Copyright
©The Author(s) 2020.
World J Cardiol. Jul 26, 2020; 12(7): 303-333
Published online Jul 26, 2020. doi: 10.4330/wjc.v12.i7.303
Published online Jul 26, 2020. doi: 10.4330/wjc.v12.i7.303
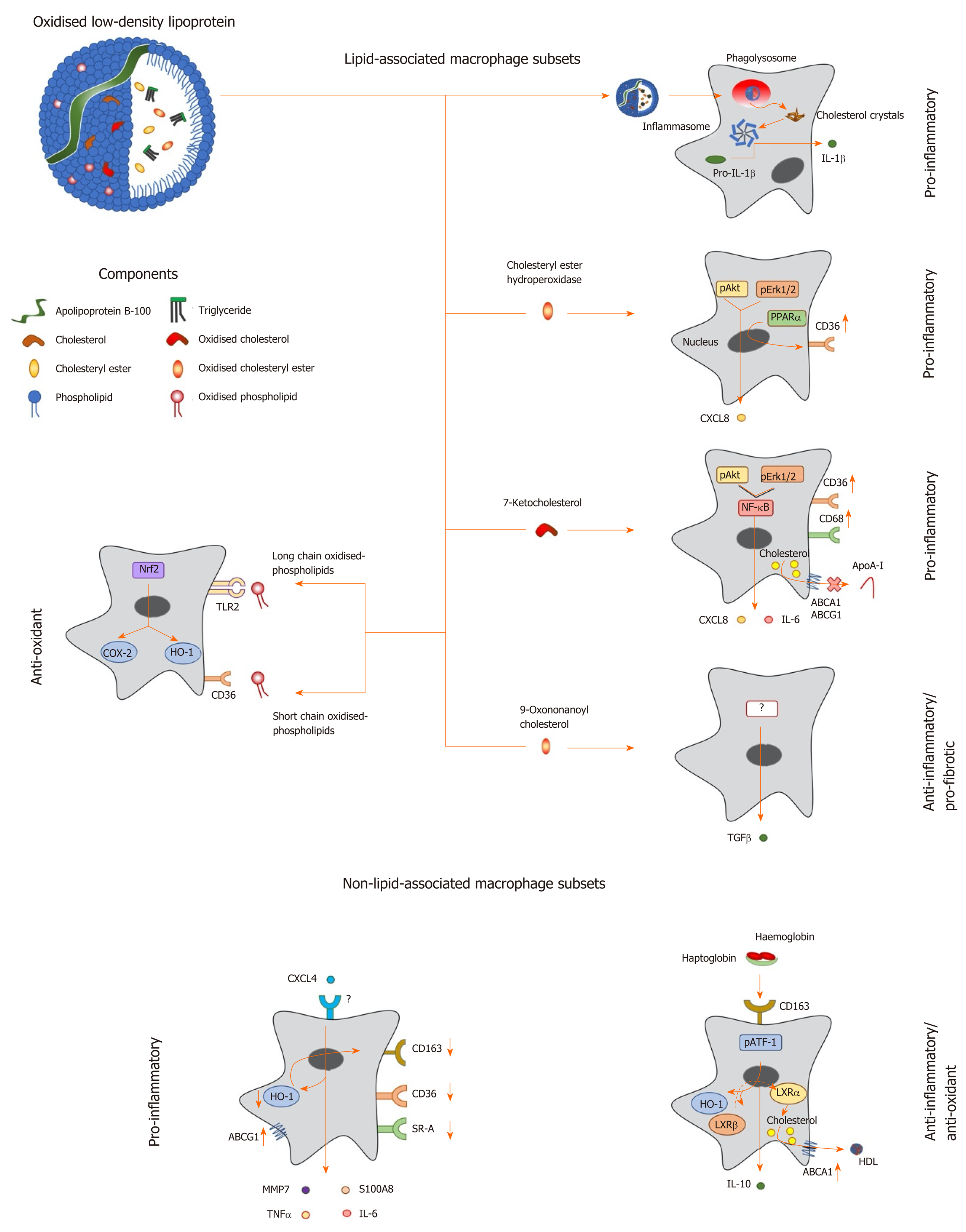
Figure 1 Macrophage phenotype in response to lipids and other factors within atherosclerotic lesions.
