Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.4
Peer-review started: September 28, 2016
First decision: October 26, 2016
Revised: December 20, 2016
Accepted: December 27, 2016
Article in press: December 28, 2016
Published online: February 26, 2017
Processing time: 151 Days and 16.1 Hours
Animal venom research is a specialized investigation field, in which a number of different methods are used and this array is constantly expanding. Thus, recently emerged omics and nanotechnologies have already been successfully applied to venom research. Animal venoms have been studied for quite a long time. The traditional reductionist approach has been to isolate individual toxins and then study their structure and function. Unfortunately, the characterization of the venom as a whole system and its multiple effects on an entire organism were not possible until recent times. The development of new methods in mass spectrometry and sequencing have allowed such characterizations of venom, encompassing the identification of new toxins present in venoms at extremely low concentrations to changes in metabolism of prey organisms after envenomation. In particular, this type of comprehensive research has become possible due to the development of the various omics technologies: Proteomics, peptidomics, transcriptomics, genomics and metabolomics. As in other research fields, these omics technologies ushered in a revolution for venom studies, which is now entering the era of big data. Nanotechnology is a very new branch of technology and developing at an extremely rapid pace. It has found application in many spheres and has not bypassed the venom studies. Nanomaterials are quite promising in medicine, and most studies combining venoms and nanomaterials are dedicated to medical applications. Conjugates of nanoparticles with venom components have been proposed for use as drugs or diagnostics. For example, nanoparticles conjugated with chlorotoxin - a toxin in scorpion venom, which has been shown to bind specifically to glioma cells - are considered as potential glioma-targeted drugs, and conjugates of neurotoxins with fluorescent semiconductor nanoparticles or quantum dots may be used to detect endogenous targets expressed in live cells. The data on application of omics and nanotechnologies in venom research are systematized concisely in this paper.
Core tip: A number of different methods are used in animal venom research, and this array is constantly expanding. The development of new methods in mass spectrometry and sequencing have allowed for the characterization of venom at different levels, ranging from identification of new toxins to profiling the changes in metabolism of an envenomed organism. The various omics technologies-proteomics, peptidomics, transcriptomics, genomics and metabolomics-have played key roles, as has nanotechnology. Nanomaterials are promising in medicine, and most studies combining venoms and nanomaterials are directed to medical applications, with conjugates of nanoparticles and venom components being proposed for use as drugs or diagnostics.
- Citation: Utkin YN. Modern trends in animal venom research - omics and nanomaterials. World J Biol Chem 2017; 8(1): 4-12
- URL: https://www.wjgnet.com/1949-8454/full/v8/i1/4.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v8.i1.4
Animal venoms are complex mixtures of toxic substances, which are used to defend against predators and/or to hunt. They are extremely rich and complex natural sources of biologically active molecules, having a variety of molecular targets and functions. Venoms are aqueous solutions containing a large number of components, mostly protein and peptide in nature. Several hundred different substances, including toxins, may be found in venom from a single animal. Exposure of an envenomed organism to a toxin leads to significant dysfunction of the nervous, cardiovascular and muscular systems. The best known venomous animals are snakes, scorpions and spiders, and their toxins are used widely in research as molecular tools. Because toxin molecules often have a very strong potential for clinical structural optimization, some have been developed into new drugs (based on the optimized structures).
In attempts to understand the basis of venom biological activity, scientists have used a reductionist approach to venom composition and aimed to isolate the active compounds. Traditionally, this activity-driven approach was applied for purification of toxins. Having been initiated several decades ago, the present biochemical venom studies have advanced a long way from use of standard chemical fractionation methods, based in particular on differences in solubility, to modern high-performance liquid chromatography methods.
Most recently, venom research was revolutionized by the introduction of new mass spectrometry (MS) methods and the development of “omics” technologies, including but not limited to genomics, proteomics and metabolomics. These English-language neologisms specify fields of study in biology that deal with very large-scale data collection and analysis, in particular characterization and quantification of pools of biological molecules[1]. In molecular biology, the suffix - ome refers to a “totality of some sort” and is used to address the objects of omics studies, such as the genome, proteome or metabolome.
Three main categories within omics technologies are genomics, proteomics, and metabolomics/metabonomics. Genomics techniques are used to analyze the structure and function of genomes, while proteomic techniques deal with cellular and tissue-wide protein expression and metabolomics techniques are concerned with the identification and quantification of all the metabolites in a biological system. Within or in addition to these main techniques, some other omics techniques exist, such as transcriptomics, peptidomics, etc., Several omics techniques have already been applied to venom studies, resulting in more comprehensive characterization of venoms and their effects on organisms.
Another modern technology that carries substantial promise for having a high impact on venom studies is nanotechnology. Nanotechnology handles materials at the nanometer size level. Nanomaterials represent physical objects possessing size between 1 nm and 150 nm, in at least one dimension. This scale reduction results in significant changes of typical physical and chemical properties of materials, making them very attractive for novel and innovative applications in various fields, and they have garnered particular interest in medicine, pharmacy and medical diagnostics. Indeed, nanomaterials have been used successfully in magnetic and fluorescent bioimaging, as carriers for drugs, and even as medicines themselves (e.g., antimicrobial agents).
