Published online Nov 26, 2015. doi: 10.4331/wjbc.v6.i4.379
Peer-review started: May 30, 2015
First decision: June 18, 2015
Revised: July 29, 2015
Accepted: September 29, 2015
Article in press: September 30, 2015
Published online: November 26, 2015
Processing time: 177 Days and 11.4 Hours
AIM: To study the binding of connective tissue growth factor (CTGF) to cystine knot-containing ligands and how this impacts platelet-derived growth factor (PDGF)-B signaling.
METHODS: The binding strengths of CTGF to cystine knot-containing growth factors including vascular endothelial growth factor (VEGF)-A, PDGF-B, bone morphogenetic protein (BMP)-4, and transforming growth factor (TGF)-β1 were compared using the LexA-based yeast two-hybrid system. EYG48 reporter strain that carried a wild-type LEU2 gene under the control of LexA operators and a lacZ reporter plasmid (p80p-lacZ) containing eight high affinity LexA binding sites were used in the yeast two-hybrid analysis. Interactions between CTGF and the tested growth factors were evaluated based on growth of transformed yeast cells on selective media and colorimetric detection in a liquid β-galactosidase activity assay. Dissociation constants of CTGF to VEGF-A isoform 165 or PDGF-BB homo-dimer were measured in surface plasma resonance (SPR) analysis. CTGF regulation in PDGF-B presentation to the PDGF receptor β (PDGFRβ) was also quantitatively assessed by the SPR analysis. Combinational effects of CTGF protein and PDGF-BB on activation of PDGFRβ and downstream signaling molecules ERK1/2 and AKT were assessed in rabbit corneal fibroblast cells by Western analysis.
RESULTS: In the LexA-based yeast two-hybrid system, cystine knot motifs of tested growth factors were fused to the activation domain of the transcriptional factor GAL4 while CTGF was fused to the DNA binding domain of the bacterial repressor protein LexA. Yeast co-transformants containing corresponding fusion proteins for CTGF and all four tested cystine knot motifs survived on selective medium containing galactose and raffinose but lacking histidine, tryptophan, and uracil. In liquid β-galactosidase assays, CTGF expressing cells that were co-transformed with the cystine knot of VEGF-A had the highest activity, at 29.88 ± 0.91 fold above controls (P < 0.01). Cells containing the cystine knot of BMP-4 expressed the second most activity, with a 24.77 ± 0.47 fold increase (P < 0.01). Cells that contained the cystine knot of TGF-β1 had a 3.80 ± 0.66 fold increase (P < 0.05) and the ones with the cystine knot of PDGF-B had a 2.64 ± 0.33 fold increase of β-galactosidase activity (P < 0.01). Further SPR analysis showed that the association rate between VEGF-A 165 and CTGF was faster than PDGF-BB and CTGF. The calculated dissociation constant (KD) of CTGF to VEGF165 and PDGF-BB was 1.8 and 43 nmol/L respectively. PDGF-BB ligand and PDGFRβ receptor formed a stable complex with a low dissociation constant 1.4 nmol/L. Increasing the concentration of CTGF up to 263.2 nmol/L significantly the ligand/receptor binding. In addition, CTGF potentiated phosphorylation of PDGFRβ and AKT in rabbit corneal fibroblast cells stimulated by PDGF-BB in tissue culture condition. In contrast, CTGF did not affect PDGF-B induced phosphorylation of ERK1/2.
CONCLUSION: CTGF has a differential binding affinity to VEGF-A, PDGF-B, BMP-4, and TGF-β. Its weak association with PDGF-B may represent a novel mechanism to enhance PDGF-B signaling.
Core tip: The relative binding strength of connective tissue growth factor (CTGF) was vascular endothelial growth factor-A > bone morphogenetic protein-4 > transforming growth factor-β1 > platelet-derived growth factor (PDGF)-B. CTGF binding could potentiate PDGF-B signaling as evidenced by enhanced phosphorylation of PDGF receptor β and downstream AKT molecules in rabbit corneal fibroblast cells. Our findings provide deep insight into CTGF action in fine-tuned regulation of extracellular signaling mediated by different cysteine knot containing growth factors.
- Citation: Pi L, Chung PY, Sriram S, Rahman MM, Song WY, Scott EW, Petersen BE, Schultz GS. Connective tissue growth factor differentially binds to members of the cystine knot superfamily and potentiates platelet-derived growth factor-B signaling in rabbit corneal fibroblast cells. World J Biol Chem 2015; 6(4): 379-388
- URL: https://www.wjgnet.com/1949-8454/full/v6/i4/379.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i4.379
Connective tissue growth factor (CTGF), also termed as CCN2, is a secreted cysteine-rich protein in the CCN (Cyr61/CTGF/Nov) family involved in a diverse variety of biological processes ranging from angiogenesis, chondrogenesis, osteogenesis, and tissue repair, to cancer development. CTGF was originally isolated from human umbilical vein endothelial cells as a 349-amino acid mitogen with biological activities similar to platelet-derived growth factor (PDGF)-B[1]. CTGF protein contains a modular architecture starting with an N-terminal signal peptide followed by an insulin-like growth factor-binding (IGFBP) domain, a von Willebrand factor type C domain, a thrombospondin type I homology domain, and a C terminal cystine knot motif[2]. These domains enable CTGF interaction with a broad repertoire of proteins including cell surface proteins, extracellular matrix proteins, and growth factors. Through these interactions, CTGF functions as an extracellular signaling modulator and regulates cell adhesion, migration, activation and differentiation[3].