Oxidized low density lipoprotein particles are composed of a number of components including (modified) apolipoprotein B, (oxidized) free cholesterol, (oxidized) cholesteryl esters and (oxidized) phospholipids, which induce a range of unique transcriptional responses in macrophages (indicated by arrows), resulting in altered expression of secreted cytokines and cell surface receptors, and differing macrophage phenotypes (highlighted in bold). Macrophages can be polarised to a pro-inflammatory phenotype by components of oxidized LDL in a number of differing ways. Cholesterol crystals activate the inflammasome to increase expression of interleukin-1 (IL-1), while cholesteryl ester hydroperoxides activate phospho-extracellular signal-regulated protein kinases 1 and 2 (pERK1/2) and phospho-protein kinase B (pAkt) pathways, resulting in induction of peroxisome proliferator activated receptor alpha and increased expression of C-X-C motif ligand 8 (CXCL8) and the scavenger receptor, cluster of differentiation 36 (CD36). The modified cholesterol metabolite, 7-ketocholesterol, also triggers pAkt and pERK1/2 signalling, resulting in activation of nuclear factor kappa B (NF-κB), and enhanced output of pro-inflammatory cytokines such as IL-6 and CXCL8; this oxysterol also increases expression of CD36 and CD68, potentiating uptake of modified lipoproteins, and represses expression of ATP binding cassette transporter A1 (ABCA1) and ABCG1, limiting cholesterol removal via apolipoprotein A-I (apoA-I) and high density lipoprotein. By contrast, other components of oxLDL can induce an anti-inflammatory phenotype: For example, 9-oxononanoyl cholesterol induces expression of transforming growth factor β and stimulates fibrosis. Short chain oxidized phospholipids interact with CD36 to activate cyclooxygenase-2 (COX-2) and heme oxygenase-1 (HO-1), while long chain oxidized phospholipids bind toll-like receptor-2, resulting in activation of nuclear factor erythroid-2-related factor 2 which enhances expression of antioxidant genes, including HO-1, to protect cells against further oxidative damage. Other factors found within atherosclerotic lesions can also influence macrophage phenotype: For example, the chemokine CXCL4 induces the expression of pro-inflammatory factors such IL-6, tumor necrosis factor α, matrix metalloproteinase 7 and S100 calcium binding protein A8, while repressing the expression of ABCG1, HO-1, CD36, scavenger receptor-A and CD163. Alternatively an anti-inflammatory and antioxidant phenotype can be induced by the interaction of hemoglobin:haptoglobin complexes with CD163, which activates phospho-activating transcription factor-1, resulting in changes in LXR expression, induction of HO-1 and increased expression of IL-10 and ABCA1. IL: Interleukin; TGF: Transforming growth factor; TNF: Tumor necrosis factor; HO-1: Heme oxygenase-1; MMP: Matrix metalloproteinase; LXR: Liver X receptor; ABCA1: ATP binding cassette transporter A1; HDL: High density lipoprotein; pAkt: Phospho-protein kinase B; NF-κB: Nuclear factor kappa B; pATF-1: Phospho-activating transcription factor-1; PPAR-α: Peroxisome proliferator activated receptor alpha; CXCL: C-X-C motif ligand; ABCG1: ATP binding cassette transporter G1.
