Published online Nov 26, 2015. doi: 10.4331/wjbc.v6.i4.281
Peer-review started: May 29, 2015
First decision: June 18, 2015
Revised: July 7, 2015
Accepted: September 29, 2015
Article in press: September 30, 2015
Published online: November 26, 2015
Processing time: 179 Days and 23.8 Hours
Recent advances in amino acid metabolism have revealed that targeting amino acid metabolic enzymes in cancer therapy is a promising strategy for the development of novel therapeutic agents. There are currently several drugs in clinical trials that specifically target amino acid metabolic pathways in tumor cells. In the context of the tumor microenvironment, however, tumor cells form metabolic relationships with immune cells, and they often compete for common nutrients. Many tumors evolved to escape immune surveillance by taking advantage of their metabolic flexibility and redirecting nutrients for their own advantage. This review outlines the most recent advances in targeting amino acid metabolic pathways in cancer therapy while giving consideration to the impact these pathways may have on the anti-tumor immune response.
Core tip: Amino acid metabolism has been a focus of increased attention by cancer researchers and immunologists due to its importance for the metabolic reprogramming of proliferating cells. Many amino acid enzymes are described as immunosuppressive in the tumor microenvironment and targeted for cancer therapy. This review addresses the metabolic control of tumor progression in the context of anti-tumor immunity and discusses current and future therapeutic approaches. Special emphasis is given to the emerging role of branched chain amino acid metabolism in cancer and immunity highlighting some recent work by our research group.
- Citation: Ananieva E. Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J Biol Chem 2015; 6(4): 281-289
- URL: https://www.wjgnet.com/1949-8454/full/v6/i4/281.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i4.281
Two functionally distinct types of cells, cancer and T cells, undergo similar metabolic reprogramming during proliferation to support their increased biosynthetic and energy demands[1,2]. To meet these demands, cancer cells need a continuous supply of nutrients that maintain abnormal growth and rapid division during cancer progression. Activated T cells also rely on a continuous nutrient supply to ensure proper differentiation and performance during cell-mediated immunity against pathogen attacks or while fighting cancer[3]. In areas with poor nutrient and oxygen access, such as the tumor microenvironment[4] and inflammatory sites[5], cellular metabolism must adapt to promote continued survival and function of the cells residing in and/or migrating to these areas. Not surprisingly, both cancer and T cells rely on high rates of glycolysis while retaining mitochondrial respiration, a phenomenon known as the Warburg effect[6,7]. Ever since Otto Warburg observed that cancer cells produced lactate from glucose even under non hypoxic conditions[7], subsequent reports demonstrated that high rates of glycolysis are beneficial for cell survival, providing energy and biosynthetic material for the synthesis of new proteins, lipids, and nucleic acids in proliferating cells[8-13]. Recent advances in cancer biology and cellular immunity reveal that not only glucose but also amino acids are essential to support the high metabolic demands of tumor and/or immune cells and different strategies for targeting amino acid metabolism and corresponding enzymes are now a focus of innovative treatment approaches[14-17]. The purpose of this review is to highlight the importance of amino acid metabolism in the tumor microenvironment and use this knowledge to generate more effective immunotherapies while keeping in mind that both cancer and immune cells have similar metabolic requirements for growth and function and therefore may compete for common nutrients.
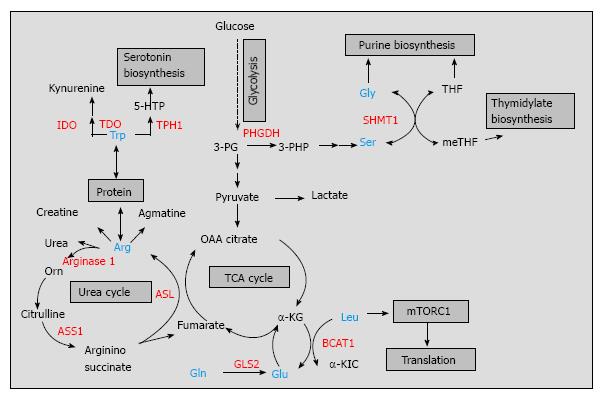