The cystine-knot motif is structurally conserved in various kinds of growth factors, ligands, and mucins[4,5]. Consensus sequences of this structure can be identified by a pattern of six cysteine amino acids with a defined space consisting of three intertwined disulfide bridges. Two of the disulfide bonds and their adjacent amino acids form a ring that is passed by the third disulfide. This structure forces exposure of many hydrophobic residues in monomers and favor the formation of homo- or hetero-dimers to decrease hydrophobicity in an aqueous environment. These dimers represent active states of the cystine knot-bearing growth factors able to elicit signal transduction via their cognate receptors. Interestingly, many members of this superfamily including transforming growth factor (TGF)-β, bone morphogenetic protein (BMP)-4, BMP-2, vascular endothelial growth factor (VEGF)-A, PDGF-B, von Willebrand factor (vWF), the neuronal guidance cue Slit3, the growth and differentiation factor 5 (GDF5), and GDF15 are able to interact with CTGF[6-11]. It has been shown that CTGF binding can sequester VEGF-A from VEGF receptor 1 and inhibit VEGF-A stimulated angiogenesis[6]. Moreover, the high affinity of CTGF inhibits the presentation of BMP-4 to BMP receptor type Ia and the phosphorylation of this receptor and downstream molecule Smad1, whereas its low affinity potentiates TGF-β1 binding to TβRII receptor and enhances phosphorylation of Smad2[7]. So far, little has been determined concerning the effect of CTGF on PDGF-B activity and signaling. In this study, we compared the binding strengths of CTGF to VEGF-A, PDGF-B, TGF-β1 and BMP-4 using the LexA-based yeast two-hybrid system. The effect of CTGF on PDGF-B presentation to PDGFRβ receptor was determined in surface plasma resonance (SPR) analysis. The modulation of CTGF on PDGF-B signaling was investigated using rabbit corneal fibroblast cells.
Yeast two-hybrid assays were performed according to the Clontech yeast protocol handbook. Synthetic dropout minimal medium (SD) and supplements used in this study were purchased from Clontech (Mountain View, CA). Dropout supplements containing nucleotides and amino acid residues as well as additional supplements of glucose, galactose (Gal), and raffinose (Raf) were prepared according to the manufacturer’s instructions. CTGF, cystine knot motifs in VEGF-A and PDGF-B were described previously[9]. The cystine knot motif of TGF-β1 was amplified using primers 5’ AGGTCGACCTCCTGGCGATACCTCAGCAACGC 3’ (sense) and 5’ TTGCGGCCGCTGCACTTGCAGGAGCGCAC 3’ (antisense) from a human cDNA clone (Genebank Accession # NM_000660.5). The cystine knot motif of BMP-4 was amplified using primers 5’ AGGTCGACC GGACACCTCATCACACGACTA 3’ (sense) and 5’ TTGCGGCCGCGGCATCCACACCCCTCTAC 3’ (antisense) from a mouse cDNA clone (Genebank Accession #NM_007554.2). All the constructs contained restriction enzyme sites Sal I and Not I that facilitated their cloning into the prey pB42AD vector carrying an activation domain (AD) of VP16 or the bait pLexA vector containing a LexA-DNA binding domain (BD).
In yeast two-hybrid analysis, Saccharomyces cerevisiae EGY48 was used as the reporter strain, which carried a wild-type LEU2 gene under the control of LexA operators and a lacZ reporter plasmid p80p-lacZ containing eight high affinity LexA binding sites. SD medium that lacked uracil (Ura) was used for the maintenance of p8op-lacZ plasmids. BD or AD plasmids containing CTGF and cystine knot motifs of tested growth factors were co-transformed into EGY48(p8op-lacZ) and selected using histidine (His; a selection marker for pLexA plasmid) and tryptophan (Trp; for selection of pB42-AD plasmids). Co-transformants were initially selected on SD medium lacking Ura, His, and Trp but supplemented with glucose, which repressed fusion protein expression. Then these co-transformed cells were grown overnight prior to dilution into SD media that contained Gal and Raf but lacked Trip, His, and Ura. Gal and Raf supplements were added to induce fusion gene expression. The diluted cells were grown to mid-log phase with optical densities of 0.5-0.8 units. One-half of each cell culture was used to determine activation of the Leu2 reporter gene by growth assay based on survival in SD medium contained Gal and Raf but lacked Trp, His, Ura, and leucine (Leu). The remaining mid-log-phased cells was pelleted, washed and resuspended in Z buffer (60 mmol/L Na2HPO4, 40 mmol/L NaH2PO4, 10 mmol/L KCl, 10 mmol/L MgSO4, pH = 7.0). β-galactosidase activity was measured by O-nitrophenyl β-D-galactopyranoside (ONPG) cleavage in a liquid assay (Pierce Biotechnology, Rockford, IL). The results were obtained by reading absorbance at 420 nm and normalized against cell density (A600) and reaction time. Values are the average folds of activation over pB42 vector alone (arbitrarily set to 1) from three independent experiments. Statistical analysis was carried out using Student t test in Microsoft Excel software.