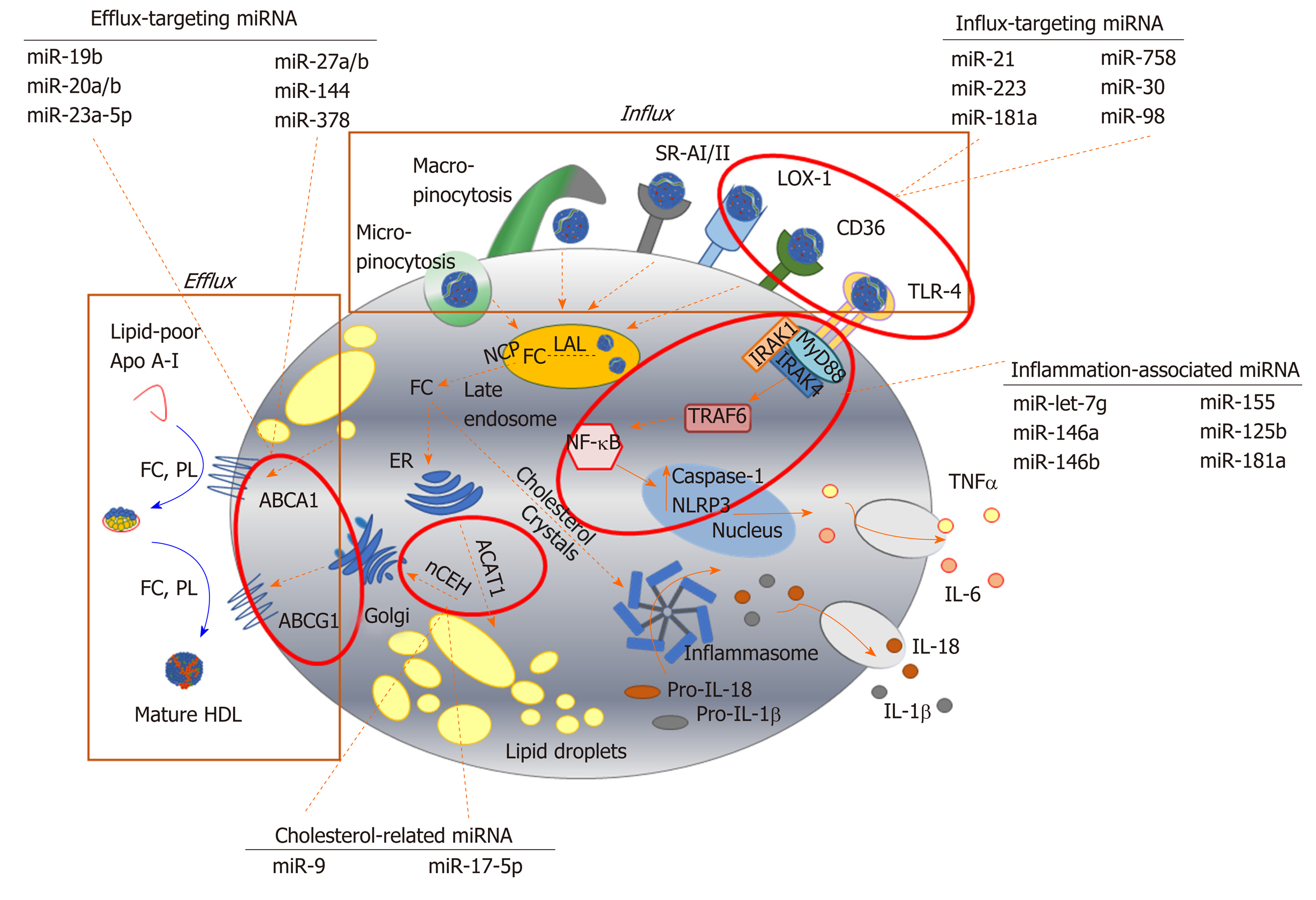
Figure 2 Key pathways involved in foam cell formation regulated by microRNA.