The amino acid arginine has considerable nutritional and physiological significance as it is recognized as an important precursor for the synthesis of proteins, urea, and creatine as well as for the synthesis of signaling molecules such as glutamate, nitric oxide, and agmatine[18] (Figure 1). Although arginine is a dispensable (nonessential) amino acid for healthy humans, it is conditionally essential under certain physiological conditions or disease state[10,19,20]. For example, many tumors are dependent on exogenous arginine for growth as they lack the enzyme argininosuccinate synthetase 1 (ASS1)[21]. ASS1 catalyzes the conversion of citrulline into argininosuccinate in an ATP-dependent manner, completing one of the last steps in the arginine biosynthetic pathway[18] (Figure 1). Loss of ASS1 prevents the production or arginine and may lead to arginine depletion. Osteosarcoma and bladder cancer cell lines expressing low levels of ASS1 failed to grow in an arginine-free medium, indicating that ASS1 behaves as a tumor suppressor[22,23]. However, arginine depletion in ASS1-negative tumor cells is usually associated with an aggressive phenotype and negative prognostic impact[22]. Kobayashi et al[22], showed that ASS1 deficiency in osteosarcoma patients correlated with the development of pulmonary metastasis in patients with osteosarcoma and can be used as a predictive biomarker for unfavorable prognosis. The aggressiveness of ASS1-negative tumors can be explained partially by the ability of these tumors to utilize more efficiently exogenous arginine coming from the tumor microenvironment. It is hypothesized that tumor associated myeloid cells (TAMCs), that consist of macrophages, monocytes, myeloid suppressor cells, and neutrophils[24], form metabolic relationship with the tumor cells in the tumor microenvironment. They provide arginine to help the tumor cells by-pass the effect of arginine deprivation[25]. In addition, TAMCs express high levels of another enzyme in the arginine metabolism, arginase 1, that hydrolyses arginine into urea and ornithine and sustains tumor growth by providing precursors for polyamine synthesis[26,27]. By producing high levels of arginase 1 in the tumor microenvironment, TAMCs reduce arginine availability for other immune cells such as T cells. However, TAMCs can arrest cytotoxic T cell proliferation and induce T cell dysfunction by more than one mechanism, including generation of nitric oxide from arginine by nitric oxide synthase[28]. Additionally, these cells can induce regulatory T cell differentiation[29] potentially promoting immune tolerance in the tumor microenvironment. Thus, TAMCs have the ability to suppress the protective anti-tumor immune response by targeting arginine metabolism and helping tumors escape immune destruction. While immunotherapeutic targeting of arginine metabolism in the tumor microenvironment is still in its infancy, several decades of substantial in vitro and in vivo studies on arginine metabolism led to the development of arginine deprivation therapy, which is currently the subject of ongoing clinical trials with several arginine depleters, such as pegylated arginine deiminase (ADI-PEG20, Polaris group) and bioengineered forms of human arginase[25,30,31] (Table 1). A small study group of patients with hepatocellular carcinoma revealed response rates to ADI-PEG20 between 25%-47% and this compound holds great potential in the arginine depravation therapy[30]. ADI-PEG20 is not limited to cancer therapy as there is ADI-PEG20 with anti-viral activity designed for treatment of hepatitis C by Polaris group.
| Amino acidmetabolism | Targeted enzyme | Drug design | Drug toxicity, adverse events | Cancer type | Response rate | Clinical studies | Ref. |
| Arginine | ASS1, Arginine deaminase | ADI-PEG20 | 1Grade 3-4: Fatigue, hyperuricemia, anemia | HCC (nonresectable and metastatic) | 31%-47% | Phase I/II | [30,94,95] |
| . | ADI-PEG20 | 1Grade 3-4: Pain in extremity, arthralgia, pruritus, lymphedema, seizure | Melanoma (stage III and IV) | 25% | Phase I/II | [96,97] | |
| Arginase 1 | rhArgIpeg5000 | 1Grade 3-4: Elevated ALT, AST, bilirubin and GGT | HCC (advanced) | 26.70% | Phase I | [31] | |
| Co-hArgI | Nude BALBc mouse model: Weight loss, hunch posture, lethargy, bone marrow injuries (> 15 mg/kg) | HCC (HepG2), pancreatic cancer (Panc-1); tumor site (right flank) | Smaller tumors | Preclinical studies | [98] | ||
| Tryptophan | IDO | Indoximod and docetaxel | Grade 1: Anemia, fatigue, hyperglycemia Grade 3-4: Headache, hypotension, infection | Various metastatic solid tumors | 41% PD | Phase I | [99] |
| 1-D-MT | Grade 1: Fatigue; Grade 2: Hypophysitis | Various metastatic solid tumors | 4P (SD), 3P (PD) | Phase I | [100] | ||
| Indoximod and anti-CTLA4 | C57BL/6 mouse model: No weight loss or acute/delayed toxicity observed | B16 F10 melanoma cell line, tumor site -flank | Delayed tumor growth | Preclinical studies | [101] | ||
| Tryptophan | TDO | Indole LM10 | DBA/2 mouse model: No liver toxicity observed for 3 mo | P815B tumor cell line, tumor site –peritoneal cavity | Delayed tumor progression | Preclinical studies | [50] |
| Serine | PHGDH | NA | NA | Melanoma and breast cancer cell lines (SkBr3, MCF7) | NA | Preclinical studies | [15] |