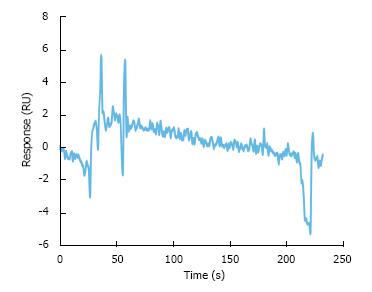
Biacore 2000 and CM4 dextran sensor chips (GE healthcare, Piscataway, NJ) were used to characterize protein-protein interactions based on SPR technology. Filtered and degassed HBS-EP buffer (pH = 7.4) containing 0.01 mol/L HEPES, 0.15 mol/L NaCl, 3 mmol/L EDTA, and 0.005% Tween 20 was used as running buffer. Prior to the kinetic measurements, the sample channel was immobilized with carrier free recombinant human CTGF protein (BioVendor, Asheville, NC), and the reference channel was blocked with ethanolamine. Then, serial concentrations of recombinant human homodimeric PDGF-BB protein (Shenandoah, Warwick, PA) and recombinant human VEGF-A variant 165 (VEGF165) (Abcam, Cambridge, MA) in HBS-EP buffer were injected over the sampling and reference channels simultaneously at 30 μL/min for 3 min. The sensorgrams acquired from the reference channels were deducted from those of CTGF-immobilized channels to eliminate the responses caused by the refractive index changes. The dissociation was then followed by injecting HBS-EP buffer for 10 min. The difference in SPR response unit (RU) between the baseline and the signal after the dissociation reflects the amount of bound analytes. Before starting the next injection, the chip surface was regenerated by exposing the chip surface with 10 mmol/L of cysteine-HCl buffer pH 2.0 at 30 μL/min for 30 s.
To determine the effect of CTGF on the binding between PDGF-BB and PDGFRβ, a mixture of PDGF-BB (2 nmol/L) and varied concentrations of CTGF (0 to 263.2 nmol/L) was injected over the PDGFRβ-immobilized channel and reference channel. Amine coupling strategy was used to immobilize the recombinant human PDGFRβ Fc chimera protein (R&D Systems, Minneapolis, MN) on the sampling channel. Briefly, a mixture of 0.4 mol/L 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and 0.1 mol/L N-hydroxysuccinimide in equal proportion was used to activate the surface of a CM4 dextran sensor chip at a flow rate of 10 μL/min for 7 min. Then, PDGFRβ was injected over separate sampling channels for 7 min to covalently immobilize on the surfaces. Remaining active groups were blocked with 1 mol/L ethanolamine-HCl at pH 8.5 on both sampling and reference channels. The sensorgrams acquired from the reference channels were deducted from those of the PDGFRβ-immobilized channels to eliminate the responses caused by the refractive index changes. All data were analyzed with the BIAevaluation software for calculating dissociation constants.
Determination phosphorylation of PDGFRβ, ERK1/2 and AKT in Western analysis
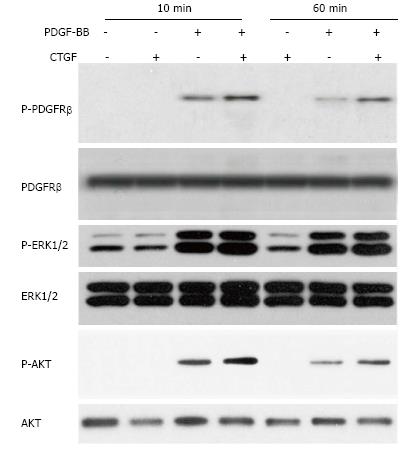
Primary rabbit corneal fibroblast cells were isolated as described previously[12]. Cells were cultured to 100% confluence and serum-starved for 20 h in DMEM with 0.1% FBS. PDGF-B (1.6 nmol/L), CTGF (6.6 nmol/L), or a combination was added to the prepared cells. At 10 min and again at 1 h after stimulation, cells were collected and lysed in RIPA buffer (150 mmol/L NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mmol/L Tris, pH = 8.0) containing proteinase inhibitor (Sigma, St Louis, MI). Total protein lysates (20 μg) were separated on 10% SDS-PAGE gels followed by transferring to polyvinylidene difluoride membranes. The primary antibodies for anti-phospho-PDGFRβ, anti-PDGFRβ, anti-phospho-ERK1/2 (Thr202/Tyr204), anti-ERK1/2, anti-phospho-AKT, and anti-AKT were purchased from Cell Signaling (Danvers, MA). HRP conjugated rabbit or mouse secondary antibodies were used followed by detection with the ECL kit (GE Health Biosciences).