Native low-density lipoprotein (LDL) and oxidized LDL is processed in the late endosome/lysosomes by lysosomal acid lipase, generating free cholesterol which is stored in the form of lipid droplets after esterification via acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1 at the endoplasmic reticulum. Cholesterol can be removed from the cell by trafficking from the late endosomes, or by hydrolysis of lipid droplets and transport to the plasma membrane to be transferred to lipid-poor apolipoprotein A-I or nascent high-density lipoprotein via ABC transporters (ATP binding cassette transporter A1, ATP binding cassette transporter G1). Inflammatory signalling pathways can be activated in foam cells via recognition of oxLDL by toll-like receptor 4 (TLR4), or by intracellular free cholesterol forming cholesterol crystals, leading to the expression and release of inflammatory cytokines. Since their initial discovery, numerous miRNA sequences have been shown to play a role in targeting each of these key steps in the formation of foam cells. For example, influx of modified lipoproteins into macrophages via oxidized low-density lipoprotein (lectin-like) receptor 1, cluster of differentiation 36 and TLR4, is reported to be regulated by miR-21, miR-30, miR-98, miR-181a, miR-223 and miR-758. The cholesterol efflux pathway is modulated by distinct sequences, including miR-19b, miR-20a/b, miR-23, miR-27a/b, mir-155 and miR-378, while miR-9 and miR-17 regulate the esterification and hydrolysis of cholesterol droplets. MicroRNA-181a regulates both lipid metabolism and inflammatory response, while let-7g, miR-146a/b, miR125b and miR-155 influence macrophage inflammatory phenotype. Dotted arrows indicate the paths for cholesterol derived from the extracellular environment within the macrophage; solid arrows indicate inflammatory signalling pathways. HDL: High density lipoprotein; ABCA1: ATP binding cassette transporter A1; ABCG1: ATP binding cassette transporter G1; ACAT-1: Acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1; ApoA-I: Apolipoprotein A-I; CD: Cluster of differentiation; ER: Endoplasmic reticulum; FC: Free cholesterol; IL: Interleukin; IRAK: Interleukin 1 receptor associated kinase 4; LAL: Lysosomal acid lipase; LOX-1: Oxidized low-density lipoprotein (lectin-like) receptor 1; MyD88: Myeloid differentiation primary response 88; nCEH: Neutral cholesteryl ester hydrolase; NPC: Niemann-Pick disease type C; NF-κB: Nuclear factor kappa beta; PL: Phospholipid; SR: Scavenger receptor; TLR: Toll-like receptor; TNF-α: Tumour necrosis factor alpha.