Among the different methodological directions being taken in venom studies, the development of new drugs based on venom components is the most prospective and rewarding. The combination of venom components with nanomaterials, and their application in treatment, diagnostics and disease prevention, will benefit the health and quality of human life. However, only the first steps considered in this Editorial have been made in this direction, thus far.
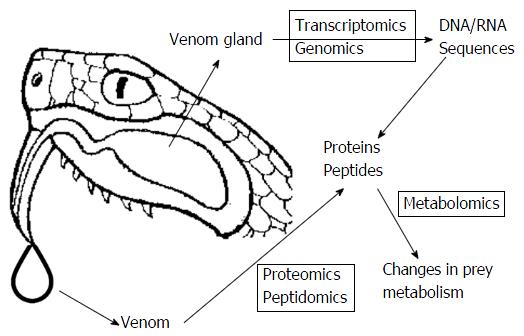
In their quest to better understand the nature of venoms, researchers are constantly looking for new analytical methods of investigation. The recently emerged powerful analytical approaches united by the term “omics” have been successfully applied for venom studies. The relationship within “omics” technologies in venom studies is summarized in Figure 1.
Among all omics technologies, proteomics has been the most frequently used in venom research. The first publication[2] on venom proteomics appeared at the beginning of the millennium, and since then the number of works using this technology for venom studies has increased steadily. The special term “venomics” was introduced[3], which refers to omics methods applied to venom studies. Considering this rapid growth, a key question emerges: What are the main advantages of proteomics over traditional methods? One of the major benefits is that proteomics gives a complete description of all the polypeptides present in a venom sample. This data may contribute to a deeper comprehension of the nature of a venom’s toxic effects. Quantitative proteomics is especially beneficial for this purpose, as it not only describes the polypeptide array but determines their amounts as well[4], yielding an exhaustive catalog of the toxins present in the venom. Information about the toxins can be used in several ways; for example, some toxins are valuable molecular tools for studying physiological processes and may be applied for basic research. Venom proteomes also represent great potential for clinical diagnosis and development of new medicines for clinical use.
Proteomics contributes greatly to the development of antivenoms, which are currently the most efficient medications available for the treatment of bites or stings. The new term “antivenomics” was invented to specify an omics method used to identify venom proteins bearing epitope(s) that is(are) recognized by an antivenom[5]. The basis of antivenomics is immunoaffinity chromatography, through which toxins bound to immobilized antivenom are identified by proteomics and in parallel the venom components that failed to raise antibodies in the antivenom, or which triggered the production of low-affinity antibodies and therefore remain unbound to antivenom, are identified. Antivenomics provides qualitative as well as quantitative data on both the toxins inducing antibodies with high affinity and those toxins exhibiting poor immunoreactivity.
It should be noted that proteomic analysis is capable of revealing many toxin variants belonging to known toxin families. In addition to this, several toxins of protein families not found in earlier venom studies have been identified and characterized by proteomics. For example, two proteins recently identified in a venom analysis of the dog-faced water snake Cerberus rynchops showed sequence homology to ficolin, a mammalian protein with collagen-like and fibrinogen-like domains[6]. These proteins were named as ryncolin 1 and ryncolin 2 (rynchops ficolin), and this new family of snake venom proteins was called veficolins (venom ficolins). The authors speculated that the ryncolins may induce platelet aggregation and/or initiate complement activation in the envenomed organism. Similarly, investigation of the venom proteome of the rear-fanged snake Thamnodynastes strigatus lead to the discovery of a new kind of matrix metalloproteinase (MMP) that is unrelated to the classical snake venom metalloproteinases found in all snake families[7]. In that study, a protein related to lactadherin was newly identified and suggested to be a venom component[7]. Finally, investigation of the venom of a cryptic Australian elapid snake Drysdalia coronoides resulted in the discovery of a new structural type of toxin from the three-finger toxin superfamily, and a new family phospholipase B was identified as well[8]. These data highlight the great potential of proteomics in the discovery of new toxins and toxin families.
As was mentioned earlier in this Editorial, animal venoms are complex mixtures of different substances, but mostly proteins and peptides. In some venoms, the content of peptides is quite high; this is especially true for spider venoms[9]. The proteomic approach is also used to study peptide components of the venom. This peptide-aimed type of study was given the name “peptidomics”. Thus far, peptidomics has been used mostly for peptide profiling of invertebrate venoms and its application has resulted in the discovery of several new toxins[10,11].
Despite having been successfully used for many studies of venoms, proteomics continues to present several unresolved challenges. The usual first step in a proteomic venom study is the application of tandem MS/MS and database searching for the identification of protein and peptide toxins. In this so-called bottom-up approach, the comparison of experimentally determined sequences produced by MS/MS analysis with those obtained by in silico digestion of proteins present in a given database is performed. The problems encountered at this step, however, are the absence of tryptic cleavage or miscleavage at sites that differ from the classical trypsin sites. The absence of tryptic cleavage might be explained by post-translational modifications proximal to a tryptic cleavage site, which may result in the loss of trypsin activity. The activity of venom proteases might also result in the loss of some trypsin cleavage sites; such activity could also produce an artificial venom peptidome[5].