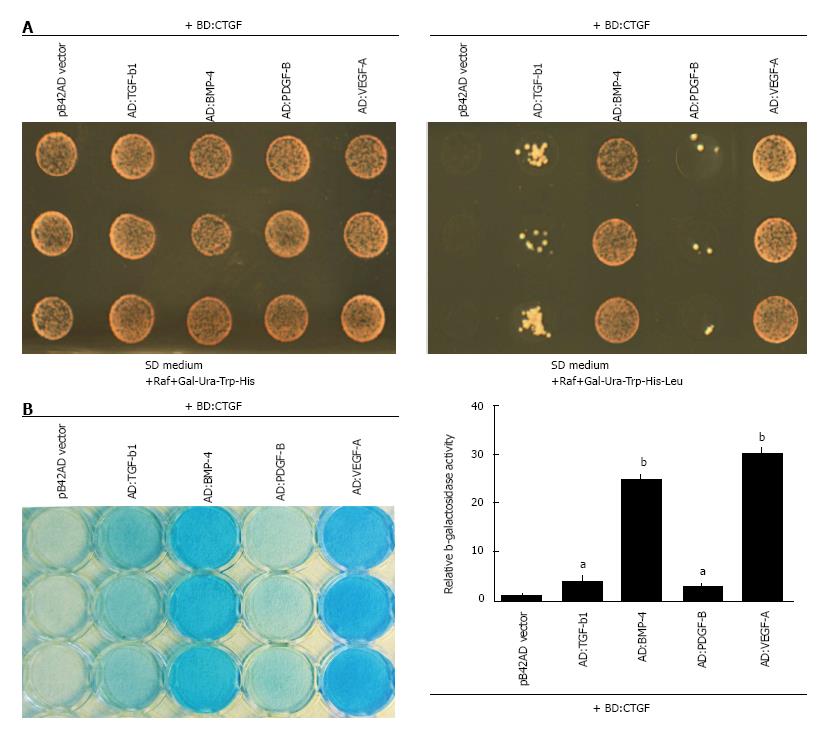
Differential binding ability of CTGF to VEGF-A, PDGF-B, TGF-β, and BMP-4 in the yeast two-hybrid LexA system
To compare the relative binding activity of CTGF to VEGF-A, PDGF-B, TGF-β1, and BMP-4, we took the yeast two-hybrid approach and expressed the cystine knot motifs of these specific growth factors as AD fusion proteins under the control of the GAL1 promoter in the pB42-AD plasmids. In addition, full-length CTGF cDNA was expressed in fusion with LexA BD under the control of an alcohol dehydrogenase promoter in pLexA plasmids. These plasmids and their corresponding empty vectors were transformed into the EGY48 (p8op-lacZ) reporter strain (Figure 1A). CTGF itself was not able to self-activate in this LexA system since yeast cells that were co-transformed with the BD:CTGF plasmid and pB42 vector did not grow without Leucine supplementation and did not turn blue in β-galactosidase assays. Nor were cystine knot motifs of tested growth factors because they also failed to trigger Leu2 and LacZ reporter genes when co-transformed with pB42-AD empty vector. In contrast, co-transformation with CTGF allowed survival of cells that expressed cystine knot motifs of all four growth factors on selective medium lacking leucine, supporting our previous observations that CTGF has broad binding capabilities to members of cystine knot superfamily[9]. In addition, we carried out an ONPG-based assay and assessed relative β-galactosidase activity in comparison to control cells that carried plasmids for CTGF and pLexA vector. As shown in Figure 1B, CTGF expressing cells that were co-transformed with the cystine knot of VEGF-A had the highest β-galactosidase activity, at 29.88 ± 0.91 fold above controls. Cells containing the cystine knot of BMP-4 expressed the second most β-galactosidase activity, with a 24.77 ± 0.47 fold increase. Cells that contained the cystine knot of TGF-β1 had a 3.80 ± 0.66 fold increase and the ones with the cystine knot of PDGF-B had a 2.64 ± 0.33 fold increase. These observations indicated that VEGF-A and BMP-4 were strong interactors whereas PDGF-B and TGF-β were weak binding proteins for CTGF.