| NA | NA | Murine mammary fat pad tumors with MDA-MB-468 cells expressing PHGDH-shRNA | Reduced tumor growth | Preclinical studies | [63] | ||
| Glycine | SHMT1 | NA | λ-Myc Shmt1-/- transgenic mice: no toxicity observed | Accelerated lymphomagenesis | NA | Preclinical studies | [102] |
| Glutamine | Glutamine –dependent enzymatic steps | L-DON2, azaserine2 | None reported | Various animal and human xenografted tumors | Tumor growth inhibition | Preclinical studies | [80] |
| Acivicin2 | Grade1-3: neurological toxicity Grade 2: vomiting, infections | High grade astrocytoma | Median 128 day survival | PhaseII | [103] | ||
| Leucine, Isoleucine, Valine | BCATc | NA | CD-1 nude mouse model bearing tumors with BCATc knockdown: lethargy and uncoordinated motor activity | U-87MG glioblastoma cells with BCATc-shRNA; tumor site-intracerebral transplantation | Smaller tumors | Preclinical studies | [16] |
| NA | NA | Nasopharyngeal carcinoma (5-8F, 6-10B), colorectal cancer | Induced cell proliferation | Preclinical studies | [92,93] |
Tryptophan is another amino acid linked to the regulation of immune tolerance and anti-tumor immune responses[32-35]. Tryptophan degradation occurs via the kynurenine pathway where two different enzymes, indoleamine-2,3-dioxygenase (IDO) and tryptophan-2,3-dioxygenase (TDO), catalyze the conversion of tryptophan into kynurenine, while tryptophan hydroxylase-1 (TPH-1) converts tryptophan to 5-hydroxytryptophan and provides precursors for serotonin biosynthesis[32,36] (Figure 1). IDO is the most studied enzyme in tryptophan metabolism expressed by both immune cells (dendritic cells, macrophages) and tumor cells[33,37-39]. Due to the fact that tryptophan readily crosses the plasma membrane, dendritic cells expressing IDO are capable of depleting tryptophan in the extracellular space, limiting tryptophan supply to surrounding T cells. T cell activation is sensitive to local tryptophan concentrations, and lack of tryptophan blocks their proliferation[40]. By degrading tryptophan, IDO inhibits T cell proliferation and plays a role in autoimmunity and anti-inflammatory responses[41-43]. For example, Apoe-/- mice treated with the IDO inhibitor, 1-methyl-Trp, showed a significant increase in atherosclerotic lesions in the aortic arch and root of their hearts along with enhanced vascular inflammation[41]. Additionally, IDO-expressing dendritic cells suppressed the allograft rejection and increased the survival time of small bowel transplanted mice[44]. In the tumor microenvironment, however, IDO limits the T cell response to tumor growth[45-47]. By using an IDO-negative subline of the mouse tumor model P815B, Uyttenhove et al[45] elegantly showed that mice transfected with this subline completely rejected the tumor challenge while mice transfected with IDO-expressing cells developed progressive tumors and died. These results suggest that effector T cells that infiltrate the tumor microenvironment are more susceptible to the effects of IDO than are tumor cells. Indeed, immunohistochemical staining for IDO expression in endometrial cancer tissues revealed significant correlation between high IDO expression and low numbers of CD3+, CD8+, and CD57+ immune cells, which possibly contributed to disease progression and impaired clinical outcome for patients with endometrial cancer[48]. In addition to IDO, tumors use other enzymes in tryptophan metabolism to resist immune destruction[36,49]. TDO, a well described liver enzyme, is another immunosuppressive enzyme found in bladder carcinomas, melanomas, and hepatocarcinomas[50]. In a preclinical model, Pilotte et al[50] demonstrated that systemic treatment of immunized mice with a TDO inhibitor prevented the growth of TDO-expressing tumor cells. Lastly, TPH-1 expressed by mast cells is also necessary for long-term graft tolerance and a suppressive anti-tumor microenvironment[51]. Therefore, more than one tryptophan catabolic enzyme contribute to the establishment of immune tolerance and could be suitable for combinatorial therapies. Thus far, IDO inhibitors, specifically designed for cancer immunotherapy, have been broadly used in preclinical and clinical trials alone or in combination with T cell checkpoint inhibitors[52-54], while systemic inhibition of TDO, although promising, may raise safety concerns[54] (Table 1). Another limitation of cancer trials targeting tryptophan metabolism is that they do not select cancer patients based on assessment of systemic IDO/TDO activities or by analysis of their metabolites in patients’ serum[54]. Nevertheless, combinatorial approaches targeting tryptophan metabolism will continue to deliver novel therapeutic avenues in cancer therapy.