Another challenge in proteomic studies is related to the absence of comprehensive databases containing venom protein sequences. To address this, several specialized toxin databases have been generated and released during the last decade. For example, the Animal Toxin Database supported by the Hunan Normal University[12] contains more than 9000 toxins, while the ArachnoServer, a manually-curated database containing information on the toxins derived from spider venom[13] contains more than 1500 entries and the most recent Kalium database of potassium channel toxins from scorpion venom[14] contains about 200 entries. Moreover, in the last UniProt release, the current number of entries for the query “snake venom” was around 29000 (UniProt release 2016_07). Nevertheless, this mass of information is still not sufficient and to describe the proteome of any particular venom completely and precisely, and as such transcriptomic and/or genomic data are necessary.
Genomics represents another versatile omics technology that is used widely in the life sciences. By definition “genomics is a discipline in genetics that applies recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyze the function and structure of genomes”[15]. Yet, genomics has not been so widely applied as proteomics in venom studies. The genomes of poisonous animals that have been sequenced so far include the honeybee Apis mellifera[16], the Chinese scorpion Mesobuthus martensii[17], the Brazilian whiteknee tarantula Acanthoscurria geniculata[18] and the King cobra Ophiophagus hannah[19]. The rapid progress in sequencing methods has resulted in cost effective high-throughput sequencing (or next-generation sequencing) technologies that allow for the obtainment of millions of sequences at once. This makes the use of genomics more attractive for scientists involved in venom studies. And, according to a search of the up-to-date publicly available literature, among the snake species, the genomes of more than 10 are currently being studied[20].
Venom proteins are produced by the venom gland, wherein translation of the information encoded by messenger (m)RNA takes place. This means that the data about venom protein composition can be obtained from an array of mRNA present in the venom gland. This approach is the basis of transcriptomic studies. Regarded sometimes as a part of genomics, transcriptomics can be defined as the study of the expression level of mRNAs in a given cell population, often by using high-throughput techniques. Transcriptomics has been embraced by venom researchers and its application has provided valuable information on the anticipated protein array in a given venom gland under a given biological condition. However, the presence of a given mRNA in the gland does not mean that it is guaranteed to be translated to a protein. Moreover, the transcriptomics approach does not give any information about the post-translational modification of a protein.
Nevertheless, compared to proteomics, which requires some initial data for protein identification, transcriptomics has a greater potential for discovery of new toxins. Thus, the transcriptomic approach has been applied to investigations of the composition of venom gland peptides from the Hadogenes troglodytes scorpion[21]. A total of 121 novel peptides were discovered, including 20 new-type peptides cross-linked with 1, 2, 3, 4, 5 or 7 disulfide bridges, 11 novel K+-channel toxin-like peptides, 2 novel ryanodine receptors-specific toxin-like peptides, a unique peptide containing the cysteine knots of spider toxins, 15 novel La1-like toxins, 3 novel TIL-domain containing peptides, 5 novel peptides with atypical cysteine patterns, 19 novel antimicrobial peptides, 6 novel cysteine-free peptides and 39 new-type cysteine-free peptides[21]. These data demonstrate the great potential of transcriptomics for discovery of toxins.
The main disadvantage of the transcriptomic approach is the necessity to sacrifice the animal under investigation. However, it has been shown that mRNA encoding toxins can be recovered from the venom of several species[22]. Subsequent studies have supported this finding and showed its applicability to RNA-based venom studies[23,24].
Venoms have a great impact on the vital system in the prey organism that should result in substantial changes in the metabolism of the envenomed organism, which in principle may be revealed by metabolomics. The aims of metabolomics are the identification and quantification of all the metabolites in a biological system (cell, tissue, organ, organism) under given conditions. Metabonomics, in contrast, focus on the dynamic metabolic response of living systems to some biological stimuli. The task therein is to understand systemic change through time in complex multicellular systems. However, only a few publications in the literature to date report on the changes in prey metabolism after envenomation. One of these studies aimed to assess the response to a honeybee venom by analyzing serum levels of 34 free amino acids[25]. To achieve this, serum samples were collected from 27 beekeepers within 3 h after they had been stung and after a minimum of 6 wk following the last sting. The two analyzed groups showed statistically significant differences in serum levels of L-glutamine, L-glutamic acid, L-methionine and 3-methyl-L-histidine. Moreover, L-glutamine and L-glutamic acid were found to be the most important metabolites for distinguishing the beekeepers who had been tested shortly after a bee sting from those who had been tested at least 6 wk later. The results obtained may contribute to better understanding of the human body’s response to the honeybee sting[25].