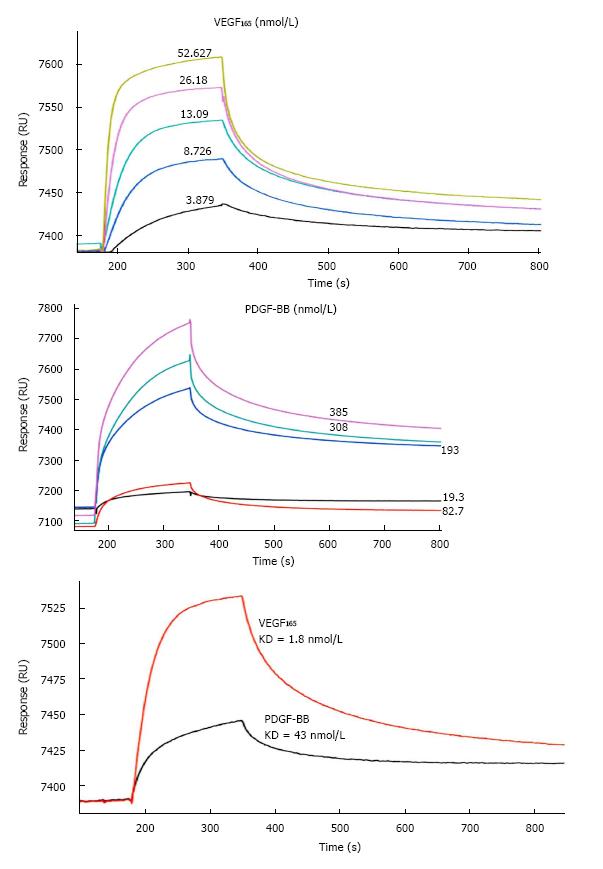
SPR analysis was performed to characterize the affinity of CTGF to VEGF165 and PDGF-BB. Initial experiments indicated that injecting homodimeric PDGF-BB over a CM5 dextran sensor chip caused a significant non-specific response on the reference channels, which may be due to the electrostatic interaction between the cationic growth factors and negatively charged carboxymethyl dextran covered on the chip. Therefore, we chose a CM4 dextran sensor chip with a lower degree of carboxymethylation to avoid the non-specific interaction. The kinetic analyses between PDGF-BB/CTGF and VEGF165/CTGF complexes were carried out by injecting different concentrations of PDGF-BB (19.3 to 385 nmol/L) or VEGF165 (3.879 to 52.267 nmol/L) over the CTGF-immobilized channel and reference channel simultaneously at 30 μL/min for 3 min. The dissociation process was followed by injecting HBS-EP buffer at 30 μL/min for 10 min. The higher flow rate was required to reduce the mass transfer effect and obtain kinetic parameters accurately. As shown in Figure 2A-C, the association rate between VEGF165 and CTGF was faster than PDGF-BB and CTGF. The calculated dissociation constant (KD) of CTGF to VEGF165 and PDGF-BB was 1.8 and 43 nmol/L respectively. These results indicated that CTGF had higher affinity to VEGF165 than PDGF-BB.
CTGF promotes the binding of PDGF-B to PDGFRβdetermined by SPR analysis
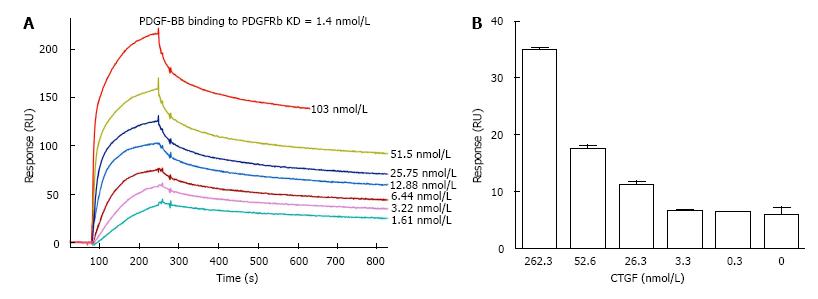
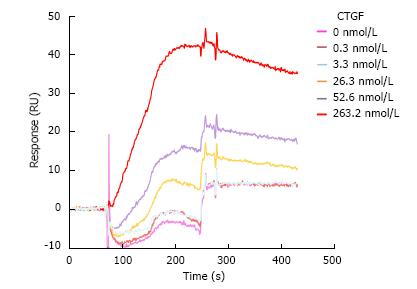
PDGF-BB ligand and PDGFRβ receptor formed a stable complex indicated by a low dissociation constant 1.4 nmol/L in our SPR analysis (Figure 3A). To determine whether CTGF protein influenced the formation of this ligand/receptor complex, we injected a mixture of CTGF and PDGF-BB over the PDGFRβ-immobilized channel and measured the SPR response unit. As shown in Figures 3B and 4, the presence of CTGF was able to potentiate the binding of PDGF-BB to its receptor. Increasing the amount of CTGF further promoted this effect, especially when the concentration of CTGF was more than 3.3 nmol/L. Specifically, two-fold more ligand/receptor binding was observed when 26.3 nmol/L CTGF was mixed with 2 nmol/L PDGF-BB in comparison to the same amount of PDGF-BB alone. Moreover, three and a half-fold more ligand/receptor binding was seen when 263.2 nmol/L CTGF was mixed with 2 nmol/L PDGF-BB. In other words, seven-fold more ligand/receptor binding response was seen when CTGF to PDGF-BB ratio in molar concentration was 131.6 in comparison to PDGF-BB alone. To validate if the binding was indeed attributable to PDGF-BB, we also injected pure 263.2 nmol/L CTGF over PDGFRβ-immobilized channels as a control. We found no binding interaction (Figure 5).