Serine and glycine metabolism are interconnected via the glycine cleavage system, a major metabolic pathway in one-carbon metabolism that provides cofactors for purine and pyrimidine nucleotide biosynthesis for proliferating lymphocytes, cancer cells, and/or fetal tissues[55-57] (Figure 1). Studies from the late 1980s to more recent years, strongly suggest that cancer cells have an increased capacity for de novo serine synthesis via the phosphoglycerate dehydrogenase (PHGDH) pathway. PHGDH oxidases around 10% of 3-phosphogycerate produced during glycolysis by converting it to 3-phosphohydroxypyruvate[58-61]. This compound is then transaminated, forming 3-phosphoserine, and dephosphorylated to yield serine (Figure 1). PHGDH along with other enzymes in the serine biosynthetic pathway were upregulated in highly metastatic breast cancer, a finding associated with overall poor patient survival[62]. Two independent studies published in 2011 reported that the gene encoding PHGDH was recurrently amplified in melanoma and breast cancers and this amplification was not associated with oncogene regulation[15,63]. Apart from PHGDH, serine hydroxymethyl transferase (SHMT), that converts serine to glycine, has also been implicated in tumorigenesis. Two isoforms of SHMT (cytoplasmic SHMT1 and mitochondrial SHMT2) were both described as targets of c-myc oncogene[64], a transcriptional factor abnormally expressed in many tumors that controls transcription of up to 15% of the genes in human cells[65]. Both SHMT1 and SHMT2 were described as downstream effectors of c-myc function and rescued the growth defects of c-myc-null cells[64]. At first glance, the role of serine and glycine metabolism in tumorigenesis may appear disadvantageous as PHGDH diverts metabolites from glycolysis to de novo synthesis of serine followed by conversion to glycine by SHMT (Figure 1). However, both serine and glycine are major sources of methyl groups for the one carbon pool required for a variety of biosynthetic pathways and/or DNA methylation that tumor cells use. Similar to cancer cells, immune cells are also shown to use glycolytic intermediates for serine/glycine biosynthesis with the ultimate goal of synthesizing building materials for cell growth and proliferation[66]. Preclinical studies are underway to test the efficacy of many one carbon metabolic enzymes, including PHGDH[67], as anti-tumor targets (Table 1). Dietary intervention is another strategy to target cancer metabolism and although preclinical studies restricting serine and glycine metabolism showed promising results[68,69], global or systematic interventions need to be carefully examined in the context of the immune system that rely on the same metabolic pathways for proper function.
As early as 1951, Mider[70] described that tumors behaved as “nitrogen traps” where glutamine was the preferred nitrogen donor[70,71]. More reports form the 1980’s, demonstrated that not only cancer cells but also rapidly dividing cells such as lymphocytes, thymocytes, and colonocytes had high glutamine consumption rates[72-74]. Despite the fact that glutamine is a nonessential amino acid, proliferating cells display addiction to glutamine implying that glutamine plays more roles in cell metabolism than simply being a nitrogen donor. As such, glutamine is a conditionally essential amino acid for the proliferating cells as well as critically ill humans[75]. Glutamine provides intermediates for the TCA cycle, restores glutathione to its reduced form suppressing oxidative stress, and maintains mitochondrial membrane integrity thus contributing to the survival of proliferating cells (reviewed by Wise and Thompson)[76]. Tumor suppressors (p53) and oncogenes (c-myc) were shown to regulate glutamine metabolism. Hu et al[77] demonstrated that p53 targeted the mitochondrial isoform of glutaminase [glutaminase 2 (GLS2)], that converts glutamine to glutamate (Figure 1). p53 increased GLS2 expression and led to enhanced mitochondrial respiration and generation of ATP along with reduction in reactive oxygen species due to increased glutathione levels[77]. Likewise, introduction of an inducible c-myc transgene in mouse embryonic fibroblasts led to induction of glutaminase 1 along with lactate dehydrogenase and glutamine transporters, suggesting that c-myc is required to support cellular dependence on glutamine[78]. Cancer “addiction” to glutamine had been explored in cancer therapeutics and three compounds, 6-diazo-5-oxo-L-norleucine (L-DON), azaserine, and acivicin, showed significant activity as glutamine analogs[79]. In early preclinical and clinical studies, these compounds showed promising results against different tumor types by inhibiting ribonucleotide biosynthesis, glutamine oxidation and reducing cell viability[80,81] (Table 1). However, due to the important role of glutamine metabolism for normal tissue physiology, these compounds were discontinued while better tumor-targeting options with less general toxicity are currently considered[76,82].
Branched chain amino acids (BCAAs, leucine, isoleucine, and valine) constitute about 40% of the essential amino acid requirements of healthy individuals[83]. They play an important role in protein synthesis and serve as major nitrogen donors for alanine and glutamine synthesis[84]. Increasing evidence shows that BCAAs, especially leucine, are not merely building blocks necessary to support biosynthetic demands but also nutrient signals regulating the mammalian target of rapamycin (mTOR) pathway[85,86]. By controlling protein translation, cell growth, proliferation, and autophagy, the mTOR pathway is recognized as a critical regulator of cellular function[87]. Immune cells are particularly sensitive to mTOR regulation as mTOR pathway responds to environmental cues and coordinates immune cell differentiation and function, accordingly[88]. For example, inhibition of mTOR pathway in T cells promoted T cell tolerance and the mechanism responsible for the maintenance of tolerance was failure of T cells to upregulate mTOR activity in the presence of metabolic inhibitors including leucine antagonists[89]. In this regard, leucine appears to be an important nutrient signal that is sensed by the immune cells via mTOR pathway and is critical for their proliferation. Leucine supply to mTOR pathway is regulated through BCAA metabolism as shown recently in T cells[90]. The cytoplasmic branched chain aminotransferase (BCATc), that catalyzes leucine transamination, was induced in activated T cells, where it regulated leucine supply to complex 1 of the mTOR pathway. Loss of BCATc expression eliminated cytosolic leucine catabolism leading to upregulation of complex 1 of the mTOR pathway and increased glycolysis[90]. While mTOR pathway is upregulated in many cancer types and mTOR-targeted cancer therapy has been a part of clinical research[91], a direct link between mTOR pathway and leucine/BCAA metabolism in the tumor microenvironment awaits to be explored. Similar to the tryptophan degrading enzymes, BCATc may play an immunosuppressive role in the tumor microenvironment possibly contributing to tumor escape mechanisms. Another mechanism of BCATc function in cancer was explored in glioblastomas[16]. Tönjes et al[16] demonstrated that BCATc and BCAA metabolism are attractive targets for the development of therapeutic approaches to treat glioma patients. The majority of gliomas show mutations in their isocitrate dehydrogenase enzyme 1 (IDH1mut) and although IDH1 mutation status is a powerful prognostic factor, it was insufficient to induce tumors in mice alone. BCATc was overexpressed in normal (wild type) IDH1wt gliomas but not in gliomas with mutated IDH1, demonstrating a connection between IDH1 mutation and BCATc[16]. Additionally, BCATc was identified as a c-myc target in nasopharyngeal carcinoma and BCATc overexpression induced cancer cell proliferation and migration[92]. Although the majority of these studies imply that cancer cells require BCAA metabolism to sustain growth, and overexpression of BCATc results in increased cell proliferation[92,93], the mechanism through which changes in BCAA metabolism affect cancer growth is currently unknown. It is possible that T cells and cancer cells share similar requirements for BCAA catabolism, where mTOR pathway is dependent on leucine regulation. Thus, future use of leucine antagonists or specific inhibitors aimed at BCATc may be suitable for targeted cancer therapies.
Research on amino acid metabolism in cancer cells in the last decades has provided valuable insights on the potential impact of metabolic control and regulation in the tumor microenvironment. Amino acids are no longer regarded solely as building materials but also as nutrient signals that regulate important signaling pathways. A number of amino acid metabolic enzymes are regulated by oncogenes and tumor suppressors and have been explored as targets for cancer therapies. Design and use of inhibitors targeting tryptophan, arginine and/or glutamine metabolism either alone or in combination with anti-tumor drugs has been introduced in clinical trials. However, cancer and immune cells share similar requirements for amino acid metabolic enzymes and often compete for the same nutrients. Therefore, therapeutic interventions in the tumor microenvironment must be cautiously explored to eliminate potential negative impacts on the anti-tumor immunity. Understanding the underlying mechanisms of metabolic interplay between tumor and immune cells will provide new directions to manipulate the tumor microenvironment and unleash the anti-tumor immune response.
The author would like to thank Dr. Wayne Wilson for a critical evaluation of the manuscript.
P- Reviewer: Hall TR, O'Connor TR, Ramana KV, Shao R S- Editor: Tian YL L- Editor: A E- Editor: Lu YJ
| 1. | Macintyre AN, Rathmell JC. Activated lymphocytes as a metabolic model for carcinogenesis. Cancer Metab. 2013;1:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 2. | Marelli-Berg FM, Fu H, Mauro C. Molecular mechanisms of metabolic reprogramming in proliferating cells: implications for T-cell-mediated immunity. Immunology. 2012;136:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844-852. [PubMed] |
| 4. | Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1877] [Cited by in RCA: 1940] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 5. | McNamee EN, Korns Johnson D, Homann D, Clambey ET. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol Res. 2013;55:58-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 182] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 6. | Warburg O. On the origin of cancer cells. Science. 1956;123:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9117] [Cited by in RCA: 9935] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 7. | Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2023] [Cited by in RCA: 2459] [Article Influence: 175.6] [Reference Citation Analysis (0)] |
| 8. | Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. Eur J Immunol. 2003;33:2223-2232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769-777. [PubMed] |
| 10. | Roos D, Loos JA. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation. Exp Cell Res. 1973;77:127-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 138] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12491] [Cited by in RCA: 11817] [Article Influence: 738.6] [Reference Citation Analysis (0)] |
| 12. | Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492-1499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 532] [Cited by in RCA: 520] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 13. | Younes M, Lechago LV, Somoano JR, Mosharaf M, Lechago J. Wide expression of the human erythrocyte glucose transporter Glut1 in human cancers. Cancer Res. 1996;56:1164-1167. [PubMed] |
| 14. | Avramis VI. Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res. 2012;32:2423-2437. [PubMed] |
| 15. | Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 884] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 16. | Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 390] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 17. | Muñoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 296] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 18. | Wu G, Morris SM. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2025] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 19. | Rose WC, Haines WJ, Warner DT. The amino acid requirements of man. V. The rôle of lysine, arginine, and tryptophan. J Biol Chem. 1954;206:421-430. [PubMed] |
| 20. | Barbul A. Arginine: biochemistry, physiology, and therapeutic implications. JPEN J Parenter Enteral Nutr. 1986;10:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 365] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 21. | Dillon BJ, Prieto VG, Curley SA, Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer. 2004;100:826-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 22. | Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Allen MD, Luong P, Hudson C, Leyton J, Delage B, Ghazaly E, Cutts R, Yuan M, Syed N, Lo Nigro C. Prognostic and therapeutic impact of argininosuccinate synthetase 1 control in bladder cancer as monitored longitudinally by PET imaging. Cancer Res. 2014;74:896-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 24. | Sica A, Porta C, Morlacchi S, Banfi S, Strauss L, Rimoldi M, Totaro MG, Riboldi E. Origin and Functions of Tumor-Associated Myeloid Cells (TAMCs). Cancer Microenviron. 2012;5:133-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: opportunities and challenges. Cancer Res Treat. 2013;45:251-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 26. | Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24:302-306. [PubMed] |
| 27. | Bronte V, Serafini P, De Santo C, Marigo I, Tosello V, Mazzoni A, Segal DM, Staib C, Lowel M, Sutter G. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170:270-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 376] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 28. | Bingisser RM, Tilbrook PA, Holt PG, Kees UR. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J Immunol. 1998;160:5729-5734. [PubMed] |
| 29. | Wakkach A, Fournier N, Brun V, Breittmayer JP, Cottrez F, Groux H. Characterization of dendritic cells that induce tolerance and T regulatory 1 cell differentiation in vivo. Immunity. 2003;18:605-617. [PubMed] |
| 30. | Izzo F, Marra P, Beneduce G, Castello G, Vallone P, De Rosa V, Cremona F, Ensor CM, Holtsberg FW, Bomalaski JS. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: results from phase I/II studies. J Clin Oncol. 2004;22:1815-1822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 194] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 31. | Yau T, Cheng PN, Chan P, Chan W, Chen L, Yuen J, Pang R, Fan ST, Poon RT. A phase 1 dose-escalating study of pegylated recombinant human arginase 1 (Peg-rhArg1) in patients with advanced hepatocellular carcinoma. Invest New Drugs. 2013;31:99-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Grohmann U, Bronte V. Control of immune response by amino acid metabolism. Immunol Rev. 2010;236:243-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 232] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 33. | Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1666] [Cited by in RCA: 1770] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 34. | Mellor AL, Munn DH. Tryptophan catabolism and regulation of adaptive immunity. J Immunol. 2003;170:5809-5813. [PubMed] |
| 35. | Mellor AL, Munn D, Chandler P, Keskin D, Johnson T, Marshall B, Jhaver K, Baban B. Tryptophan catabolism and T cell responses. Adv Exp Med Biol. 2003;527:27-35. [PubMed] |
| 36. | Huang L, Mellor AL. Metabolic control of tumour progression and antitumour immunity. Curr Opin Oncol. 2014;26:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Brandacher G, Perathoner A, Ladurner R, Schneeberger S, Obrist P, Winkler C, Werner ER, Werner-Felmayer G, Weiss HG, Göbel G. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144-1151. [PubMed] |
| 38. | Inaba T, Ino K, Kajiyama H, Yamamoto E, Shibata K, Nawa A, Nagasaka T, Akimoto H, Takikawa O, Kikkawa F. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol Oncol. 2009;115:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 39. | Godin-Ethier J, Hanafi LA, Piccirillo CA, Lapointe R. Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin Cancer Res. 2011;17:6985-6991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 318] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 40. | Mellor AL, Munn DH. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation? Immunol Today. 1999;20:469-473. [PubMed] |
| 41. | Polyzos KA, Ovchinnikova O, Berg M, Baumgartner R, Agardh H, Pirault J, Gisterå A, Assinger A, Laguna-Fernandez A, Bäck M. Inhibition of indoleamine 2,3-dioxygenase promotes vascular inflammation and increases atherosclerosis in Apoe-/- mice. Cardiovasc Res. 2015;106:295-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 42. | Swanson KA, Zheng Y, Heidler KM, Mizobuchi T, Wilkes DS. CDllc+ cells modulate pulmonary immune responses by production of indoleamine 2,3-dioxygenase. Am J Respir Cell Mol Biol. 2004;30:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 43. | Sakurai K, Zou JP, Torres NI, Tschetter JR, Kim HS, Shearer GM. Study of the effect of indoleamine 2,3-dioxygenase on murine mixed lymphocyte reactions and skin allograft rejection. Transplant Proc. 2002;34:3271-3273. [PubMed] |
| 44. | Xie FT, Cao JS, Zhao J, Yu Y, Qi F, Dai XC. IDO expressing dendritic cells suppress allograft rejection of small bowel transplantation in mice by expansion of Foxp3(+) regulatory T cells. Transpl Immunol. 2015;33:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 45. | Uyttenhove C, Pilotte L, Théate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1642] [Cited by in RCA: 1742] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 46. | Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101:151-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 282] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 47. | Munn DH, Sharma MD, Hou D, Baban B, Lee JR, Antonia SJ, Messina JL, Chandler P, Koni PA, Mellor AL. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114:280-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 516] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 48. | Ino K, Yamamoto E, Shibata K, Kajiyama H, Yoshida N, Terauchi M, Nawa A, Nagasaka T, Takikawa O, Kikkawa F. Inverse correlation between tumoral indoleamine 2,3-dioxygenase expression and tumor-infiltrating lymphocytes in endometrial cancer: its association with disease progression and survival. Clin Cancer Res. 2008;14:2310-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 49. | Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435-5440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 545] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 50. | Pilotte L, Larrieu P, Stroobant V, Colau D, Dolusic E, Frédérick R, De Plaen E, Uyttenhove C, Wouters J, Masereel B. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci USA. 2012;109:2497-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 473] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 51. | Nowak EC, de Vries VC, Wasiuk A, Ahonen C, Bennett KA, Le Mercier I, Ha DG, Noelle RJ. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J Exp Med. 2012;209:2127-2135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 52. | Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, Mellor AL, Prendergast GC, Munn DH. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792-801. [PubMed] |
| 53. | Meininger D, Zalameda L, Liu Y, Stepan LP, Borges L, McCarter JD, Sutherland CL. Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and discovery of selective IDO1 inhibitors. Biochim Biophys Acta. 2011;1814:1947-1954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 54. | Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer Immunotherapy by Targeting IDO1/TDO and Their Downstream Effectors. Front Immunol. 2014;5:673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 229] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 55. | Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J. 1987;245:609-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 56. | Snell K. Liver enzymes of serine metabolism during neonatal development of the rat. Biochem J. 1980;190:451-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 57. | Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1:169-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 256] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 58. | Snell K. Enzymes of serine metabolism in normal and neoplastic rat tissues. Biochim Biophys Acta. 1985;843:276-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Snell K, Weber G. Enzymic imbalance in serine metabolism in rat hepatomas. Biochem J. 1986;233:617-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 60. | Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 794] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 61. | DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14:285-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 62. | Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 209] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 63. | Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1354] [Cited by in RCA: 1317] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 64. | Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793-5800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 168] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 65. | Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 2590] [Article Influence: 199.2] [Reference Citation Analysis (0)] |
| 66. | Maciolek JA, Pasternak JA, Wilson HL. Metabolism of activated T lymphocytes. Curr Opin Immunol. 2014;27:60-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 67. | Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1038] [Cited by in RCA: 1211] [Article Influence: 100.9] [Reference Citation Analysis (0)] |
| 68. | Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542-546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 615] [Cited by in RCA: 781] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 69. | Tavana O, Gu W. The Hunger Games: p53 regulates metabolism upon serine starvation. Cell Metab. 2013;17:159-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Mider GB. Some aspects of nitrogen and energy metabolism in cancerous subjects: a review. Cancer Res. 1951;11:821-829. [PubMed] |
| 71. | Medina MA, Núñez de Castro I. Glutaminolysis and glycolysis interactions in proliferant cells. Int J Biochem. 1990;22:681-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 72. | Ardawi MS, Newsholme EA. Glutamine metabolism in lymphocytes of the rat. Biochem J. 1983;212:835-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 326] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 73. | Ardawi MS, Newsholme EA. Fuel utilization in colonocytes of the rat. Biochem J. 1985;231:713-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 156] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 74. | Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J. 1985;228:353-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 150] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 75. | Lacey JM, Wilmore DW. Is glutamine a conditionally essential amino acid? Nutr Rev. 1990;48:297-309. [PubMed] |
| 76. | Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1408] [Cited by in RCA: 1343] [Article Influence: 89.5] [Reference Citation Analysis (0)] |
| 77. | Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455-7460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 673] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 78. | Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782-18787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1606] [Cited by in RCA: 1544] [Article Influence: 90.8] [Reference Citation Analysis (0)] |
| 79. | Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol Ther. 1990;46:243-271. [PubMed] |
| 80. | Ovejera AA, Houchens DP, Catane R, Sheridan MA, Muggia FM. Efficacy of 6-diazo-5-oxo-L-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice. Cancer Res. 1979;39:3220-3224. [PubMed] |
| 81. | Griffiths M, Keast D, Patrick G, Crawford M, Palmer TN. The role of glutamine and glucose analogues in metabolic inhibition of human myeloid leukaemia in vitro. Int J Biochem. 1993;25:1749-1755. [PubMed] |
| 82. | Thornburg JM, Nelson KK, Clem BF, Lane AN, Arumugam S, Simmons A, Eaton JW, Telang S, Chesney J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10:R84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 230] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 83. | Hutson SM, Sweatt AJ, Lanoue KF. Branched-chain [corrected] amino acid metabolism: implications for establishing safe intakes. J Nutr. 2005;135:1557S-1564S. [PubMed] |
| 84. | Garber AJ, Karl IE, Kipnis DM. Alanine and glutamine synthesis and release from skeletal muscle. II. The precursor role of amino acids in alanine and glutamine synthesis. J Biol Chem. 1976;251:836-843. [PubMed] |
| 85. | Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1387] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 86. | Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592-E602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 286] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 87. | Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589-3594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1542] [Cited by in RCA: 1729] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 88. | Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 327] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 89. | Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009;183:6095-6101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 218] [Article Influence: 13.6] [Reference Citation Analysis (1)] |
| 90. | Ananieva EA, Patel CH, Drake CH, Powell JD, Hutson SM. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. J Biol Chem. 2014;289:18793-18804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 91. | Advani SH. Targeting mTOR pathway: A new concept in cancer therapy. Indian J Med Paediatr Oncol. 2010;31:132-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 92. | Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W, Liu Z, Li Z, Zeng L, Wang L. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 93. | Yoshikawa R, Yanagi H, Shen CS, Fujiwara Y, Noda M, Yagyu T, Gega M, Oshima T, Yamamura T, Okamura H. ECA39 is a novel distant metastasis-related biomarker in colorectal cancer. World J Gastroenterol. 2006;12:5884-5889. [PubMed] |
| 94. | Glazer ES, Piccirillo M, Albino V, Di Giacomo R, Palaia R, Mastro AA, Beneduce G, Castello G, De Rosa V, Petrillo A. Phase II study of pegylated arginine deiminase for nonresectable and metastatic hepatocellular carcinoma. J Clin Oncol. 2010;28:2220-2226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 95. | Yang TS, Lu SN, Chao Y, Sheen IS, Lin CC, Wang TE, Chen SC, Wang JH, Liao LY, Thomson JA. A randomised phase II study of pegylated arginine deiminase (ADI-PEG 20) in Asian advanced hepatocellular carcinoma patients. Br J Cancer. 2010;103:954-960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 96. | Ott PA, Carvajal RD, Pandit-Taskar N, Jungbluth AA, Hoffman EW, Wu BW, Bomalaski JS, Venhaus R, Pan L, Old LJ. Phase I/II study of pegylated arginine deiminase (ADI-PEG 20) in patients with advanced melanoma. Invest New Drugs. 2013;31:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 97. | Ascierto PA, Scala S, Castello G, Daponte A, Simeone E, Ottaiano A, Beneduce G, De Rosa V, Izzo F, Melucci MT. Pegylated arginine deiminase treatment of patients with metastatic melanoma: results from phase I and II studies. J Clin Oncol. 2005;23:7660-7668. [PubMed] |
| 98. | Glazer ES, Stone EM, Zhu C, Massey KL, Hamir AN, Curley SA. Bioengineered human arginase I with enhanced activity and stability controls hepatocellular and pancreatic carcinoma xenografts. Transl Oncol. 2011;4:138-146. [PubMed] |
| 99. | Jackson E, Minton SE, Ismail-Khan R, Han H, Neuger A, Antonia S, Sullivan D, Soliman HH. A phase I study of 1-methyl-d-tryptophan in combination with docetaxel in metastatic solid tumors. J Clin Oncol. 2012;30 Suppl:abstrTPS2620. |
| 100. | Soliman HH, Antonia S, Sullivan D, Vanahanian N, Link C. Overcoming tumor antigen anergy in human malignancies using the novel indeolamine2,3- dioxygenase(IDO) enzyme inhibitor, 1-methyl-d-tryptophan(1MT). J Clin Oncol. 2009;27:abstr3004. |
| 101. | Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J Exp Med. 2013;210:1389-1402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 450] [Cited by in RCA: 528] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 102. | Nilsson LM, Forshell TZ, Rimpi S, Kreutzer C, Pretsch W, Bornkamm GW, Nilsson JA. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet. 2012;8:e1002573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 103. | Olver IN, Green M, Millward MJ, Bishop JF. Phase II study of acivicin in patients with recurrent high grade astrocytoma. J Clin Neurosci. 1998;5:46-48. [PubMed] |