In other work, the toxic mechanism of scorpion Hemiscorpius lepturus venom was investigated by metabolome profiling of the victims using proton nuclear magnetic resonance (commonly known as 1H NMR)[26]. In parallel, the physiological effects of this venom on biochemical pathways and organs were investigated. The results obtained showed that the most affected pathways were pyrimidine, histidine and tyrosine metabolisms, and steroid hormone biosynthesis. The crude venom was found to affect mostly the pancreas and spleen. Mitochondria and nerve cells were attacked, resulting in acute seizures that resembled the early markers of myocardial injury and seizure disorders[26].
The data discussed above indicate that omics technologies have contributed greatly to increasing our understanding of different aspects of venom’s impact on a living organism. The application of these technologies in venom studies is constantly increasing, and the popularity of omics in venom study is evidenced by the fact that, to date, among the snake venoms alone over 150 species have already been examined by these techniques.
Most investigations of venom composition using analytical “omics” methods, as discussed in the previous section, are not goals in and of themselves, but are aimed at finding new compounds that can be of practical use. From this perspective, any enhancements that may improve the useful properties and applicability of these compounds will be of great value. One of these is the combination of venoms or toxins with nanomaterials.
Nanomaterials have numerous applications and are used in many areas, including research, technology and medicine. They can be used alone or in combination with other materials, such as venoms or toxins, as has been accomplished in magnetic and fluorescent bioimaging, as carriers for drugs, and as medicines.
The unusual properties of nanoparticles (NPs) can be exploited to improve the pharmacologic and therapeutic properties of drugs. While larger molecules may be eliminated from the body in a relatively short period of time, the cells can capture NPs very efficiently. To study the effectiveness of NP-based drug delivery to a biological target, hydrophilic NPs such as chitosan, nanogold, nanosilver and dendrimers, to name a few, were conjugated to potent animal toxins. Both crude venoms and isolated toxins have been used for this purpose.
Cobra Naja naja oxiana venom was encapsulated in chitosan NPs, and such NPs were evaluated for their antigen delivery potential[27]. The authors concluded by suggesting the possible application of chitosan NPs as an alternative to the currently used adjuvants.
In another study, poly(D,L-Lactide)-based NPs were combined with toxic fractions obtained from Androctonus australis hector and Buthusoccitanus tunetanus scorpions’ venoms in order to safely vaccinate animals to induce production of sera with neutralizing activity and achieve an immunoprotection outcome[28]. The results obtained showed that polylactide NPs coated with toxic venom fraction preserve antigenicity and enhance immune response to weakly immunogenic toxins. This method can be easily extrapolated for horse therapeutic immune sera production, thereby reducing production costs. Moreover, PLA-NPs could be used for prophylactic purposes, as a biodegradable adjuvant to promote long-lasting immune protection against accidental envenomation of humans in hazardous areas[28].
NPs loaded with anticancer agents have also shown great promise for the treatment of cancer. To study the effects of venom extracted from the cobra Walterinnesia aegyptia (WEV) on the proliferation and apoptosis of human breast cancer cells, either the venom alone or in combination with silica NPs (WEV+NP) were investigated through monitoring caspase activity and free radical levels[29]. Cells that had been isolated from female patients clinically diagnosed with breast cancer were used. It was found that both WEV and WEV+NP inhibited the growth of breast cancer cells in dose- and time-dependent manners, with the WEV+NP combination showing greater efficacy against the cancer cells. Thus, the combination of NPs with WEV significantly increased the antitumor effects of the venom.
To enhance the medicinal activity of bee venom (BV) acupuncture, it was loaded into biodegradable poly(D,L-Lactide-co-glycolide) NPs (BV-PLGA-NPs). The BV-PLGA-NPs were found to prolong the analgesic effect of BV in formalin-induced pain test in rats. PLGA-encapsulation of BV was also found to be effective in alleviating edema induced by allergens in BV[30].
The above data indicate that NPs can significantly improve therapeutic potential of animal venoms. This improvement was more evident when individual toxins were used.
As was mentioned in the previous section, crude venoms are very complex mixtures of different compounds, some of which may show beneficial effects while others may show adverse effects. Thus, it is evident that the use of isolated compounds with beneficial effects may greatly enhance their potential.
Bombesin, the peptide isolated from the poisonous skin of the frog Bombina bombina, shows high affinity for the gastrin-releasing peptide (GRP)-receptors, which are widely represented in prostate cancer cells, breast cancer cells and small cell lung cancer in vivo. When bombesin was conjugated with gold NPs and radioisotope label was attached to these conjugates[31], a high degree of specific binding to GRP-receptor and a high selectivity for the GRP-receptors of prostate tumor cells in mice was attained. The intraperitoneal drug delivery method was found to be effective for the bombesin-gold conjugate, avoiding absorption by the reticuloendothelial system of healthy organs and facilitating a concomitant increase in uptake of the drug by tumor cells.
The cytolytic peptide melittin, which is derived from BV, is considered a good candidate for cancer control. To overcome its main disadvantages - toxicity to non-targeted cells, non-specificity and unfavorable pharmacokinetic - the melittin nanoconjugate was developed, whereby the toxin has been inserted into the outer monolayer of perfluorocarbon NPs[32]. This nano-carrier approach allows melittin to accumulate in tumors in vivo in mice and significantly reduce the tumor growth without any visible signs of envenomation. Moreover, such nanocarriers are capable of delivering melittin selectively to several tumor targets through the polydiffusion mechanism when the integrity of the cell membrane is maintained.