CTGF potentiates activation of PDGFRβand downstream signaling molecule AKT
PDGFRβ is recognized by PDGF-B ligand and deletion of this receptor gene affects key fibroblast functions in wound healing[13,14]. PDGF-BB binding to the extracellular region of PDGFRβ induces dimerization and activation of intracellular kinase domain of this receptor. As a result, PDGFRβ receptors undergo auto-phosphorylation in critical tyrosine residues and gain full enzymatic activity to phosphorylate substrates. These phosphorylation regions can function as docking sites and recruit signal molecules containing SH2 and SH3 domains. For instances, Tyr751 in the kinase-insert region of PDGFRβ is the docking site for PI3 kinase, which in turn activates downstream molecules including the serine/threonine kinase AKT[15]. We used cultured rabbit corneal fibroblast cells and detected that PDGF-BB (1.6 nmol/L) quickly stimulated PDGFRβ phosphorylation at Tyr751 within minutes (Figure 6). This phosphorylated PDGFRβ gradually decreased probably due to a rapid turnover of this receptor in cultured rabbit corneal fibroblast cells. This pattern was correlated with phosphorylation of AKT and ERK1/2 in PDGF-BB stimulated cells. The phosphorylation of both PDGFRβ and AKT induced by 1.6 nmol/L PDGF-BB was enhanced by the addition of CTGF (6.6 nmol/L) to rabbit corneal fibroblast cells. In contrast, CTGF alone was unable to cause significant changes of PDGFRβ receptor and AKT levels in the cultured cells. Moreover, phosphorylation of ERK1/2 stimulated by PDGF-BB was not influenced by presence of CTGF. These results indicated that CTGF could potentiate some PDGF-BB mediated signaling including activation of PDGFRβ and AKT in rabbit corneal fibroblast cells.
The yeast two-hybrid system is designed to measure protein-protein interaction by reconstitution of a functional transcription factor that activates receptor genes. Previously, we used a GAL4 system and revealed CTGF’s broad ability to bind with multiple cystine knot-bearing factors using His3 reporter[9]. This is a qualitative assay with potential variation dependent upon the expression levels of the fusion proteins under investigation. In this study, we took advantage of LexA-responsive LEU2 and lacZ reporter genes, and carried out quantitative assessments according to both colony survival and β-galactosidase activity to discriminate high- and low-affinity interactions. Our results suggested that CTGF strongly bound to VEGF-A and BMP-4 but loosely interacted with PDGF-B and TGF-β1. These observations were consistent with published results about measurement of dissociation constants for BMP-4, TGF-β1, PDGF-BB, and VEGF-A using SPR or I125 labeled solid phase protein binding assays[7,10]. This study further supported a previous report that the degree of protein interaction determined by yeast two-hybrid approaches correlates well with those examined in biochemical analysis[16].
The cystine-knot superfamily is composed of several subgroups of proteins involved in a diverse variety of pathophysiological functions including cell growth, organogenesis, embryonic development, cell-to-cell communication, differentiation, tissue repair and cancer progression. In mammals, the PDGF subfamily has at least 10 members (i.e., VEGF-A, VEGF-B, VEGF-C, VEGF-D, and VEGF-E from the VEGF family; PDGF-A, PDGF-B, PDGF-C, and PDGF-D from the PDGF family; and placenta growth factors PlGF-1 and PlGF-2)[14]. The TGF-β subfamily has about 37 members (BMP2-7, BMP8A, BMP8B, BMP10, BMP15, GDF1-3, GDF5-11, GDF15, Activin βE, Inhibin α, Inhibin βA, Inhibin βB, Inhibin βC, TGF-β1-3, Nodal, Anti-Mullerian hormone, LEFTY-A, LEFTY-B, GDNF, Neurturin, Persephin, and Artemin-2I)[17]. The Slit subfamily has three members (Slit1-3). The mucin-like subgroup include nine BMP antagonists (Follistatin, FSTL-3, Cerberus, Chordin, Gremlin, Noggin, SOST/Sclerostin, TSG, and USAG-1)[18], five mucin related protein (Norrin, mucin-2, MUC5AC protein, secretory mucin), and vWF[4]. So far, CTGF is found to interact with at least one member in each subgroup of the cystine knot superfamily. This broad binding spectrum of CTGF may be due to unique structure organization of members of the CCN family. Based on small angle X-ray scattering data in CCN3 and CCN5, CCNs seem to form long extended structures able to move and flex to allow multiple domains to bind a ligand[19]. It is conceivable that the four modular structures of CTGF protein form long extended and flexible scaffolds that “glue” the cystine knot proteins from destruction by cleavage in extracellular space.
Differential binding activity of CTGF to cystine knot-bearing ligands appears to give rise to disparate regulatory mechanisms for the ligand presentation to their receptors. One mechanism is through a high affinity/interaction with CTGF. Examples are VEGF-A, BMP-4, and BMP-2, which are strong interactors for CTGF. Our study, together with others[10] show that VEGF-A has a dissociation constant for CTGF in SPR assays around 1.8-4.8 nmol/L. BMP-4 and BMP-2 have dissociation constants of 5 and 0.77 nmol/L with CTGF respectively[7,10]. CTGF binding sequesters VEGF-A from its receptor and inhibits downstream signaling leading to anti-angiogenesis[6]. Similarly, CTGF binding inhibits BMP-4 from BMPRIa receptor and downstream signaling leading to altered development of Xenopus embryo[7]. BMP-2 activity on the proliferation of chondrocytes is also inhibited by CTGF[12]. Hence, we speculate that strong interaction with CTGF causes sequestration of the bound ligands from their receptors thereby preventing signal transduction.