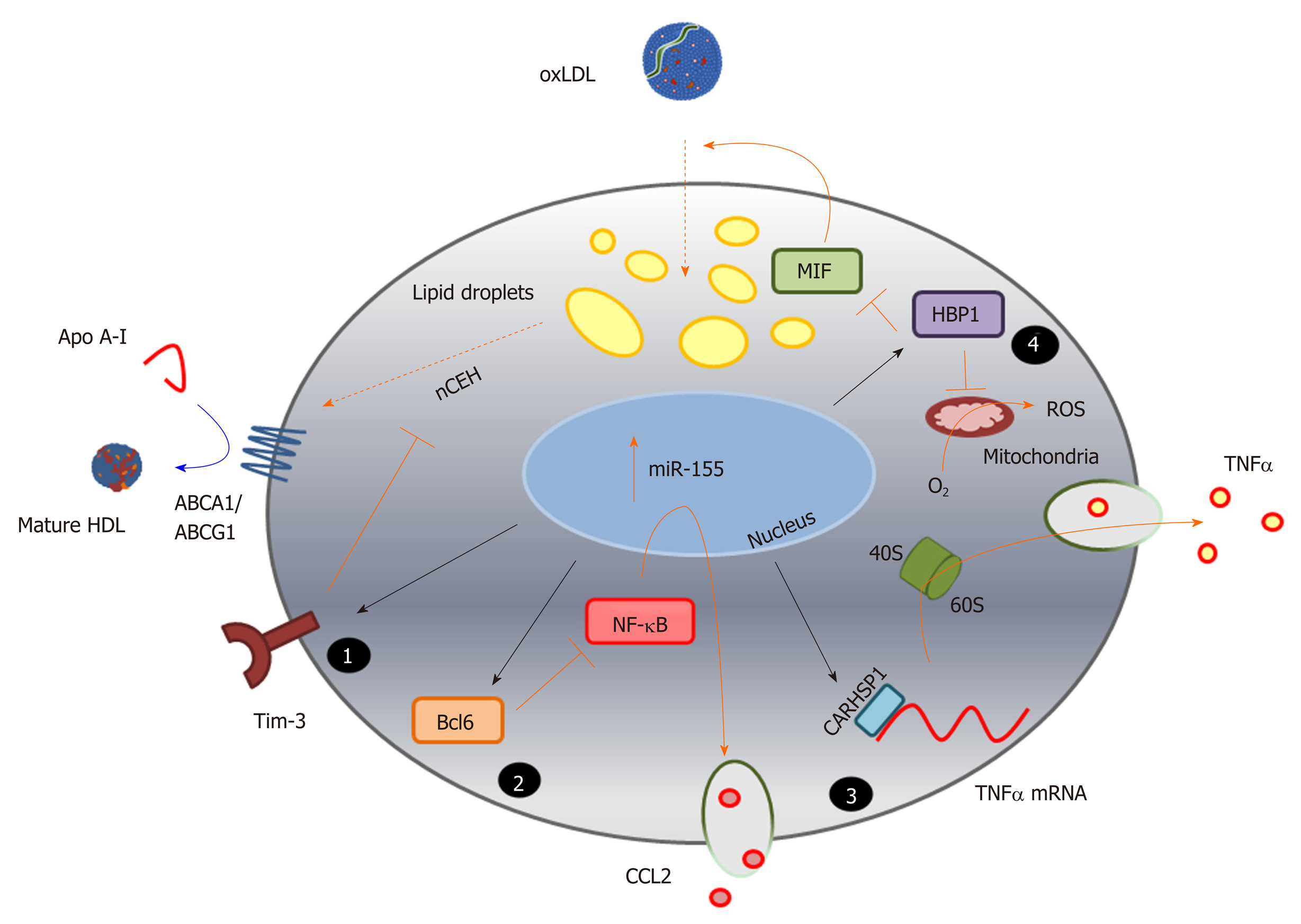
Figure 3 Identification of novel pathways associated with foam cell formation.
MicroRNA sequences altered during macrophage foam cell formation reveal novel pathways involved in this process, and highlight the complexity of miRNA function in targeting multiple gene pathways, exemplified here by miR-155 (Table 1). Inhibition of expression of the cell surface immunoregulatory glycoprotein, T-cell immunoglobulin and mucin-domain containing-3 (Tim-3), by miR-155 (1) increases hydrolysis of cholesteryl esters by neutral cholesteryl ester hydrolase, and promotes efflux of this lipid via ATP binding cassette transporter A1 and ATP binding cassette transporter G1, to apolipoprotein A-I and high density lipoprotein respectively. MicroRNA-155 also represses expression of the transcriptional repressor B-cell lymphoma 6 protein[236], which increases nuclear factor kappa beta activity and enhances production of the chemokine C-C motif ligand 2 (2); Repression of the cytoplasmic protein, calcium-regulated heat stable protein (CARHSP1) by miR-155, results in reduced binding of this protein to the 3’UTR of the TNFα gene transcript, and to reduced mRNA stability and decreased output of this cytokine (3); MicroRNA-155 also directly targets (represses) HMG-Box transcription factor 1, thereby increasing production of reactive oxygen species and loss of inhibition of macrophage migration inhibitory factor, leading to increased oxidatively modify proteoglycan-bound low density lipoprotein uptake and lipid accumulation (4). The black arrows represent direct targeting by miR-155; orange bars and arrows represent the functions of miR-155 targets in the absence of this miRNA sequence. HDL: High density lipoprotein; ABCA1: ATP binding cassette transporter A1; ABCG1: ATP binding cassette transporter G1; ACAT-1: Acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1; ApoA-I: Apolipoprotein A-I; Bcl6: B-cell lymphoma 6 protein; CARHSP1: Calcium-regulated heat stable protein; CCL2: C-C motif chemokine ligand 2; HBP1: HMG-Box transcription factor 1; MIF: Macrophage migration inhibitory factor; NF-κB: Nuclear factor kappa beta; ROS: Reactive oxygen species; TNF-α: Tumour necrosis factor alpha.
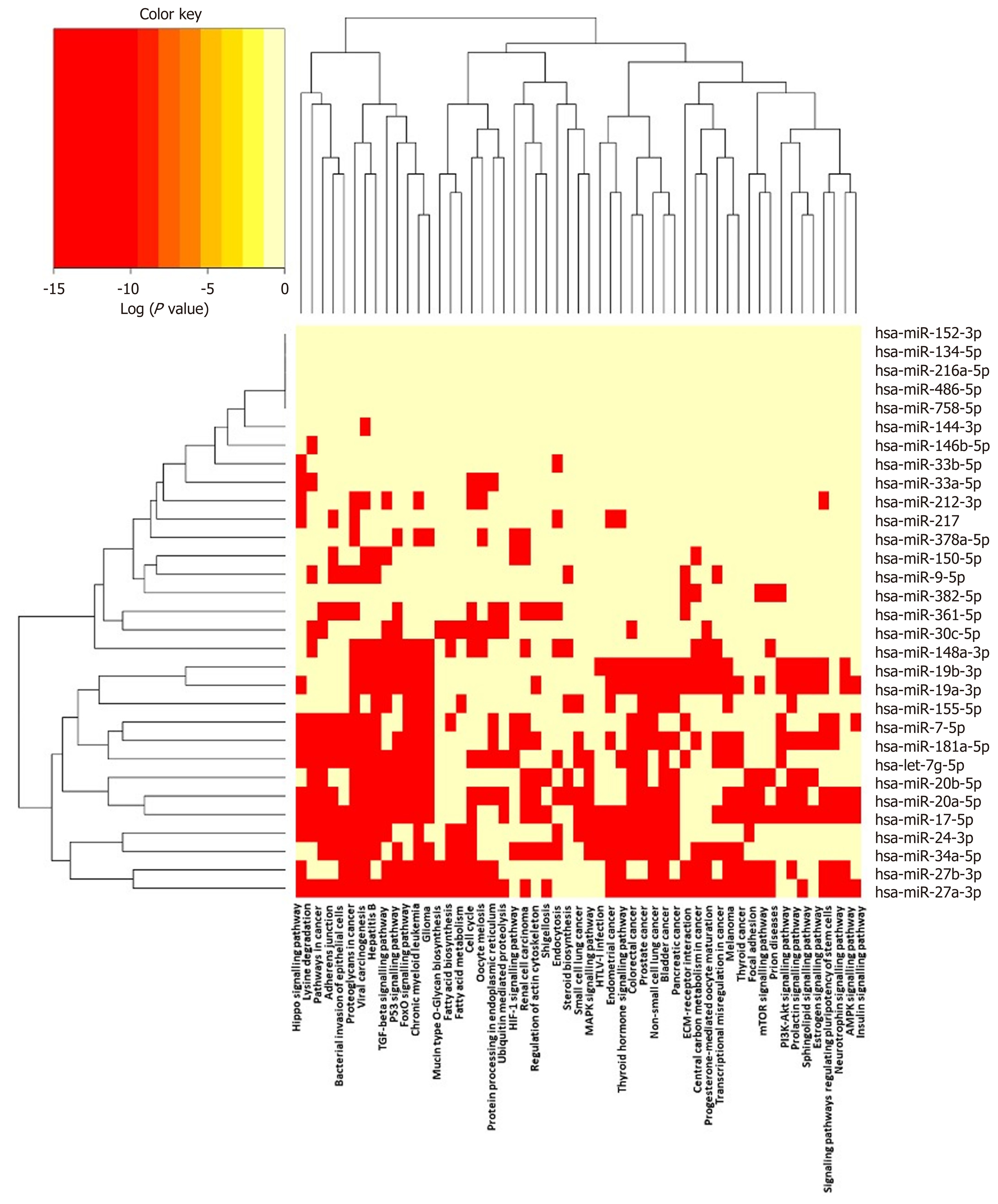
Figure 4 Pathways targeted by miRNA sequences altered in human macrophage “foam” cells: DIANA/KEGG predictive analysis.
The human miRNA sequences described in Table 1 were analysed using DIANA-miRPATH v3.0, yielding a heat map of miRNA vs Gene Ontology/GOSlim/Kyoto Encyclopedia for Genes and Genomes entries.
- Citation: Lightbody RJ, Taylor JMW, Dempsie Y, Graham A. MicroRNA sequences modulating inflammation and lipid accumulation in macrophage “foam” cells: Implications for atherosclerosis. World J Cardiol 2020; 12(7): 303-333
- URL: https://www.wjgnet.com/1949-8462/full/v12/i7/303.htm
- DOI: https://dx.doi.org/10.4330/wjc.v12.i7.303