Nicotinic acetylcholine receptor of alpha7 type (α7-nAChR) is among several receptors that are regarded as tumor targets owing to expression specificity and significancy for cancer. The venom isolate alpha-conotoxin ImI from the cone snail Conus imperialis possesses high affinity for α7-nAChR and has been used to increase targeted drug delivery to α7-nAChR-overexpressing tumors. In particular, the toxin was conjugated to the polyethylene glycol-grafted distearoylphosphatidylethanolamine micelles (ImI-PMs), forming spherical NPs of about 20 nm size with high drug encapsulation potential[33]. To evaluate the targeting efficacy, the anti-tumor drug paclitaxel, which is frequently used in clinical for the treatment of breast cancer, was loaded in ImI-PMs. It was found that paclitaxel-loaded ImI-PMs exhibited greater cytotoxicity and induced more cell apoptosis in vitro, as well as displayed stronger antitumor efficacy in MCF-7 tumor-bearing mice. Thus, the alpha-conotoxin ImI-modified nanocarrier showed great potential for targeting α7-nAChR-overexpressing tumors.
Snake venom toxin NKCT1 (GNP-NKCT1) tagged to gold NPs was studied as a potential anticancer agent using the human leukemic U937 and K562 cell lines as targets. Treatment of cells with GNP-NKCT1 resulted in the loss of mitochondrial membrane potential and a strong increase in reactive oxygen species (ROS). The conjugate induced apoptosis in leukemic cells, but upon suppression of apoptosis an alternative cell death pathway (in the form of autophagy) was observed[34].
Fluorescent nanocrystals, otherwise known as quantum dots (QDs), have received great attention during the last year due to their outstanding properties. Recent advances in QD technology have made it possible to track the motion of individual molecules. Thus, the QDs have been used to monitor the receptors and ion channels in neuronal synapses, in particular in tracking movements of diffuse acetylcholine receptors on the surface of cultured myocytes[35]. In cultured myocytes, acetylcholine receptors often form clusters in the absence of stimulation. These clusters can be easily visualized by alpha-bungarotoxin (a neurotoxin isolated from the venom of the Bungarus multicinctus krait) that is fluorescently labeled with organic fluorophores. However, this method was unable to detect the individual acetylcholine receptors that are diffusely distributed throughout the cell surface. To visualize such diffuse receptors, the myocytes were labeled with biotinylated alpha-bungarotoxin and then with streptavidin-conjugated QDs. In this manner, both clusters and individual acetylcholine receptors on the surface of the myocytes were finally able to be observed.
Chlorotoxin (CTX) found in the Israeli Leiurus quinquestriatus scorpion’s venom binds preferentially to glioma cells, as compared with non-neoplastic cells or normal brain cells. This finding has allowed for the development of new methods for the treatment and diagnosis of several types of cancer with CTX-targeted NPs now being used fairly often for imaging and therapy of gliomas[36].
Using supermagnetic iron oxide as a nano-vector, a CTX conjugate with methotrexate was obtained[37]. This conjugate demonstrated preferential accumulation in and high cytotoxicity against glioma cells in vitro. Moreover, the prolonged retention of this NP conjugate was observed in tumor cells in vivo.
In another study, CTX conjugated with an amine-functionalized polysilane and supermagnetic iron oxide NPs was developed[38]. As a result, there was an increased uptake of the toxin conjugate into cancer cells and the tumor invasiveness was retarded compared to cells treated with the unconjugated CTX (98% vs 45% respectively). Moreover, the CTX-conjugates deactivated membrane-bound MMP2 and caused an increase in the internalization of lipid rafts. Because of its therapeutic action, this type of conjugate is considered a possible candidate for use both in non-invasive diagnosis and in treatment of various tumors.
A CTX-conjugate with near-infrared fluorophore and iron oxide particles coated with a biocompatible polyethylene glycol-modified chitosan was obtained[39]. This conjugate was able to cross the blood-brain barrier, capable of mainly targeting brain tumor cells. The obtained compound showed no toxic properties and remained for a long time in the tumor cells.
Polyethyleneglycol-mediated synthesis was used to create highly stable iron oxide NPs. These NPs were conjugated to CTX and Cy5.5 fluorescent dye of near-infrared range. Near-infrared fluorescence imaging showed the specific accumulation of this conjugate in mice glioblastoma cells[40].
CTX was also used to develop the upconversion nanoprobes that have proven useful for tumor targeting and visualization in living animals. Polyethylenimine-coated hexagonal-phase NaYF(4):Yb,Er/Ce NPs were prepared and conjugated with recombinant CTX[41]. The resultant conjugates were visualized by laser scanning upconversion fluorescence microscopy. Animal studies provided high-contrast images, demonstrating highly specific tumor binding and direct tumor visualization with bright red fluorescence under 980 nm near-infrared irradiation. The high sensitivity and specificity of the CTX nanoconjugate represent substantial improvements that will benefit the diagnostic and therapeutic approaches for patients suffering from cancer.