In contrast, other cystine knot-bearing growth factors such as TGF-β1 and PDGF-B are weak interactors and their signal transduction pathways are not inhibited by CTGF[7]. The dissociation constant for CTGF binding to TGF-β1 is 30 or 64.7 nmol/L measured by two different groups[7,10]. We found that CTGF bound to PDGF-BB with a dissociation constant 43 nmol/L. Much lower binding kinetics between CTGF and PDGF-BB (KD = 230 nmol/L) is observed by Khattab et al[10]. Variable factors including purity of tested recombinant proteins and variation of Biacore SPR systems may contribute to these discrepancies. Nevertheless, the measured dissociation constants for these weak interactors are often more than 10-10 mol/L. Despite of this low affinity, CTGF seems to increase the activity of TGF-β[7]. We found that CTGF was able to potentiate PDGF-BB binding to PDGFRβ and enhance activation of PDGFRβ and downstream AKT. Given that CTGF is a matricellular protein and can trigger adhesive signaling through association with cell surface proteins such as integrins[3], its binding to TGF-β1 or PDGF-B may be part of crosstalk for integration of these different signal pathways. Lack of overlapping in some downstream signaling between CTGF and PDGF-BB may explain little enhancement of ERK1/2 activation in cultured rabbit corneal fibroblast cells.
TGF-β1, PDGF-B and CTGF are known profibrotic partners for fibroblast activation[20]. Low affinity of CTGF may account for its synergistic effect on PDGF-BB and TGF-β in promoting chronic fibrosis. In particular, TGF-β1 can transcriptionally activate CTGF expression through SMAD responsive elements in the CTGF promoter[21]. In mice, co-injection of CTGF and TGF-β results in sustained, persistent fibrosis, whereas subcutaneous injection of just TGF-β causes a transient fibrotic response[22]. In addition, PDGF is a major regulator for vascular cells and its deficiency causes vascular abnormalities including capillary dilation and microaneurysms during mouse embryonic development[23]. VEGF-A is critical for the proliferation and survival of vascular endothelial cells. Loss of VEGF-A expression results in severe abnormalities in the vasculature and causes embryonic death in heterozygous mice[24]. Given the importance of PDGF-B and VEGF-A in the assembly of blood vessels, the inherent stickiness of CTGF to PDGF-B and VEGF-A may represent a fine-tuned regulation of vascular smooth muscle cells, pericytes, and vascular endothelial cells during angiogenesis. Future studies about the nature of CTGF binding to VEGF-A, PDGF-B, and TGF-β may help the development of therapeutic strategies for pathological processes involving angiogenesis and fibrosis.
We thank Marda Jorgessen for editorial assistance in preparation of this manuscript.
Connective tissue growth factor (CTGF), also termed CCN2, is one prototype member in the Cyr62/CTGF/Nov family of matricellular proteins. It contains a four conserved modular structure and regulates cell proliferation, migration, differentiation, apoptosis, and survival through interaction with extracellular matrix protein, growth factors, and cell surface proteins. CTGF exhibits a broad binding ability to a structurally conserved motif characteristic of the cystine knot superfamily. However, molecular action of CTGF in modulation of the cystine knot-containing growth factors has not been fully understood.
CTGF binding can sequester some of the cystine knot-containing growth factors such as vascular endothelial growth factor (VEGF)-A and bone morphogenetic protein (BMP)-4 from their cognate receptors x and inhibit their signaling. For other growth factors such as transforming growth factor (TGF)-β1, CTGF has distinct effects and potentiates TGF-β1 mediated downstream signal transduction pathway. Correlation between the binding strength of CTGF and its action on the majority of cystine knot-containing growth factors has not been investigated.
This paper combined LexA-based yeast two-hybrid system with surface plasma resonance (SPR) analysis and quantitatively assesses the binding strengths of CTGF to VEGF-A, platelet-derived growth factor (PDGF)-B, BMP-4, and TGF-β1. CTGF had high affinities with VEGF-A and BMP-4 but only weakly interacted with TGF-β1 and PDGF-B. These binding strengths were correlated with inhibition or potentiation by CTGF on signal transduction pathways mediated by these cystine knot-containing growth factors, implicating that stickiness to CTGF results in sequestration of the ligands from their cognate receptors while weak association facilitates ligand/receptor presentation. Our findings support that CTGF is a critical modulator of extracellular signaling and functions to integrate different pathways into signaling networks resulting in tight regulation and fine-tuning of cellular responses during diverse physiological and pathological processes.
Many cystine knot-containing growth factors are key regulators in angiogenesis and tissue repair. Understanding of the binding of CTGF to these growth factors may hold promise as a strategy of rational therapeutic design for the prevention or treatment of pathological processes in cancer, vascular and fibrotic disorders of many organs.
Cystine knots refer to a common overall topology that was initially discovered in structure of nerve growth factor solved by Tome Blundell using X-ray crystallography in 1991. This tertiary structure favors dimer formation and is unique in extracellular signaling molecules of multicellular organisms since it is not found in the unicellular yeast genome. All cysteine knot growth factors are believed to be homo- or hetero- dimers and initiate signal transduction through dimerization of their cognate receptors.
CTGF is known as an important factor in regulating diverse biological functions, including cell adhesion, migration, tissue wound repair, fibrotic diseases, and cancers. In this paper, the author demonstrated that CTGF had different binding strengths to VEGF-A, PDGF-B, BMP-4, and TGF-β, which regulated these growth factors and triggered downstream signaling pathways. The topic of this work is of high interest. The authors approach to the study of the interactions between CTGF and a number of its interactors using the LexA-based yeast two-hybrid system and SPR analysis, and tried to explain the results using some functional aspects.
P- Reviewer: Febbraio F, Hegardt FG, Wang P, Zhang HT S- Editor: Tian YL L- Editor: A E- Editor: Lu YJ
| 1. | Bradham DM, Igarashi A, Potter RL, Grotendorst GR. Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol. 1991;114:1285-1294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 649] [Cited by in RCA: 679] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 2. | Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 501] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 3. | Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 459] [Cited by in RCA: 455] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 4. | Vitt UA, Hsu SY, Hsueh AJ. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 2001;15:681-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 192] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | McDonald NQ, Hendrickson WA. A structural superfamily of growth factors containing a cystine knot motif. Cell. 1993;73:421-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 389] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y. Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J. 2002;16:219-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 285] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 7. | Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4:599-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 730] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 8. | Whitson RJ, Lucia MS, Lambert JR. Growth differentiation factor-15 (GDF-15) suppresses in vitro angiogenesis through a novel interaction with connective tissue growth factor (CCN2). J Cell Biochem. 2013;114:1424-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | Pi L, Shenoy AK, Liu J, Kim S, Nelson N, Xia H, Hauswirth WW, Petersen BE, Schultz GS, Scott EW. CCN2/CTGF regulates neovessel formation via targeting structurally conserved cystine knot motifs in multiple angiogenic regulators. FASEB J. 2012;26:3365-3379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Khattab HM, Aoyama E, Kubota S, Takigawa M. Physical interaction of CCN2 with diverse growth factors involved in chondrocyte differentiation during endochondral ossification. J Cell Commun Signal. 2015;Apr 19; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Maeda A, Nishida T, Aoyama E, Kubota S, Lyons KM, Kuboki T, Takigawa M. CCN family 2/connective tissue growth factor modulates BMP signalling as a signal conductor, which action regulates the proliferation and differentiation of chondrocytes. J Biochem. 2009;145:207-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 12. | Robinson PM, Chuang TD, Sriram S, Pi L, Luo XP, Petersen BE, Schultz GS. MicroRNA signature in wound healing following excimer laser ablation: role of miR-133b on TGFβ1, CTGF, SMA, and COL1A1 expression levels in rabbit corneal fibroblasts. Invest Ophthalmol Vis Sci. 2013;54:6944-6951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Gao Z, Sasaoka T, Fujimori T, Oya T, Ishii Y, Sabit H, Kawaguchi M, Kurotaki Y, Naito M, Wada T. Deletion of the PDGFR-beta gene affects key fibroblast functions important for wound healing. J Biol Chem. 2005;280:9375-9389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 88] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1567] [Cited by in RCA: 1746] [Article Influence: 102.7] [Reference Citation Analysis (0)] |
| 15. | Gao BB, Hansen H, Chen HC, Feener EP. Angiotensin II stimulates phosphorylation of an ectodomain-truncated platelet-derived growth factor receptor-beta and its binding to class IA PI3K in vascular smooth muscle cells. Biochem J. 2006;397:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820-5829. [PubMed] |
| 17. | Galat A. Common structural traits for cystine knot domain of the TGFβ superfamily of proteins and three-fingered ectodomain of their cellular receptors. Cell Mol Life Sci. 2011;68:3437-3451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Rider CC, Mulloy B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 165] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Holbourn KP, Malfois M, Acharya KR. First structural glimpse of CCN3 and CCN5 multifunctional signaling regulators elucidated by small angle x-ray scattering. J Biol Chem. 2011;286:22243-22249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Leask A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ Res. 2010;106:1675-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 554] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 21. | Grotendorst GR, Okochi H, Hayashi N. A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 1996;7:469-480. [PubMed] |
| 22. | Mori T, Kawara S, Shinozaki M, Hayashi N, Kakinuma T, Igarashi A, Takigawa M, Nakanishi T, Takehara K. Role and interaction of connective tissue growth factor with transforming growth factor-beta in persistent fibrosis: A mouse fibrosis model. J Cell Physiol. 1999;181:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Wilkinson-Berka JL, Babic S, De Gooyer T, Stitt AW, Jaworski K, Ong LG, Kelly DJ, Gilbert RE. Inhibition of platelet-derived growth factor promotes pericyte loss and angiogenesis in ischemic retinopathy. Am J Pathol. 2004;164:1263-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2997] [Cited by in RCA: 2871] [Article Influence: 99.0] [Reference Citation Analysis (0)] |