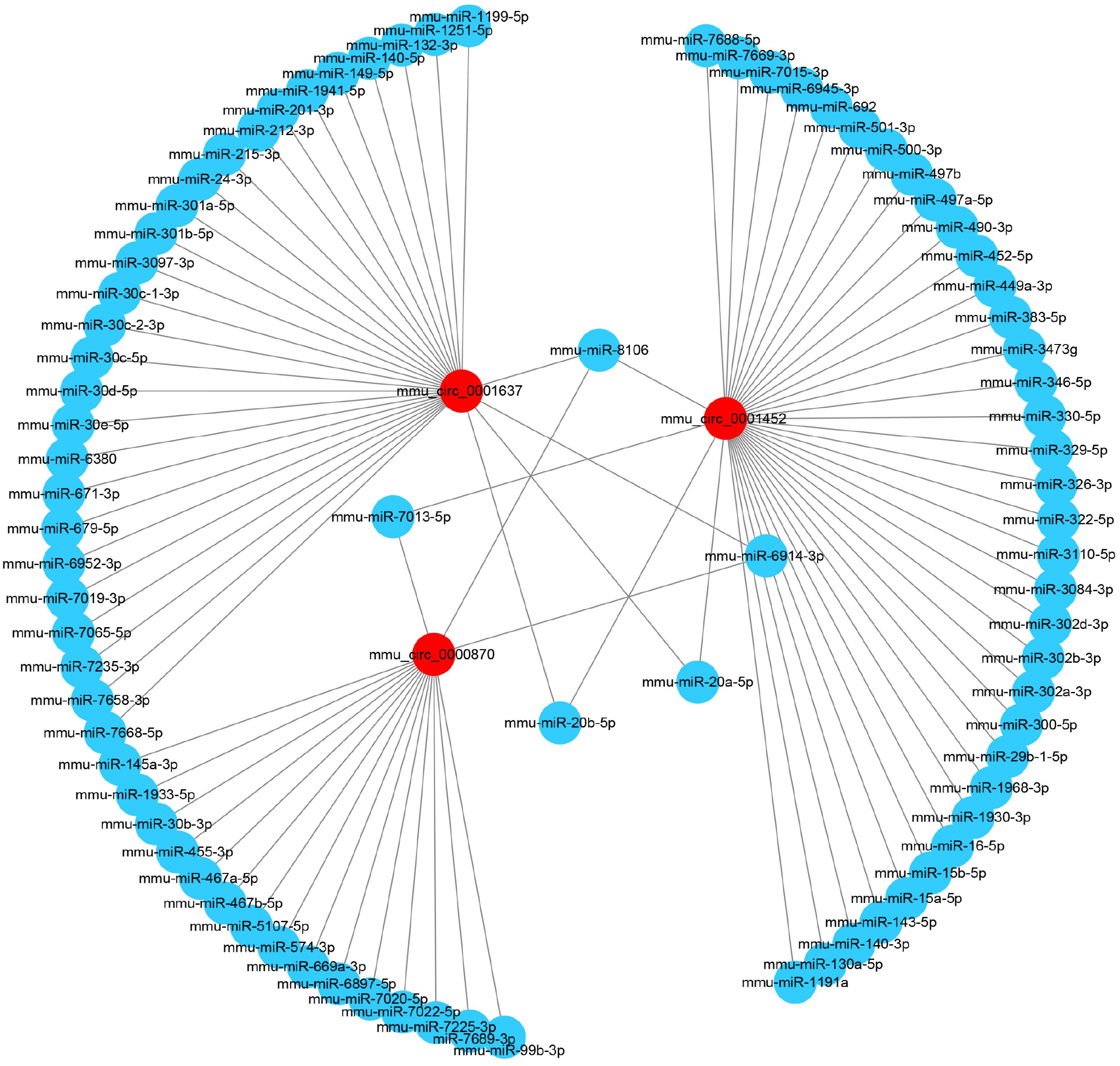
Figure 5 Construction of circular RNA–miRNA interaction networks.
This network was based on the expression profile results and the related software. The 3 dysregulated circular RNAs, mmu_circ_0001637, mmu_circ_0001452, and mmu_circ_0000870 (purple red nodes) having the highest magnitude of change, were predicted to be functionally connected with their targeted microRNAs in the network.
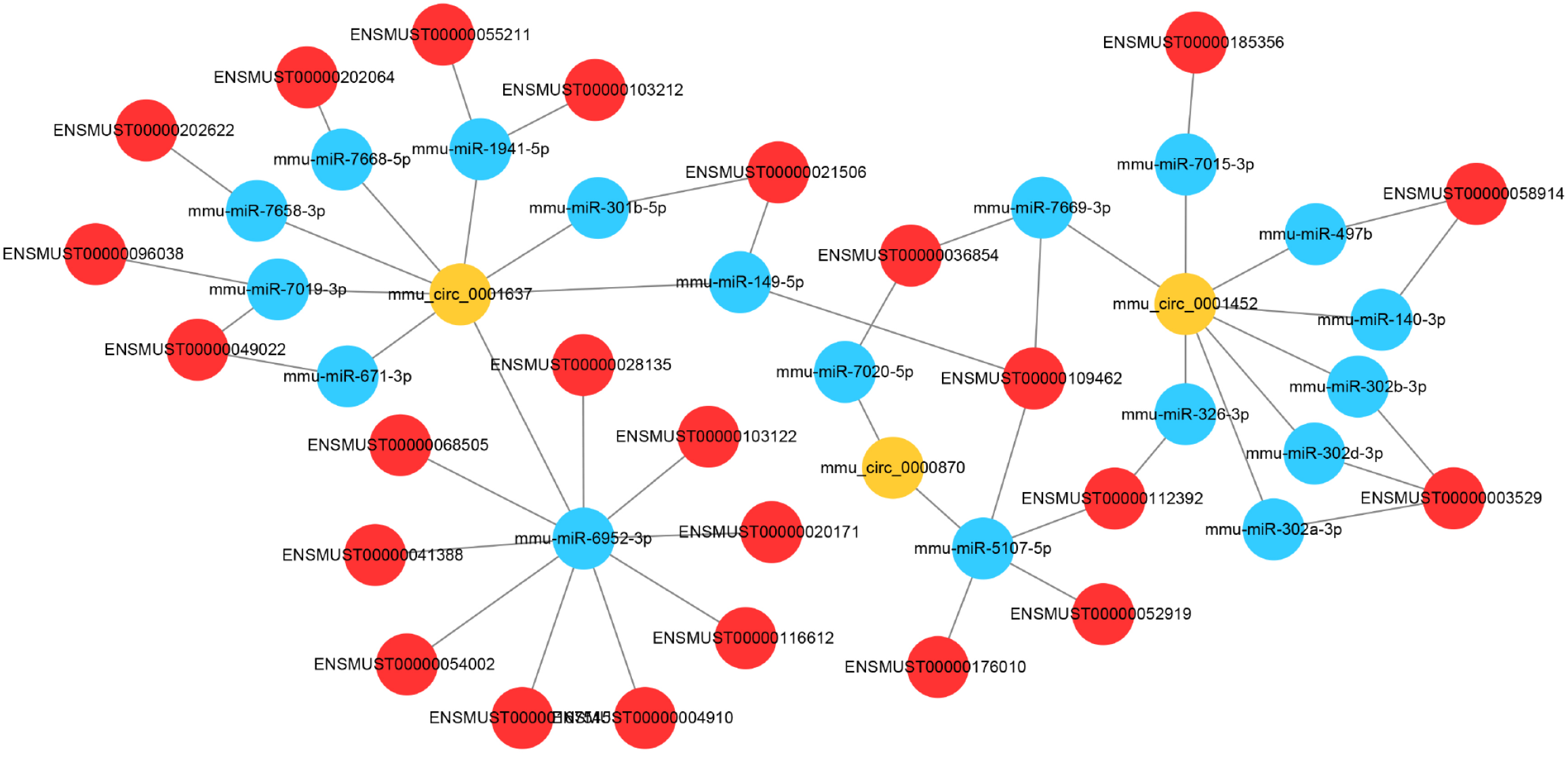
Figure 6 CeRNA network.
Circular RNA (circRNA)- microRNAs (miRNA)-mRNA interaction pairs were calculated using miRanda. The network was generated using Cytoscape software. The yellow nodes represent for circRNAs, the red nodes represent for mRNAs, the blue nodes represent for miRNA.
- Citation: Wang JN, Zhou YY, Yu YW, Chen J. Profiling and bioinformatics analyses of circular RNAs in myocardial ischemia/reperfusion injury model in mice. World J Cardiol 2025; 17(1): 102147
- URL: https://www.wjgnet.com/1949-8462/full/v17/i1/102147.htm
- DOI: https://dx.doi.org/10.4330/wjc.v17.i1.102147