Thus, using different conjugation methods, CTX can be tethered to iron oxide NPs, QDs, and upconversion NPs for targeted imaging of gliomas. In addition, CTX nanoconjugates can also be used as carriers to deliver anticancer drugs to gliomas.
The data about application of nanomaterials for the venom research are summarized in Table 1.
| Nanomaterial | Toxin/Venom | Application |
| Polyethylenimine-coated hexagonal-phase NaYF(4):Yb,Er/Ce NPs | CTX from Leiurus quinquestriatus scorpion’s venom | Glioma visualization by laser scanning upconversion fluorescence microscopy |
| Highly stable iron oxide NPs conjugated to CTX and Cy5.5 fluorescent dye | CTX | Near-infrared fluorescence imaging |
| Amine-functionalized polysilane and supermagnetic iron oxide NPs | CTX | Antitumor drug retarding tumor invasiveness and deactivating membrane-bound MMP2 |
| Supermagnetic iron oxide conjugated with methotrexate and CTX | CTX | Antitumor drug with high cytotoxicity against glioma cells in vitro |
| Streptavidin-conjugated QDs | Biotinylated alpha-bungarotoxin from Bungarus multicinctus krait’s venom | Visualization of nicotinic acetylcholine receptor in the myocytes |
| Gold NPs | Snake venom toxin NKCT1 | Potential anticancer agent |
| Polyethylene glycol-grafted distearoylphosphatidylethanolamine NPs conjugated with ImI and loaded with paclitaxel | Alpha-conotoxin ImI from the cone snail Conus imperialis | Targeted antitumor drug |
| Perfluorocarbon NPs | The cytolytic peptide melittin, derived from bee venom | Antitumor drug with reduced acute toxicity |
| Gold NPs with radioisotope label | Bombesin from the frog Bombina bombina | Increase in uptake of the drug by tumor cells |
| Biodegradable poly(D,L-Lactide-co-glycolide) NPs | Bee venom | Prolonged analgesic effect of the venom |
| Silica NPs | Walterinnesia aegyptia cobra’s venom | Significantly increased antitumor effects of the venom |
| Poly(D,L-Lactide)-based NPs | Toxic fractions obtained from Androctonus australis hector and Buthusoccitanus tunetanus scorpions’ venoms | Enhanced immune response to weakly immunogenic toxins |
| Chitosan NPs | Cobra Naja naja oxiana venom | Alternative to the currently used adjuvants |
Although nanomaterials have numerous applications and possess great advantages over the traditional materials, they can have dangerous properties, which have not yet been completely studied and can cause adverse effects in humans. NPs can enter the human body through several routes, including inhalation, ingestion, skin penetration or injection[42,43]. After entering the body, NPs can interact with different components and localize in various organs, wherein they may remain intact or be subjected to modification or metabolism. NPs can cross cell boundaries, accumulating within the cells. Once in the cell, they may bind to DNA or proteins and interfere with normal cell functions or trigger an inflammatory response. The production of excess ROS[44], including free radicals, which can cause oxidative stress, inflammation and other cellular damage, is one of the main known toxicity mechanisms of NPs. Similar to the toxicity of the NPs’ parent bulk materials, the NPs themselves have toxicity that is determined by their chemical composition; however, size, surface chemistry, shape, and/or surface smoothness or roughness may enhance the toxicity profile of an NP, and all of these features can be altered substantially. While some negative NP effects have become understandable through detailed research, considerable efforts are still needed to study the physiological effects of acute and chronic exposure to NPs. Concerning the safety of NP conjugates with venoms or toxins, their application in vivo should be carried out with great care, keeping in mind the toxicity of starting materials.
Modern technologies, namely omics and nanotechnology, have made a great impact on the development of different branches of science and have contributed substantially to recent advancements in the field of venom research. Application of omics technologies have led to a better understanding of venom composition and of the roles played by different toxins in a given venom’s effects. Preparation of conjugates of animal toxins with NPs opens a new path to creation of novel and innovative effective drugs, having better therapeutic potential and biocompatibility, as well as to the generation of more advanced systems to better deliver these drugs. However, NPs can generate ROS and induce apoptosis in the affected cells. These effects, on the one hand, may kill cancer cells, while, on the other hand, they may produce negative effects on normal cells. Thus, the possible adverse effects of NPs on human health should be taken into account when considering their application.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: Russia
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Cairo G, Carter WG, Chang KA, Liu SQ S- Editor: Song XX L- Editor: A E- Editor: Li D
| 1. | Omics: Diagram illustrating genomics, 2016. Available from: http://en.wikipedia.org/wiki/Omics. |
| 2. | Pimenta AM, Stöcklin R, Favreau P, Bougis PE, Martin-Eauclaire MF. Moving pieces in a proteomic puzzle: mass fingerprinting of toxic fractions from the venom of Tityus serrulatus (Scorpiones, Buthidae). Rapid Commun Mass Spectrom. 2001;15:1562-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 87] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Juárez P, Sanz L, Calvete JJ. Snake venomics: characterization of protein families in Sistrurus barbouri venom by cysteine mapping, N-terminal sequencing, and tandem mass spectrometry analysis. Proteomics. 2004;4:327-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 96] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Kovalchuk SI, Ziganshin RH, Starkov VG, Tsetlin VI, Utkin YN. Quantitative Proteomic Analysis of Venoms from Russian Vipers of Pelias Group: Phospholipases A2 are the Main Venom Components. Toxins (Basel). 2016;8:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM. Venoms, venomics, antivenomics. FEBS Lett. 2009;583:1736-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 276] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 6. | OmPraba G, Chapeaurouge A, Doley R, Devi KR, Padmanaban P, Venkatraman C, Velmurugan D, Lin Q, Kini RM. Identification of a novel family of snake venom proteins Veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J Proteome Res. 2010;9:1882-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 7. | Ching AT, Paes Leme AF, Zelanis A, Rocha MM, Furtado Mde F, Silva DA, Trugilho MR, da Rocha SL, Perales J, Ho PL. Venomics profiling of Thamnodynastes strigatus unveils matrix metalloproteinases and other novel proteins recruited to the toxin arsenal of rear-fanged snakes. J Proteome Res. 2012;11:1152-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 8. | Chatrath ST, Chapeaurouge A, Lin Q, Lim TK, Dunstan N, Mirtschin P, Kumar PP, Kini RM. Identification of novel proteins from the venom of a cryptic snake Drysdalia coronoides by a combined transcriptomics and proteomics approach. J Proteome Res. 2011;10:739-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Vassilevski AA, Kozlov SA, Grishin EV. Molecular diversity of spider venom. Biochemistry (Mosc). 2009;74:1505-1534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 10. | Dias NB, de Souza BM, Gomes PC, Brigatte P, Palma MS. Peptidome profiling of venom from the social wasp Polybia paulista. Toxicon. 2015;107:290-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Abreu TF, Sumitomo BN, Nishiyama MY, Oliveira UC, Souza GH, Kitano ES, Zelanis A, Serrano SM, Junqueira-de-Azevedo I, Silva PI. Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J Proteomics. 2016;151:232-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | He QY, He QZ, Deng XC, Yao L, Meng E, Liu ZH, Liang SP. ATDB: a uni-database platform for animal toxins. Nucleic Acids Res. 2008;36:D293-D297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Herzig V, Wood DL, Newell F, Chaumeil PA, Kaas Q, Binford GJ, Nicholson GM, Gorse D, King GF. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2011;39:D653-D657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 14. | Kuzmenkov AI, Krylov NA, Chugunov AO, Grishin EV, Vassilevski AA. Kalium: a database of potassium channel toxins from scorpion venom. Database (Oxford). 2016;2016:pii: baw056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Genomics: Biology portal Molecular and cellular biology portal, 2016. Available from: http://en.wikipedia.org/wiki/Genomics. |
| 16. | The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931-949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1533] [Cited by in RCA: 1279] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 17. | Cao Z, Yu Y, Wu Y, Hao P, Di Z, He Y, Chen Z, Yang W, Shen Z, He X. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 2013;4:2602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 18. | Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V, Jiang X, Cheng L, Fan D, Feng Y. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 2014;5:3765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 19. | Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110:20651-20656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 348] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 20. | Richardson MK, Kini RM. Commentary: The Promise of Snake Genomics. Snake Venoms and Envenomation: Modern Trends and Future Prospects. Hauppauge: Nova Science Publishers 2016; 283-287. |
| 21. | Zhong J, Zeng XC, Zeng X, Nie Y, Zhang L, Wu S, Bao A. Transcriptomic analysis of the venom glands from the scorpion Hadogenes troglodytes revealed unique and extremely high diversity of the venom peptides. J Proteomics. 2017;150:40-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Chen T, Bjourson AJ, Orr DF, Kwok H, Rao P, Ivanyi C, Shaw C. Unmasking venom gland transcriptomes in reptile venoms. Anal Biochem. 2002;311:152-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Modahl CM, Mackessy SP. Full-Length Venom Protein cDNA Sequences from Venom-Derived mRNA: Exploring Compositional Variation and Adaptive Multigene Evolution. PLoS Negl Trop Dis. 2016;10:e0004587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Whiteley G, Logan RA, Leung KY, Newberry FJ, Rowley PD, Dunbar JP, Wagstaff SC, Casewell NR, Harrison RA. Stabilising the Integrity of Snake Venom mRNA Stored under Tropical Field Conditions Expands Research Horizons. PLoS Negl Trop Dis. 2016;10:e0004615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Matysiak J, Dereziński P, Klupczyńska A, Matysiak J, Kaczmarek E, Kokot ZJ. Effects of a honeybee sting on the serum free amino acid profile in humans. PLoS One. 2014;9:e103533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Arjmand M, Akbari Z, Taghizadeh N, Shahbazzadeh D, Zamani Z. NMR-based metabonomics survey in rats envenomed by Hemiscorpius lepturus venom. Toxicon. 2015;94:16-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Mohammadpourdounighi N, Behfar A, Ezabadi A, Zolfagharian H, Heydari M. Preparation of chitosan nanoparticles containing Naja naja oxiana snake venom. Nanomedicine. 2010;6:137-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Ayari-Riabi S, Trimaille T, Mabrouk K, Bertin D, Gigmes D, Benlasfar Z, Zaghmi A, Bouhaouala-Zahar B, Elayeb M. Venom conjugated polylactide applied as biocompatible material for passive and active immunotherapy against scorpion envenomation. Vaccine. 2016;34:1810-1815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Badr G, Sayed D, Maximous D, Mohamed AO, Gul M. Increased susceptibility to apoptosis and growth arrest of human breast cancer cells treated by a snake venom-loaded silica nanoparticles. Cell Physiol Biochem. 2014;34:1640-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Jeong I, Kim BS, Lee H, Lee KM, Shim I, Kang SK, Yin CS, Hahm DH. Prolonged analgesic effect of PLGA-encapsulated bee venom on formalin-induced pain in rats. Int J Pharm. 2009;380:62-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Chanda N, Kattumuri V, Shukla R, Zambre A, Katti K, Upendran A, Kulkarni RR, Kan P, Fent GM, Casteel SW. Bombesin functionalized gold nanoparticles show in vitro and in vivo cancer receptor specificity. Proc Natl Acad Sci USA. 2010;107:8760-8765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 32. | Soman NR, Baldwin SL, Hu G, Marsh JN, Lanza GM, Heuser JE, Arbeit JM, Wickline SA, Schlesinger PH. Molecularly targeted nanocarriers deliver the cytolytic peptide melittin specifically to tumor cells in mice, reducing tumor growth. J Clin Invest. 2009;119:2830-2842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 228] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 33. | Mei D, Lin Z, Fu J, He B, Gao W, Ma L, Dai W, Zhang H, Wang X, Wang J. The use of α-conotoxin ImI to actualize the targeted delivery of paclitaxel micelles to α7 nAChR-overexpressing breast cancer. Biomaterials. 2015;42:52-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 34. | Bhowmik T, Gomes A. NKCT1 (purified Naja kaouthia protein toxin) conjugated gold nanoparticles induced Akt/mTOR inactivation mediated autophagic and caspase 3 activated apoptotic cell death in leukemic cell. Toxicon. 2016;121:86-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Geng L, Zhang HL, Peng HB. The formation of acetylcholine receptor clusters visualized with quantum dots. BMC Neurosci. 2009;10:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Zhao L, Shi X, Zhao J. Chlorotoxin-conjugated nanoparticles for targeted imaging and therapy of glioma. Curr Top Med Chem. 2015;15:1196-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Sun C, Fang C, Stephen Z, Veiseh O, Hansen S, Lee D, Ellenbogen RG, Olson J, Zhang M. Tumor-targeted drug delivery and MRI contrast enhancement by chlorotoxin-conjugated iron oxide nanoparticles. Nanomedicine (Lond). 2008;3:495-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 142] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 38. | Veiseh O, Gunn JW, Kievit FM, Sun C, Fang C, Lee JS, Zhang M. Inhibition of tumor-cell invasion with chlorotoxin-bound superparamagnetic nanoparticles. Small. 2009;5:256-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 39. | Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, Du K, Pullar B, Lee D, Ellenbogen RG. Specific targeting of brain tumors with an optical/magnetic resonance imaging nanoprobe across the blood-brain barrier. Cancer Res. 2009;69:6200-6207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 758] [Reference Citation Analysis (0)] |
| 40. | Sun C, Du K, Fang C, Bhattarai N, Veiseh O, Kievit F, Stephen Z, Lee D, Ellenbogen RG, Ratner B. PEG-mediated synthesis of highly dispersive multifunctional superparamagnetic nanoparticles: their physicochemical properties and function in vivo. ACS Nano. 2010;4:2402-2410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 194] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 41. | Yu XF, Sun Z, Li M, Xiang Y, Wang QQ, Tang F, Wu Y, Cao Z, Li W. Neurotoxin-conjugated upconversion nanoprobes for direct visualization of tumors under near-infrared irradiation. Biomaterials. 2010;31:8724-8731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Gwinn MR, Vallyathan V. Nanoparticles: health effects--pros and cons. Environ Health Perspect. 2006;114:1818-1825. [PubMed] |
| 43. | Sharifi S, Behzadi S, Laurent S, Forrest ML, Stroeve P, Mahmoudi M. Toxicity of nanomaterials. Chem Soc Rev. 2012;41:2323-2343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 984] [Cited by in RCA: 859] [Article Influence: 61.4] [Reference Citation Analysis (0)] |
| 44. | Syed S, Zubair A, Frieri M. Immune response to nanomaterials: implications for medicine and literature review. Curr Allergy Asthma Rep. 2013;13:50-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |