Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.249
Peer-review started: February 4, 2015
First decision: April 10, 2015
Revised: May 21, 2015
Accepted: June 9, 2015
Article in press: June 11, 2015
Published online: August 26, 2015
Processing time: 204 Days and 23.2 Hours
AIM: To identify alkyl hydroperoxide reductase subunit C (AhpC) homologs in Bacillus subtilis (B. subtilis) and to characterize their structural and biochemical properties. AhpC is responsible for the detoxification of reactive oxygen species in bacteria.
METHODS: Two AhpC homologs (AhpC_H1 and AhpC_H2) were identified by searching the B. subtilis database; these were then cloned and expressed in Escherichia coli. AhpC mutants carrying substitutions of catalytically important Cys residues (C37S, C47S, C166S, C37/47S, C37/166S, C47/166S, and C37/47/166S for AhpC_H1; C52S, C169S, and C52/169S for AhpC_H2) were obtained by site-directed mutagenesis and purified, and their structure-function relationship was analyzed. The B. subtilis ahpC genes were disrupted by the short flanking homology method, and the phenotypes of the resulting AhpC-deficient bacteria were examined.
RESULTS: Comparative characterization of AhpC homologs indicates that AhpC_H1 contains an extra C37, which forms a disulfide bond with the peroxidatic C47, and behaves like an atypical 2-Cys AhpC, while AhpC_H2 functions like a typical 2-Cys AhpC. Tryptic digestion analysis demonstrated the presence of intramolecular Cys37-Cys47 linkage, which could be reduced by thioredoxin, resulting in the association of the dimer into higher-molecular-mass complexes. Peroxidase activity analysis of Cys→Ser mutants indicated that three Cys residues were involved in the catalysis. AhpC_H1 was resistant to inactivation by peroxide substrates, but had lower activity at physiological H2O2 concentrations compared to AhpC_H2, suggesting that in B. subtilis, the enzymes may be physiologically functional at different substrate concentrations. The exposure to organic peroxides induced AhpC_H1 expression, while AhpC_H1-deficient mutants exhibited growth retardation in the stationary phase, suggesting the role of AhpC_H1 as an antioxidant scavenger of lipid hydroperoxides and a stress-response factor in B. subtilis.
CONCLUSION: AhpC_H1, a novel atypical 2-Cys AhpC, is functionally distinct from AhpC_H2, a typical 2-Cys AhpC.
Core tip: Two alkyl hydroperoxide reductase subunit C (AhpC) homologs (AhpC_H1 and AhpC_H2) were identified by searching the Bacillus subtilis database. Sequence homology and phylogenetic analyses revealed that AhpC_H1 is an ortholog of Escherichia coli (E. coli) AhpC, a representative of bacterial AhpC. AhpC_H1 forms dimers consisting of atypical 2-Cys subunits, while AhpC_H2 behaves like a typical 2-Cys AhpC. These AhpC homologs may perform their respective physiological functions at different peroxide levels. Structural and catalytic differences between the enzymes indicate that AhpC_H1 is not an ortholog of E. coli AhpC, but a novel type of atypical 2-Cys AhpC.
-
Citation: Cha MK, Bae YJ, Kim KJ, Park BJ, Kim IH. Characterization of two alkyl hydroperoxide reductase C homologs alkyl hydroperoxide reductase C_H1 and alkyl hydroperoxide reductase C_H2 in
Bacillus subtilis . World J Biol Chem 2015; 6(3): 249-264 - URL: https://www.wjgnet.com/1949-8454/full/v6/i3/249.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.249
Peroxiredoxins (Prxs) are versatile and predominantly thiol-dependent peroxidases (EC 1.11.1.15) that detoxify hydrogen peroxide and various hydroperoxides[1,2]. They are ubiquitously expressed, with multiple isoforms present in most organisms (e.g., three isoforms in Escherichia coli, five in Saccharomyces cerevisiae, six in Homo sapiens, and nine in Arabidopsis thaliana)[3-13]. Prxs exhibit high reactivity with hydrogen peroxide and organic hydroperoxides and are involved in peroxide detoxification and signal transduction[14-20].
All Prxs share a common catalytic mechanism, in which an active-site cysteine (the peroxidatic cysteine) is oxidized to a sulfenic acid by a peroxide substrate, forming a disulfide linkage with a second resolving Cys, which is reduced back by the thioredoxin system consisting of thioredoxin 1 (Trx1), Trx reductase, and NADPH[1,2]. According to the number of functional Cys residues, Prxs are divided into two families: 1-Cys Prxs and 2-Cys Prxs[11,15].
The 2-Cys family contains typical and atypical groups of enzymes[21]. The disulfide bridge formed in typical 2-Cys Prxs after hydroperoxide reduction is intermolecular, whereas in atypical Prxs, it is intramolecular[22]. The founding members of the 2-Cys Prx family, the Prx1/alkyl hydroperoxide reductase C (AhpC) subfamily, including human Prxs I-IV and bacterial AhpC proteins are well-studied and broadly distributed enzymes that form inter-subunit disulfide bonds during catalysis. The amino acid sequence and physiological function of this type of thioredoxin-dependent peroxidases (thiol-specific antioxidants) were first determined in Saccharomyces cerevisiae[23], and later in humans[24] and E. coli[3]. The members of these subfamilies are typically dimeric and form intra-subunit disulfide bonds. Another subfamily (the BCP/PrxQ group) is predominantly bacterial, although present in some eukaryotes and plants, but is absent in animals[6,12]. Most enzymes of the BCP/PrxQ group are reportedly monomeric[25], but there are two examples of dimeric proteins containing an A-type dimer (the TPx and Prx5 subfamilies). Current information suggests that BCP-PrxQ subfamily members can function as either atypical 2-Cys (α-group) or 1-Cys Prxs[26].
Moreover, there are also high molecular weight multimeric enzymes[27]. One atypical 2-Cys Prx exists as a hexamer in oxidized conditions, but dissociates to dimers upon reduction[28]; the presence of the peroxidatic Cys is critical for hexamer formation, whereas substitution of the resolving Cys does not affect oligomerization[29]. By analogy with the dimer-decamer transition of the typical 2-Cys Prx previously reported[27], the dimer-hexamer transition in atypical Prx displays a functional switch, which could be involved in signaling[28,30].
All aerobic bacteria have evolved a variety of antioxidant enzymes such as superoxide dismutases, catalases, and Prxs to cope with damaging endogenous reactive oxygen species (ROS) generated by the aerobic respiratory chain and metal-catalyzed oxidative reactions[31]. E. coli contains three Prxs: AhpC (Prx1/AhpC subfamily)[9], TPx (p20)[4,5], and BCP (BCP/PrxQ subfamily)[6,25].
Gene sequencing and phylogenetic studies have revealed that B. subtilis encodes four putative Prx enzymes with two conserved Cys residues, suggesting that they belong to different subgroups of the 2-Cys Prx family: there are two genes encoding putative AhpC-like proteins (AhpC_H1 and AhpC_H2), and two encoding TPx and BCP, respectively. AhpC_H1 has an additional N-terminal Cys residues at positions 37 (Cys37) besides two conserved Cys47 and Cys166, while ahpC_H2 possesses two conserved Cys residues (Cys52 and Cys169).
The present comparative study was designed to characterize two AhpC homologs and reveal the biological significance of the existence of two homologous AhpC enzymes in B. subtilis. This paper presents the first report of a detailed structure-functional analysis of the bacillus 2-Cys AhpC homologs AhpC_H1 and AhpC_H2.
Bacterial culture media were purchased from Difco Laboratories (Detroit, MI, United States). Restriction enzymes and DNA ligase were obtained from Promega (Madison, WI, United States). Acrylamide-Bis (40% solution) was purchased from Bio-Rad (Hercules, CA, United States). Sodium dodecyl sulfate (SDS), ultrapure glycine, EDTA, dithiothreitol (DTT), Tris base, NADPH, ampicillin powder, isopropyl-b-D-thiogalactopyranoside (IPTG), H2O2, tert-Butyl hydroperoxide (t-BOOH) and cumene hydroperoxide (CMOOH) were from Sigma Aldrich (St. Louis, MO, United States). Iodoacetamide (IAA), N-ethyl maleimide (NEM), tris (2-carboxyethyl) phosphine (TCEP), 4-acetamide-4′-maleimidylstilbene-2,2′-disulfonate (AMS), and sulfhydryl-reactive DyLight 405 maleimide were purchased from Thermo Scientific, Pierce (Rockford, IL, United States).
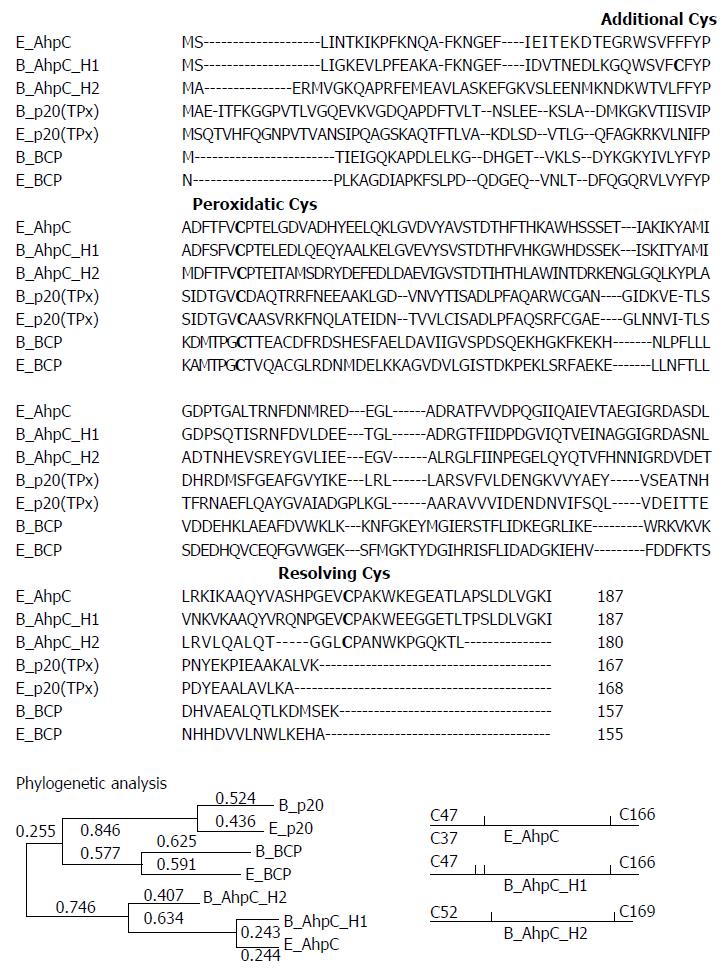
The NCBI BLAST tool (http://www.ncbi.nlm.nih.gov) was used to search for Prx amino acid homologs in the updated GenBank/EMBL and Swiss-Prot databases. Multiple sequence alignments of B. subtilis and E. coli Prxs were performed using the ClustalW 2.1 program. Phylogenetic and molecular evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) version 6. A phylogenetic tree was generated by using the Maximum Likelihood method based on the JTT matrix-based model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (next to the branches).
B. subtilis Cu1065 (subtilis strain 168 attSPβ trpC2, a derivative of B. subtilis subsp. subtilis str. 168) and E. coli XL1-blue and BL21 (DE3) strains were grown in Luria Broth (LB) medium under vigorous agitation at 37 °C. Bacterial growth was monitored by the absorbance at 600 nm (OD600), and exponentially growing B. subtilis cultures were exposed to different stress conditions for 30 min.
The entire coding regions of B. subtilis genes encoding AhpC_H1, AhpC_H2, B_BCP, and B_TPx were amplified by PCR using the High Expand Fidelity kit (Roche Life Science, United States) and primers listed in Table 1. The entire coding region of the E. coli ahpC gene was amplified by PCR using the following primers: forward, 5′-GGG ATC CCA TAT GTC CTT AAT TAA CAC-3′; reverse, 5′-CCT CGA GTT AGA TTT TAC CAA CCA GGT-3′. To obtain Cys→Ser substitutions in AhpC_H1 and AhpC_H2, point mutations in the ahpC_H1 and ahpC_H2 genes were generated by site-directed mutagenesis by using complementary primers (Table 1). Gel-purified ahpC genes were digested with NdeI and XhoI, ligated into the pET-21a(+) expression vector by using T4 DNA ligase (Promega), and used to transform competent E. coli XL1-blue. Bacteria were selected on LB agar containing ampicillin (100 μg/mL), and purified plasmids were used for the sequencing of the cloned coding region by automated DNA sequencing.
| Sequence of primer for point mutation | |
| AhpC_H1 | |
| C37S F | AGCGTATTCTCTTTCTACCCA |
| C37S R | TGGGTAGAAAGAGAATACGCT |
| C47S F | TTCTCTTTCGTATCTCCAACTGAGCTT |
| C47S R | AAGCTCAGTTGGAGATACGAAAGAGAA |
| C166S F | CCAGGTGAAGTTTCTCCGGCTAAATGG |
| C166S R | CCATTTAGCCGGAGAAACTTCACCTGG |
| AhpC_H2 | |
| C52S F | TTCACTTTTGTTTCTCCGACAGAAATT |
| C52S R | AATTTCTGTCGGAGAAACAAAAGTGAA |
| C169S F | ACTGGCGGACTCTCTCCGGCTAACTGG |
| C169S R | CCAGTTAGCCGGAGAGAGTCCGCCAGT |
| Sequence of primer for protein expression | |
| AhpC_H1 | |
| Forward | CCGCATATGTCTTTAATCGGTAAAGAAGATC |
| Reverse | CCGGTCGACTTAGATTTTACCTACTAGATCAAGG |
| AhpC_H2 | |
| Forward | CCGCATATGGCAGAACGTATGGTAGGTAA |
| Reverse | CCGGTCGACTTAAAGTGTTTTTTGGCCTGGTTT |
| B_BCP | |
| Forward | CCGCATATGACTATAGAAATCGGACAAAAAC |
| Reverse | CCGGTCGACTTACTTTTCAGACATTTTGAGG |
| B_TPx | |
| Forward | CCGCATATGGCTGAAATTACATTCAAAGGC |
| Reverse | CCGGTCGACTTACTTTCTAATGCAGCAGC |
The plasmids containing the confirmed sequences were used to transform E. coli BL21 (DE3) cells, which were then cultured at 37 °C overnight in 100 μg/mL ampicillin-containing LB. For protein expression, bacteria were diluted 1:250 in fresh medium, grown until OD600 = 0.4, and induced with 0.5 mmol/L isopropyl-L-D-thiogalactose (IPTG) for 4 h. The soluble recombinant proteins were produced in E. coli. Cells were harvested by centrifugation, suspended in 50 mmol/L Tris-HCl (pH 7.4) containing 2 mmol/L phenylmethylsulfonyl fluoride (PMSF) and 1 mmol/L EDTA, disrupted by sonication, and centrifuged at 16000 × g for 30 min. The clarified supernatant was subjected to ammonium sulfate fractionation, size exclusion chromatography, and anion exchange chromatography. Size exclusion chromatography was performed using a Pharmacia chromatography system (Pharmacia Biotech). Protein solution was loaded onto a Sephacryl S-200 column equilibrated with 200 mmol/L NaCl-containing Tris-HCl, pH 8.0, at a flow rate of 0.2 mL/min at 4 °C, and protein elution was monitored by the absorbance at 280 nm. Anion exchange chromatography was performed using DEAE-cellulose DE52 equilibrated with Tris-HCl, pH 8.0; protein was eluted with a linear salt gradient (0-500 mmol/L NaCl) in Tris-HCl, pH 8.0. Protein concentration was determined using the Bradford protein assay kit (Bio-Rad, United States).
The B. subtilis Prx-encoding genes were disrupted by the short flanking homology method[32]. The 5′ and 3′ regions of the prx genes were amplified using primers designed to obtain 0.2-kb DNA fragments (Table 2). A kanamycin-resistance determinant used as a selection marker for ΔPrx strains was amplified from the pDG780 vector using specific primers (forward, 5′-CAG CGA ACC ATT TGA GGT GAT AG-3′ and reverse, 5′-CGA GCG CCT ACG AGG AAT TTG TAT-3′). The marker was flanked by the two 0.2-kb terminal regions of the Prx-encoding genes using two-step fusion PCR. The first step was performed by fusing the 5′-terminal Prx gene fragment to the kanamycin-resistance gene. The amplified product was gel-purified and used as a template for the second round of PCR to fuse the 3′ terminal Prx gene fragment. The final PCR product was gel-purified and used for the disruption of each Prx-encoding gene in B. subtilis Cu1065. The null mutants were selected on 20 μg/mL kanamycin-containing LB agar, and chromosomal DNA was isolated and analyzed by PCR to confirm the replacement of the target gene.
| Prx null mutants | Primer names | Sequences of paired degenerated primers (5’→3’) |
| ΔAhpC_H1 | H1_Up F | CCTTATTTCACAGATAAGCTCCA |
| Kan/H1 R’ | CCTATCACCTCAAATGGTTCGCTG AATGTATATTCCTCCTAAAAATGTAT | |
| Kan/H1 F’ | CGAGCGCCTACGAGGAATTTGTATGGAGTGCATTCAATTGGTACTTG | |
| H1_Down R | ACGTGCAGTGTAGATCGCTGC | |
| ΔAhpC_H2 | H2_Up F | AAGCTGCGGATTTGAGTTGTCC |
| Kan/H2 R’ | CCTATCACCTCAAATGGTTCGCTGATGTATCCCTCCAATTTATTGTTTG | |
| Kan/H2 F’ | CGAGCGCCTACGAGGAATTTGTAT TTCTTTCCAAGAACGAAAAGCGG | |
| H2_Down R | ATCTAAAATTGAGGAAAAACATCAA | |
| ΔB_BCP | BCP_Up F | AATGGTACTGTGATCACTTCGTTT |
| Kan/BCP R’ | CCTATCACCTCAAATGGTTCGCTG TACGTTACCTCCGGATGTTTTTTT | |
| Kan/BCP F’ | CGAGCGCCTACGAGGAATTTGTAT ATCTCTATGAGCCTATGCTTACTT | |
| BCP_Down R | AGTAGTCGATATGTGCATGCATT | |
| ΔB_TPx | TPx_Up F | TTTTTCGCTTTGGACATGCTAT |
| Kan/TPx R’ | CCTATCACCTCAAATGGTTCGCTGTATAATTCCTCCCTTTTGTATGTAT | |
| Kan/TPx F’ | CGA GCGCCTACGAGGAATTTGTAT GCAGGGAAAAAAGCTCCAGGC | |
| TPx_Down R | CGTCAGCTTTGAGCTCAAACG |
Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in 14% acrylamide gels. In reducing conditions, proteins were dissolved in loading buffer containing 100 mmol/L DTT and boiled for 3 min prior to loading; in non-reducing conditions, proteins were loaded in DTT-free buffer without boiling. After electrophoresis, gels were stained with Coomassie Brilliant Blue (Sigma-Aldrich) or processed for Western blotting. For this, PAGE-separated proteins were transferred to nitrocellulose membranes (Bio-Rad), blocked with 5% (w/v) skim milk in PBS containing 0.1% Tween 20 (PBST) at room temperature for 2 h, and probed with a manufactured rabbit polyclonal anti-AhpC_H1 (1:3000) antibodies (Young In Frontier, Seoul, South Korea) for 3 h. After three washes with PBST, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at room temperature, washed three times with PBST, and developed using the TMB immunoblotting detection system (Kem-En-Tec).
Further protein characterization was performed by 2D electrophoresis. Briefly, the B. subtilis protein extract was loaded onto 7-cm immobilized pH gradient (IPG) strips (Bio-Rad, United States) with 4-7 pH gradient (left to right), and the first-dimension separation was conducted; in the second-dimension separation, 16% reducing SDS-PAGE gels were used. For protein detection, the gels were stained with Coomassie Brilliant Blue G-250 or subjected to immunoblotting.
AhpC_H1 was purified from B. subtilis and a 25-μg sample in phosphate buffer, pH 7.4, containing 150 mmol/L KCl was loaded onto a ZORBAX® Pro 10/300 GF450 column equilibrated in phosphate buffer using an automatic injector (SIL-20AC, Shimadzu, Japan). Chromatography was performed on a Shimadzu high-performance liquid chromatography (HPLC) system at the flow rate of 0.5 mL/min at room temperature, and the absorbance was monitored using a diode array detector (SPD M10A, Simadzu). The column was calibrated using protein molecular mass standards under the same conditions, and a calibration curve was obtained by plotting the Ve/Vo ratio (where Ve is the elution volume and Vo is the void volume) vs the logarithms of the molecular masses.
For the reduction of protein disulfide groups, protein was reacted with chemical reducing agents such as DTT or TCEP or subjected to enzymatic reduction with E. coli Trx1, Trx1 reductase, and NADPH. After 30-min reaction at 30 °C, thiol-specific modification reagent 4-acetamide-4′-maleimidylstilbene-2,2′-disulfonate (AMS) was added for another 30 min at 30 °C.
For HPLC analysis of Cys-containing peptides, sulfhydryl-reactive DyLight 405 maleimide (excitation and emission wavelengths 557 and 572 nm, respectively) was used to fluorescently label Cys residues in the intact or TCEP-denatured AhpC_H1. Proteins digested with trypsin treated with L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK, an inhibitor of contaminating chymotryptic activity) in the presence of 2 mol/L urea were analyzed by reversed-phase HPLC. Briefly, proteins were incubated in 50 mmol/L NH4CO3 containing 2 mol/L urea and trypsin at the ratio 1:20 (w/w, trypsin/protein) at 30 °C for 12 h, and then loaded into a C18 HPLC column (2.1 mm × 25 cm). Peptides were eluted with a 5%-100% gradient of acetonitrile containing 0.2% trifluoroacetic acid (TFA) at the flow rate of 1 mL/min for 120 min. The elution of fluorescent peptides was monitored at 572 nm using a fluorescence detector (RF-10AXL, Shimadzu).
Enzymatic assays with the Trx system (NADPH, E. coli Trx reductase, and Trx1) and peroxides as substrates were conducted to detect peroxidase activity of the purified AhpC proteins by the decrease of absorbance at 340 nm. Briefly, the enzymes were added to the reaction mixture containing 0.14 mmol/L NADPH, 100 nmol/L E. coli Trx reductase, 5 μmol/L E. coli Trx1, 50 mmol/L Tris-HCl (pH 7.4), 0.1 mmol/L EDTA, and hydroperoxide substrates (H2O2 and t-butyl hydroperoxide) at room temperature and the absorbance was continuously monitored at 340 nm using a diode array spectrophotometer (HP 7452A). Stock solutions of t-butyl hydroperoxide were prepared in DMSO prior to the reaction.
Four prx-homologous sequences in B. subtilis genome were identified by searching the Data Release R16.1 (the SubtiList World-Wide Web Server, http://genolist.pasteur.fr/SubtiList/). Figure 1 shows multiple sequence alignment of three E. coli Prxs (E_AhpC, E_BCP, and E_TPx) and four B. subtilis Prxs using the ClustalW 2.1 program, and a resulting phylogenetic tree. The hallmark of Prxs is a conserved peroxidatic Cys residue located near the N-terminus. The analysis of multiple protein alignment revealed that all B. subtilis Prxs possessed a highly conserved peroxidatic Cys, suggesting that these enzymes belong to the Prx family. The cladogram reveals that, with the exception of B_AhpC_H2, all Bacillus Prxs displayed sequence similarity with the corresponding E. coli Prxs and were therefore named after the respective E. coli enzymes. Thus, E_TPx and B_TPx showed 41.5% identity in 164 overlapping amino acids and E_BCP and B_BCP demonstrated 39.7% identity in 151 overlapping amino acids. E_AhpC and B_AhpC_H1 had 65.24% identity in a 187-amino acid region, while E_AhpC and B_AhpC_H2 had 38.4% identity in a 159-amino acid sequence. Between B_AhpC_H1 and B_AhpC_H2, there was 37.0% identity in a 181-amino acid sequence.
Sequence similarity analysis indicates that AhpC_H1 is a homologue of E. coli AhpC, a representative of the AhpC/Prx1 subfamily. Sequence alignment of AhpC homologs revealed two conserved Cys residues, catalytic and resolving, in the N-terminal and C-terminal regions, respectively, suggesting that B_AhpC_H2 also belongs to the AhpC/Prx1 family, although its similarity to E_AhpC (38.4%) is much lower than that of B_AhpC_H1 to E_AhpC (65.2%).
Taken together, these data suggest that B. subtilis encodes one member of the BCP/PrxQ, one member of the bacterial TPx, and two members of the AhpC/Prx1 subfamilies.
AhpC_H2 and AhpC_H1 each contain a pair of conserved Cys residues (Cys52/Cys169 and Cys47/Cys166, respectively); in addition, AhpC_H1 possesses an extra Cys37 (Figure 1). The members of the AhpC/Prx1 subfamily (including E. coli AhpC) are typical 2-Cys Prxs, which exist as homodimers formed via an intermolecular disulfide bond between two subunits.
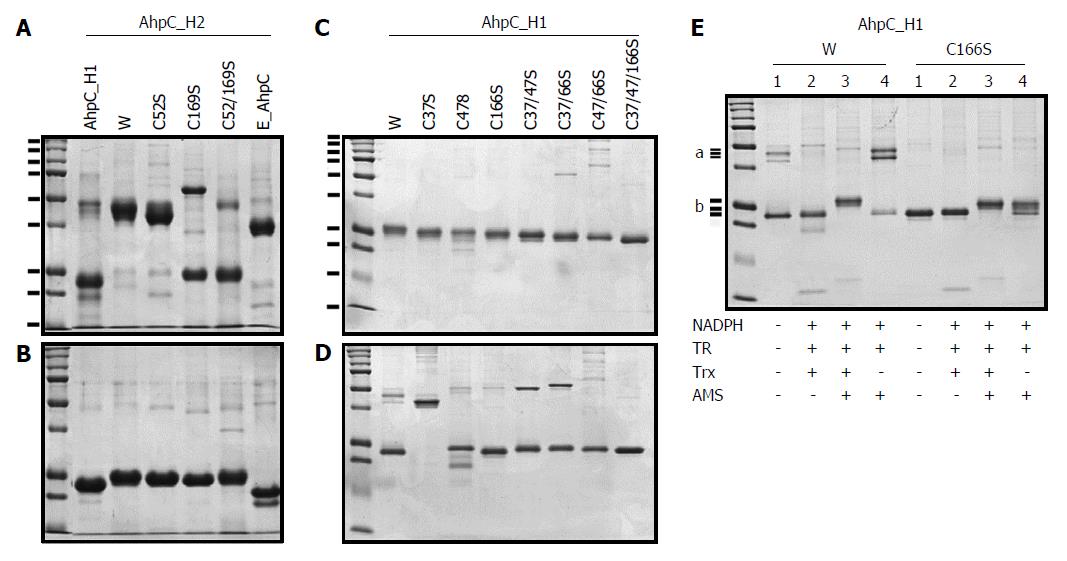
To examine the possible formation of a disulfide bond between the two conserved Cys residues in B. subtilis AhpC enzymes, Cys was replaced with Ser by site-directed mutagenesis and the purified mutant AhpC_H1 and AhpC_H2 proteins were analyzed by their migration patterns in SDS-PAGE.
In non-reducing SDS-PAGE gels, wild-type AhpC_H2 and E_AhpC appeared as single bands with the molecular masses about 48 kDa and about 40 kDa, respectively (Figure 2A), whereas single approximately 24.5-kDa and 22-kDa bands, respectively, were observed in reducing gels (Figure 2B), suggesting that in natural conditions, these enzyme exist as dimers likely held together by disulfide linkages. However, most AhpC_H1 appeared as a single band with a molecular mass of the monomer (approximately 23-kDa) under both non-reducing and reducing conditions (Figure 2A and B, respectively), suggesting the presence of monomers even in the oxidized state.
The migration pattern of AhpC_H2 mutant proteins (C52S, C169S, and C52/169S) in non-reducing SDS-PAGE gels were quite different from that of the wild-type counterpart (Figure 2A). C169S mostly existed in a monomeric form, presenting the evidence of intermolecular disulfide linkage between the two identical subunits. In non-reducing conditions, the migration of the wild-type dimer appeared to be different from that of the mutant C52S and C169S dimers, suggesting the formation of inter-subunit disulfide bonds characteristic for typical 2-Cys Prxs such as E_AhpC.
In contrast to AhpC_H2 and E_AhpC, most of the AhpC_H1 proteins existed as monomers even in the oxidized state. The migration patterns of AhpC_H1 Cys→Ser mutants (C37S, C47S, C166S, C37/47S, C37/166S, C47/166S, and C37/47/166S) indicated that the distance of protein migration in reducing conditions depended on the number of substituted Cys residues (Figure 2C), thus confirming that all three Cys residues are susceptible to AMS modification. In non-reducing conditions, the C37S protein migrated as a dimer (Figure 2D). Considering that, in non-reducing conditions, the wild-type protein was predominantly monomeric (lane W in Figure 2D), dimerization of the C37S mutant suggests that Cys37 could participate in the formation of an intramolecular disulfide bond. In non-reducing gels, most of the proteins carrying C47S and C166S substitutions were detected at the positions slightly higher than that of the wild-type protein; however, a single C166S mutant migrated similarly to the wild-type protein, suggesting the existence of intramolecular disulfide linkage between Cys37 and Cys47 in both proteins.
Taken together, these results indicate a possibility that AhpC_H1 is an atypical 2-Cys enzyme, which forms the intramolecular disulfide linkage between Cys37 and Cys47 rather than between Cys47 and Cys166. To examine the presence of Cys37-Cys47 intramolecular disulfide bond, the wild-type AhpC_H1 and its C166S mutant were reduced using the Trx system, and analyzed by non-reducing SDS-PAGE. As shown in Figure 2E, Trx-treated C166S occupied a slightly higher position than the wild-type protein; AMS modification shifted the position of both proteins toward higher molecular weight (Figure 2E, lane 3). These results reveal that the disulfide bond can be specifically reduced by the Trx system. Furthermore, AMS modification of the wild-type AhpC_H1 in the absence of Trx1 (Figure 2E, lane 4) or TCEP (data not shown) resulted in the formation of two different dimers, but did not cause dimerization of C166S, which confirms the presence of free Cys166 in AhpC_H1. Other sulfhydryl-modifying agents such as NEM and IAA also induced AhpC_H1 dimerization (data not shown). Overall, the effect of thiol-specific modification of free Cys166 on the tertiary structure of AhpC_H1 is worth noting, although such modification may not occur in vivo.
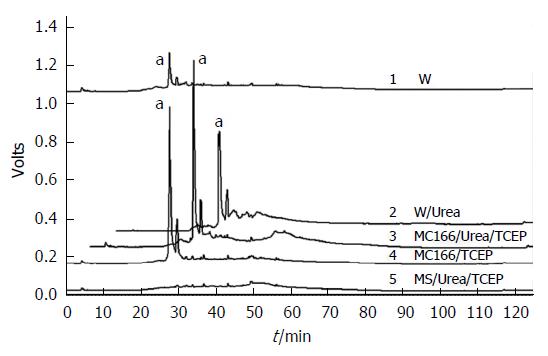
Finally, we directly demonstrated the presence of free Cys166 residue in the oxidized AhpC_H1. AhpC_H1 was modified with fluorescent sulfhydryl-reactive maleimide and digested with trypsin in the presence or absence of 2 mol/L urea. The reversed-phase HPLC chromatogram of trypsin-digested native AhpC_H1 demonstrates a unique peak of a fluorescent peptide (a in Figure 3, W), which was significantly increased in the trypsin-digested denatured (2 mol/L urea-treated) AhpC_H1 (a in Figure 3, W/Urea). The same peak appeared in the trypsin-digested denatured and native C37/47S double mutant (Figure 3, MC166/Urea/TCEP and M166/TCEP, respectively), which has only one free Cys166 residue. The triple mutant C37/47/166S (MS) did not produce any fluorescent peak. Taken together, these data suggest that the peak represents the peptide with free Cys166 residue, indicating the existence of free Cys166 and the intramolecular disulfide linkage between Cys37 and Cys47.
The modification of Trx-preincubated AhpC_H1 and AhpC_H2 with AMS, a thiol-specific reagent, resulted in the shift of the monomer to a slightly higher position (Figure 4A and B, respectively), suggesting that the Trx system (Trx1, Trx reductase, and NADPH) is able to reduce protein disulfide bonds in both AhpC_H1 and AhpC_H2.
Pre-incubation of AhpC_H1 with the Trx system containing increasing concentrations of E. coli Trx1 resulted in an increase of the AMS-modified AhpC_H1 in a Trx 1 concentration-dependent manner (Figure 4C), indicating that the intramolecular disulfide bond can be reduced by the Trx treatment. The dimerization of AMS-modified AhpC_H1 was also confirmed (Figure 4C). Similarly, the dimeric AhpC_H2 was gradually converted to a corresponding AMS-modified monomer in a Trx1 dose-dependent manner (Figure 4D).
To ensure functional activity of the Trx1 system to reduce disulfide bonds, we tested the Trx system in the reaction with Bacillus B_BCP and B_TPx as well as with Trx-supported peroxidases from other species such as E. coli AhpC and human Prx1 and Prx2 (Figure 4E-I, respectively). The results indicate that the reduced forms of all Prxs increased in a Trx1 concentration-dependent manner, indicating that all the tested enzymes were sensitive to the Trx system. More importantly, in non-reducing conditions, AhpC_H2, but not AhpC_H1, demonstrated a pattern very similar to that of E. coli AhpC (E_AhpC) (Figure 4G), suggesting that AhpC_H2 is structurally similar to E_AhpC in that they exist as intermolecular dimers linked by a disulfide bond, although amino acid sequence similarity between AhpC_H1 and E_AhpC is much higher than that between AhpC_H2 and E_AhpC (Figure 1). Taken together, these results indicate the presence of a redox-sensitive disulfide linkage between Cys37 and Cys47 of AhpC_H1, characterizing it as a member of the atypical 2-Cys subfamily.
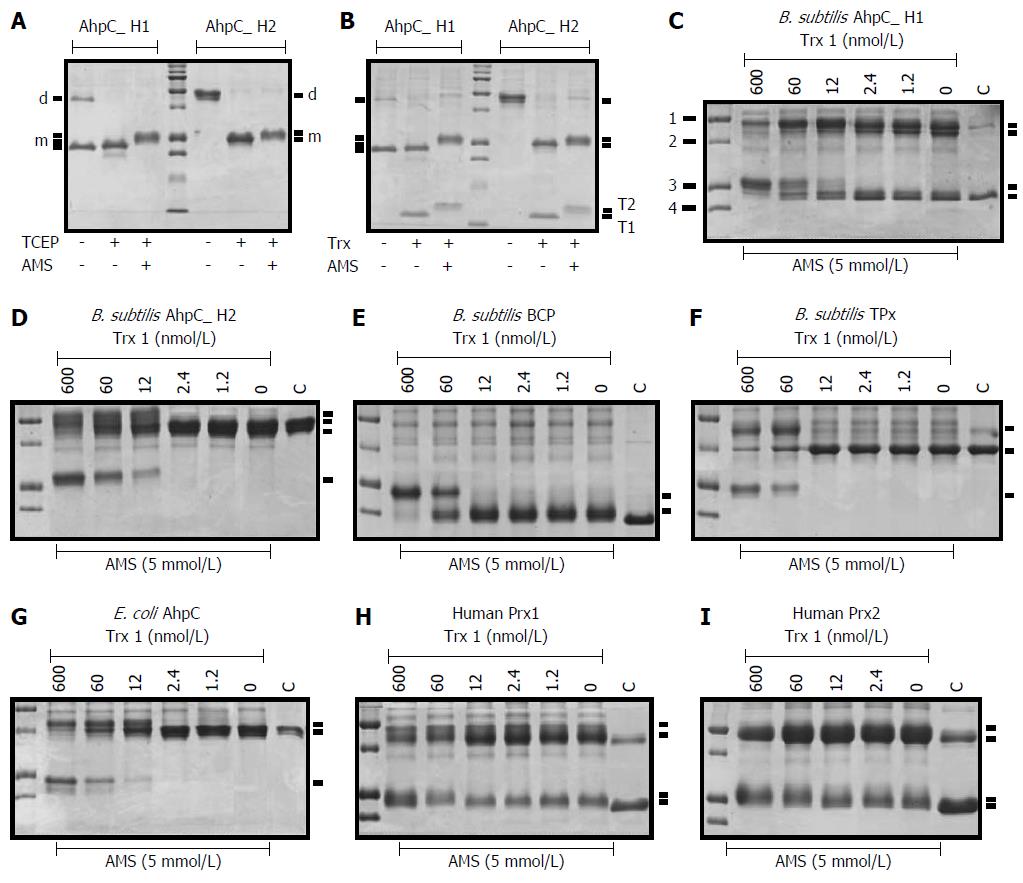
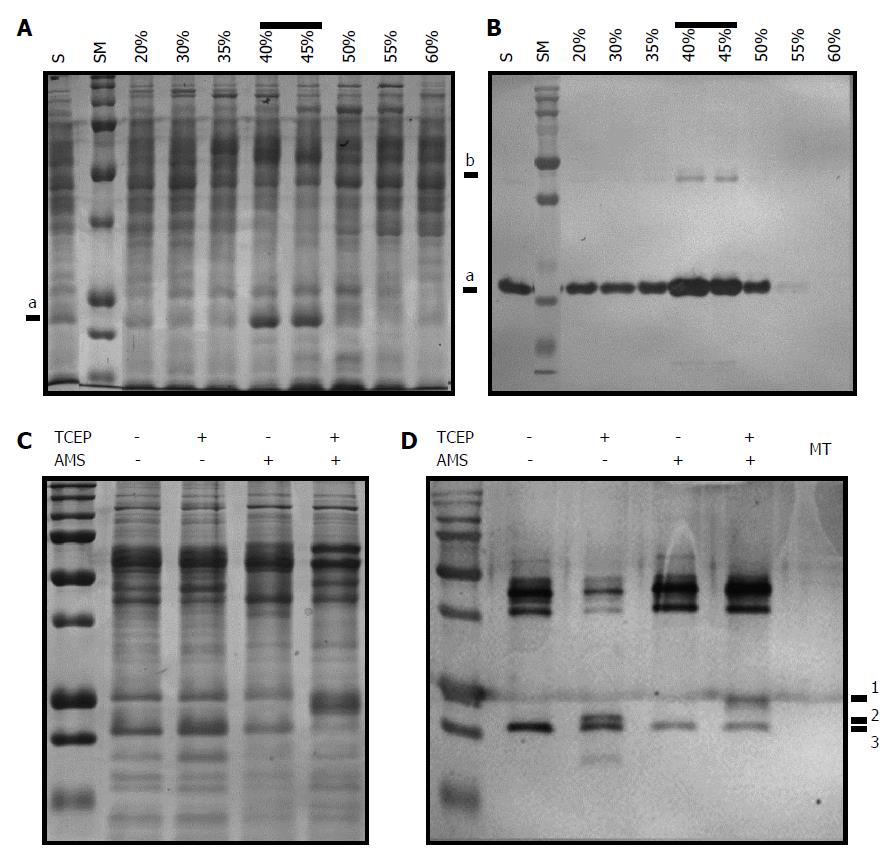
In the previous experiments, we demonstrated that, unlike AhpC_H2, AhpC_H1 formed the intramolecular Cys37-Cys47 disulfide bond. However, it may be possible that it is characteristic only for the recombinant protein and not for the native B. subtilis AhpC_H1. To test this possibility, we examined the presence of the disulfide linkage in the native B. subtilis-purified AhpC_H1. Crude protein extract was fractionated using ammonium sulfate gradient (20% to 60%). Figure 5A shows that a major protein band of approximately 22-kDa was detected in the 40% and 45% fractions. Western blot analysis with the AhpC_H1 antibody demonstrated that the predominant band was AhpC_H1 (Figure 5B). In the 40% fraction, AhpC_H1 was reduced with TCEP, and the resulting product was specifically modified with AMS (Figure 5C). The analysis of the modified proteins by non-reducing SDS-PAGE and western blotting (Figure 5D) revealed that AMS could modify only TCEP-reduced AhpC_H1 (Figure 5D). Taken together, these results demonstrate that B. subtilis expresses a significant amount of oxidized AhpC_H1 with the Cys37-Cys47 intramolecular disulfide linkage.
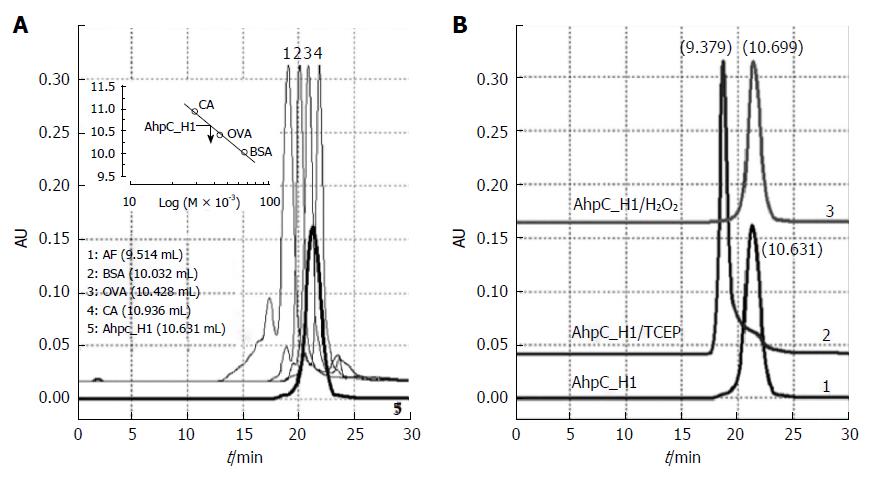
Previous studies have demonstrated that the Cys37-Cys47 bond in AhpC_H1 is redox-sensitive. AhpC_H1 molecular mass was determined by HPLC. Figure 6A demonstrates that AhpC_H1 was eluted between ovalbumin (OVA, 44 kDa) and carbonic anhydrase (CA, 29 kDa); therefore, the apparent AhpC_H1 molecular mass was estimated to be approximately 38.6 kDa. Given that a theoretical AhpC_H1 molecular mass is approximately 20.5 kDa, this result indicates that AhpC_H1 exists as a homodimer. Considering that AhpC_H1 mostly appears as a 22-kDa band in non-reducing SDS-PAGE gels, this result suggests that the dimer is likely formed via non-covalent interactions. On the other hand, AhpC_H2 was eluted at 9.116 mL (data not shown). Given that the elution volume of apoferritin (AF, 443 kDa) was 9.514 mL, it suggests that AhpC_H2 oligomer has a higher molecular mass compared to AhpC_H1.
It has been reported that in some cases, Prx oligomerization is reversible and depends on the redox status of Cys residues[27-30]. To examine AhpC_H1 redox-dependent oligomerization, the molecular mass of TCEP-reduced AhpC_H1 was determined by HPLC. Figure 6B shows that the elution volume of reduced AhpC_H1 was 9.379 mL (Figure 6B, AhpC_H1/TCEP), which, considering the elution volume of 443-kDA apoferritin (9.514 mL) indicates that the apparent molecular mass of reduced AhpC_H1 was above 443 kDa. AhpC_H1 incubation with H2O2 did not significantly affect the molecular mass (Figure 6B, AhpC_H1/H2O2).
Taken together, these results suggest that AhpC_H2 exists as a high molecular mass oligomer, while AhpC_H1 is a homodimer. Oxidized AhpC_H1 monomers are dimerized via noncovalent interactions; however, after reduction, the dimers tend to assemble into high molecular mass oligomers.
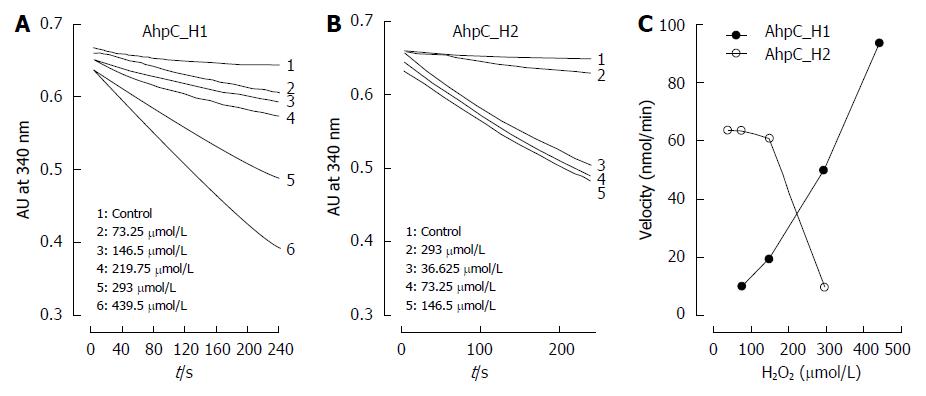
Because some Prxs, especially of eukaryotic origin, are known to be inactivated by high substrate concentrations[1,2], we tested the response of AhpC_H1 and AhpC_H2 to elevated concentrations of their preferred substrate H2O2 by using the Trx1 system. Peroxidase activity was continuously monitored by the absorbance at 340 nm indicating NADPH concentration. Figure 7A and B shows a time-dependent decrease in NADPH concentration due to the peroxidase activity of AhpC_H1 and AhpC_H2, respectively. At low H2O2 concentrations (< 100 μmol/L), AhpC_H2 was more active than AhpC_H1, but was significantly inhibited by higher H2O2 concentrations, whereas AhpC_H1 was resistant even to high H2O2 levels, which completely inhibited AhpC_H2 activity (Figure 7C), indicating that AhpC_H1 is a much more robust enzyme compared to AhpC_H2.
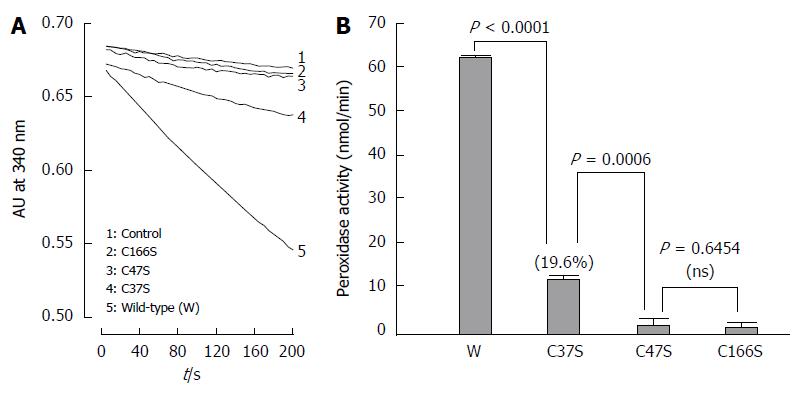
Figure 8 shows Trx-dependent peroxidase activity of the wild-type AhpC_H1 and its C37S, C47S, C166S mutants toward t-butyl hydroperoxide substrate. Compared to almost inactive C47S and C166S mutants, the C37S protein showed a substantial enzymatic activity (approximately 19.6% of that of the wild-type protein), indicating that, in addition to Cys47 and Cys166, Cys37 is also required for the peroxidation activity of AhpC_H1.
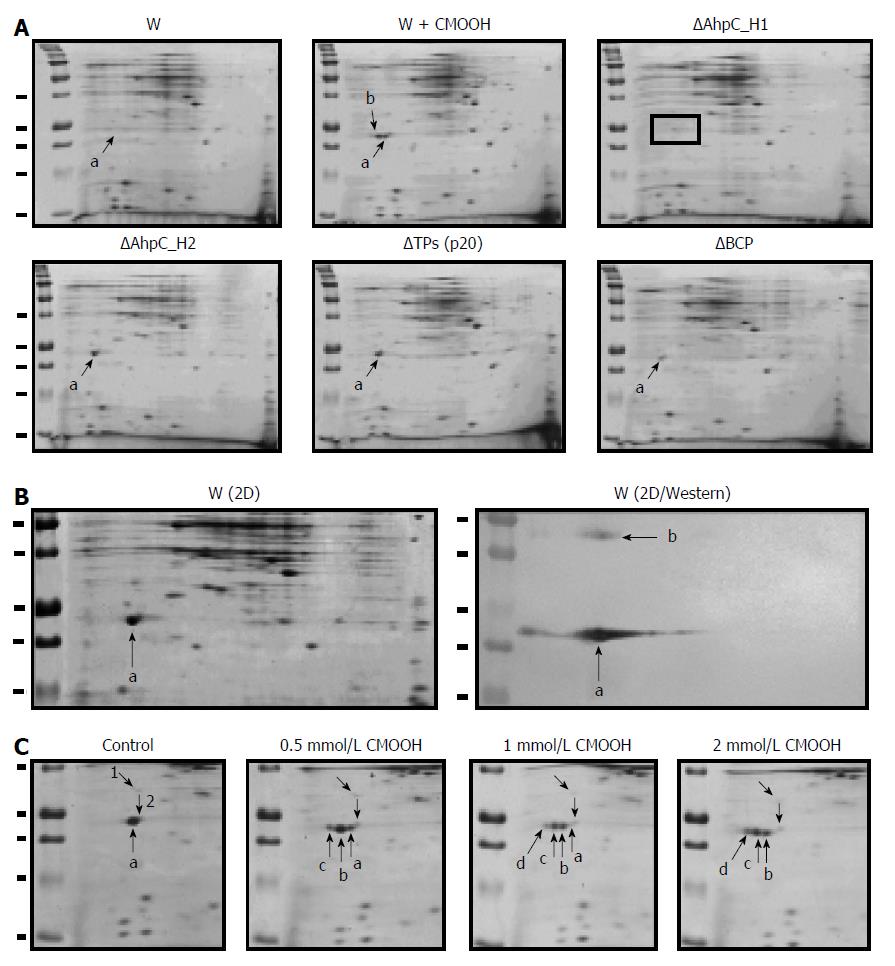
B. subtilis genome encodes four Prxs (B_BCP, B_TPx, B_AhpC_H1, and B_AhpC_H2) (Figure 1), and their expression as antioxidant proteins should be regulated by growth conditions in a gene-specific manner. SDS-PAGE analysis of B. subtilis crude extracts indicates high expression of AhpC_H1 (Figure 5). To further evaluate the regulation of Prx enzymes in B. subtilis, mutant strains deficient in B_BCP (ΔBCP), B_TPx (ΔTPx), B_AhpC_H1 (ΔAhpC_H1), or B_AhpC_H2 (ΔAhpC_H2) were constructed and analyzed by 2D electrophoresis. Protein patterns visualized by Coomassie Brilliant Blue staining indicate that one of the predominant spots represented the AhpC_H1 monomer (Figure 9A, a), whereas the other spot detected in the cumene hydroperoxide (CMOOH)-stressed wild-type strain (Figure 9A, W + CMOOH, b) is suggested to be the AhpC_H1 dimer.
Except for AhpC_H1, other Prxs were not detected by 2D electrophoresis (Figure 9A). As expected, AhpC_H1 (Figure 9A, a) was seen in all mutants but ΔAhpC_H1. It is worth noting that another spot corresponding to the AhpC_H1 dimer was detected by western blotting analysis using anti-AhpC_H1 antibodies (Figure 9B, W/2D Western, b). Taken together, these results suggest that the expression of AhpC_H1 in B. subtilis is much higher than that of B_BCP, B_TPx, and AhpC_H2.
In oxidative stress-exposed B. subtilis, the position of the AhpC_H1-corresponding spot at a more acidic area (Figure 9A, W+CMOOH, b) suggests that Cys residue(s) could be hyperoxidized to Cys-SO2H or Cys-SO3H[34]. To further investigate AhpC_H1 response to hyperoxidation, exponentially growing B. subtilis was treated with increasing concentrations of CMOOH, and protein migration was analyzed by 2D electrophoresis. Figure 9C shows that CMOOH treatment stimulated, at a concentration-dependent manner, the appearance of two different protein spots (b and c) at more acidic positions; at the same time, the original protein spot (a) gradually disappeared, while spot с increased. Considering that sulfinic acid (Cys-SO3H) is more negatively charged than sulfenic acid (Cys-SO2H), spots b and c seem to correspond to the sulfenic and sulfinic forms of AhpC_H1. The predominant appearance of the more acidic AhpC_H1 form in the oxidative stress-exposed B. subtilis might be considered as evidence that Cys166 residue is the primary site for hyperoxidation, provided that only Cys166 is reduced among the three Cys residues of AhpC_H1.
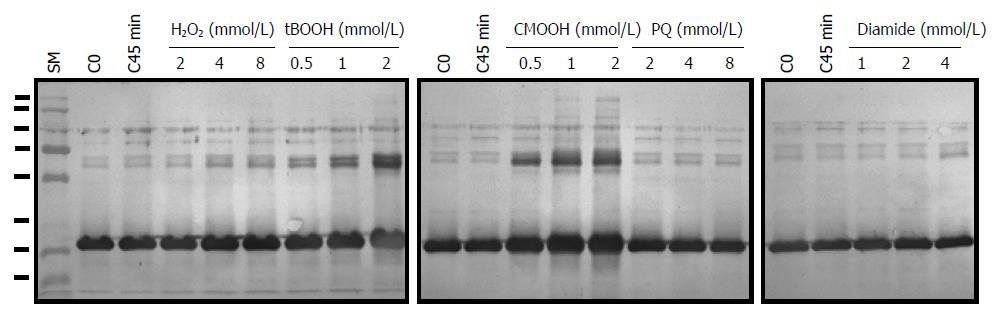
AhpC_H1 expression in B. subtilis in response to oxidative stress was analyzed in exponentially growing cultures treated with different concentrations of H2O2, t-butyl hydroperoxide (tBOOH), CMOOH, paraquat (PQ), and diamide for 45 min. Western blotting analysis (Figure 10) indicates that AhpC_H1 levels were not significantly affected by H2O2, paraquat, and diamide; however, organic peroxides, especially CMOOH, markedly increased AhpC_H1 expression in B. subtilis, suggesting the function of AhpC_H1 as an antioxidant scavenger of lipid hydroperoxides.
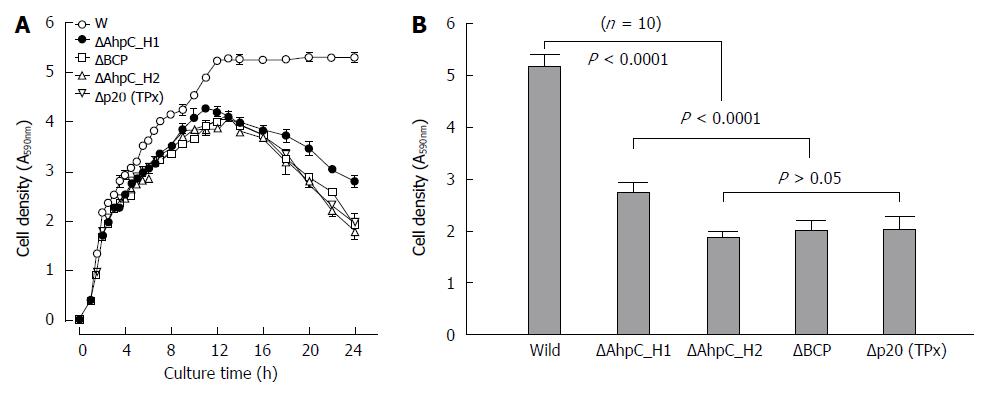
To determine functional significance of four Bacillus Prxs, we compared the growth of the ΔAhpC_H1, ΔAhpC_H2, ΔB_BCP, and ΔB_TPx deletion mutants with that of the parental wild-type strain. All of the mutants exhibited gradual reduction of cell density in the stationary phase (Figure 11A), which could be caused by a decrease of cell viability. Figure 11B shows that among the four null mutants, ΔAhpC_H1 exhibited better survival in the stationary phase, despite much higher levels of AhpC_H1 expression in the wild-type B. subtilis compared to other Prxs.
Sequence homology and phylogenetic analysis performed in this study reveal that AhpC_H1 is closely related to E. coli AhpC (E_AhpC), a typical representative of bacterial AhpC, suggesting that AhpC_H1 may be an E_AhpC ortholog. AhpC_H1 has the highest sequence similarity to E_AhpC among the four analyzed B. subtilis Prxs (65.24% identity in a 187-amino acid region), although it possesses an extra Cys37 in addition to two conserved residues, Cys47 and Cys166. AhpC_H2, another AhpC enzyme in B. subtilis, showed 38.4% identity with E_AhpC in a 159-amino acid sequence. Our results suggest that unlike AhpC_H1, AhpC_H2 is a member of the typical 2-Cys AhpC/Prx1 subfamily, which includes E. coli alkyl hydroperoxide reductase. Comparative structural analysis and evaluation of peroxidase activity, resistance to inactivation by peroxide substrates, and phenotypes of null mutants have demonstrated that AhpC_H1 is a new type of bacterial AhpC, with the structure and functional activity distinct from those of AhpC_H2. Thus, our data indicate that AhpC_H2, not AhpC_H1, is an ortholog of E. coli AhpC. AhpC_H1 (GenBank accession number BAA11268) was first suggested to be an AhpC subunit of the B. subtilis alkyl hydroperoxide reductase induced in response to stressful conditions such as heat and high salinity or growth inhibition at the stationary phase[35-37]. We found a gradual decrease in the viability of Prx-deficient mutant strains in the stationary phase, which is consistent with previous reports, suggesting a functional role of these enzymes in B. subtilis stress response during the stationary growth phase. Together with AhpC_H1, other three B. subtilis Prxs are likely to be involved in bacterial defense system against toxic conditions created by increased oxidative stress[38].
Bacterial AhpC enzymes are known as electron acceptors from the Trx system, although they show much higher reactivity with their specialized flavoprotein reductase AhpF[9]. AhpC proteins from Mycobacterium tuberculosis[39] and Helicobacter pylori[40] have been identified as Trx-dependent alkyl hydroperoxide reductases. In this study, we observed that, regardless of the nature of disulfide linkages (intra- or intermolecular), all four B. subtilis Prxs could be easily reduced by the Trx system consisting of Trx1, Trx reductase, and NADPH, although in the oxidized AhpC_H1, the intramolecular disulfide bond was formed between Cys37 and Cys47 and not between the conserved Cys47 and Cys166 residues. Conversion of the reduced AhpC_H1 from a dimeric form to a high-molecular mass oligomer raises a possibility that the disulfide bond in AhpC_H1 has a more complex role as a functional motif, which promotes conformational transitions unrelated to disulfide linkage. In the case of typical 2-Cys Prx members belonging to the AhpC/Prx1 subfamily, hyperoxidation of the conserved Cys residue(s) has been known as a covalent modification, which stabilizes higher order structures[33,41]. We have also demonstrated the existence of the hyperoxidized AhpC_H1 as a predominant form in oxidative-stress-exposed B. subtilis. Based on the observation of oxidation-induced dimerization (Figure 6B), we could speculate that the reduction of the disulfide bond between Cys37 and Cys47 may be the first step in the oligomerization process. Covalent modification of Cys residues could stabilize higher order structures as is the case with typical 2-Cys Prx members that lack the intramolecular disulfide bond.
Similar to AhpC_H1, M. tuberculosis AhpC has been reported to possess three Cys residues, two at positions 174 and 176 in the C-terminal region, and the conserved Cys61 at the N-terminus. Both Cys174 and Cys176 can form an intermolecular disulfide bond with the conserved N-terminal Cys61 during cyclic catalysis, and all three Cys residues are shown to be important for enzymatic activity[42]. In AhpC_H1, the additional Cys37 was also observed to be involved in peroxidation catalysis. Distinct catalytic activities of AhpC_H1 and AhpC_H2 toward H2O2 (Figure 7C) may provide a rationale for the existence of two different AhpC enzymes in B. subtilis, which may function at different concentrations of H2O2.
Peroxiredoxins, a class of ubiquitously expressed cysteine-dependent peroxidases, serve not only as ROS detoxifiers but also as regulators of signal transduction pathways and peroxide responses. Thus, an intriguing feature of AhpC_H1 is redox-dependent oligomerization, which regulates enzyme activity. Other functional consequences of this dimer-oligomer conversion remain to be solved, but we hypothesize that the reduction of the intra-subunit disulfide linkage between Cys37 and Cys47 by the Trx system or other electron donors serves to stabilize the oligomeric structure, and further regulates some physiological processes. Given that AhpC_H1 is the most abundant protein among the four types of B. subtilis Prxs, the detailed in vivo function of AhpC_H1 is worth further investigation.
In summary, structural and functional differences between AhpC_H1 and AhpC_H2 revealed in this study clearly demonstrate that AhpC_H1 is not an ortholog of E. coli AhpC, but a novel type of atypical 2-Cys AhpC. The present data also suggest that AhpC_H2, as a typical 2-Cys enzyme, is a true member of the AhpC/Prx1 subfamily, although AhpC_H1 has the highest sequence similarity to E. coli AhpC among the four B. subtilis Prxs.
Peroxiredoxins (Prxs) are versatile and predominantly thiol-dependent peroxidases that detoxify hydrogen peroxide and various hydroperoxides. They are ubiquitously expressed, with multiple isoforms present in most organisms. Prxs exhibit high reactivity with hydrogen peroxide and organic hydroperoxides and are involved in peroxide detoxification and signal transduction. The present comparative study was designed to characterize two alkyl hydroperoxide reductase (AhpC) homologs and reveal the biological significance of the existence of two homologous AhpC enzymes in B. subtilis.
This paper is the first report of a detailed structure-functional analysis of the bacillus 2-Cys AhpC homologs AhpC_H1 and AhpC_H2.
AhpC_H1 is not an ortholog of E. coli AhpC, but a novel type of atypical 2-Cys AhpC, and AhpC_H2, as a typical 2-Cys enzyme, is a true member of the AhpC/Prx1 subfamily, although AhpC_H1 has the highest sequence similarity to E. coli AhpC among the four B. subtilis Prxs.
Peroxiredoxins, a class of ubiquitously expressed cysteine-dependent peroxidases, serve not only as ROS detoxifiers but also as regulators of signal transduction pathways and peroxide responses. Thus, an intriguing feature of AhpC_H1 is redox-dependent oligomerization, which regulates enzyme activity. The reduction of the intra-subunit disulfide linkage between Cys37 and Cys47 by the Trx system or other electron donors serves to stabilize the oligomeric structure, and further regulates some physiological processes. Given that AhpC_H1 is the most abundant protein among the four types of B. subtilis Prxs, the detailed in vivo function of AhpC_H1 is worth further investigation.
Peroxiredoxins (Prxs): a ubiquitous family of antioxidant enzymes that convert a harmful peroxide to water and oxygen. Peroxiredoxins are frequently referred to as alkyl hydroperoxide reductase (AhpC) in bacteria. Other names include thiol specific antioxidant (TSA). This family contains AhpC and TSA, as well as related proteins; Phylogenetic analysis: An analysis of relationships between collection of genes and proteins that are derived from a common ancestor; Orthologous proteins: any proteins found in two or more species that can be traced to a common ancestor; specif. one of two homologous genes that is descended from a common ancestor, but which has evolved in a different way; Homologous proteins: proteins having a very similar primary, secondary, and tertiary structure.
The paper examines the two proteins in an attempt to discover the function of these two homologous enzymes in the same organism, both of which are postulated to protect against oxidative damage in cells. These two proteins were initially identified by sequence homologies, cloned, expressed, and purified. The authors have also generated a number of site-directed mutants of the proteins and examined the role of Cys residues in the proteins. Overall, the work in the manuscript carefully examines both the biochemistry and genetics of the AhpC H1 and H2 proteins. AhpC H1levels increase in response to alky peroxides. The authors have indicated that at least AhpC H1 protects against oxidative stress. It appears that there is a significant drop in cell density. Using the mutant proteins and complementation with the AhpC H1 or H2 would have provided further proof of the role of these proteins. Also, showing actual survival of the cells in response to alkyl peroxides or hydrogen peroxide would have been useful.
P- Reviewer: Chen XH, O'Connor TR S- Editor: Ji FF L- Editor: A E- Editor: Wang CH
| 1. | Flohé L, Toppo S, Cozza G, Ursini F. A comparison of thiol peroxidase mechanisms. Antioxid Redox Signal. 2011;15:763-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 2. | Poole LB. The Catalytic Mechanism of Peroxiredoxins. Peroxiredoxin Systems. NY: Springer 2007; 61-81. |
| 3. | Cha MK, Kim HK, Kim IH. Thioredoxin-linked „thiol peroxidase” from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635-28641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Cha MK, Kim HK, Kim IH. Mutation and Mutagenesis of thiol peroxidase of Escherichia coli and a new type of thiol peroxidase family. J Bacteriol. 1996;178:5610-5614. [PubMed] |
| 5. | Cha MK, Kim WC, Lim CJ, Kim K, Kim IH. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J Biol Chem. 2004;279:8769-8778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924-2930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 7. | Cha MK, Hong SK, Kim IH. Four thiol peroxidases contain a conserved GCT catalytic motif and act as a versatile array of lipid peroxidases in Anabaena sp. PCC7120. Free Radic Biol Med. 2007;42:1736-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Cha MK, Hong SK, Lee DS, Kim IH. Vibrio cholerae thiol peroxidase-glutaredoxin fusion is a 2-Cys TSA/AhpC subfamily acting as a lipid hydroperoxide reductase. J Biol Chem. 2004;279:11035-11041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Jönsson TJ, Ellis HR, Poole LB. Cysteine reactivity and thiol-disulfide interchange pathways in AhpF and AhpC of the bacterial alkyl hydroperoxide reductase system. Biochemistry. 2007;46:5709-5721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Park SG, Cha MK, Jeong W, Kim IH. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J Biol Chem. 2000;275:5723-5732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 181] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Knoops B, Loumaye E, Van der Eecken V. Evolution of the peroxiredoxins: Taxonomy, homology and characterization. Peroxiredoxin Systems. NY: Springer 2007; 27-40. |
| 12. | Dietz KJ. Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal. 2011;15:1129-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Lim YS, Cha MK, Kim HK, Kim IH. The thiol-specific antioxidant protein from human brain: gene cloning and analysis of conserved cysteine regions. Gene. 1994;140:279-284. [PubMed] |
| 14. | Lim YS, Cha MK, Kim HK, Uhm TB, Park JW, Kim K, Kim IH. Removals of hydrogen peroxide and hydroxyl radical by thiol-specific antioxidant protein as a possible role in vivo. Biochem Biophys Res Commun. 1993;192:273-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 149] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1077] [Cited by in RCA: 1068] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 16. | Flohé L. Changing paradigms in thiology from antioxidant defense toward redox regulation. Methods Enzymol. 2010;473:1-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Fomenko DE, Marino SM, Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells. 2008;26:228-235. [PubMed] |
| 18. | Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. FEBS J. 2009;276:2469-2477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 382] [Cited by in RCA: 366] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 19. | Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 347] [Cited by in RCA: 312] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 20. | Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid Redox Signal. 2011;15:781-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 355] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 21. | Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017-7021. [PubMed] |
| 22. | Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346-20354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Chae HZ, Kim IH, Kim K, Rhee SG. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268:16815-16821. [PubMed] |
| 24. | Knoops B, Clippe A, Bogard C, Arsalane K, Wattiez R, Hermans C, Duconseille E, Falmagne P, Bernard A. Cloning and characterization of AOEB166, a novel mammalian antioxidant enzyme of the peroxiredoxin family. J Biol Chem. 1999;274:30451-30458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 194] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 25. | Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 148] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 26. | Ellis HR, Poole LB. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry. 1997;36:13349-13356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 157] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 27. | Evrard C, Capron A, Marchand C, Clippe A, Wattiez R, Soumillion P, Knoops B, Declercq JP. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J Mol Biol. 2004;337:1079-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lázaro JJ, Dietz KJ. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J Exp Bot. 2008;59:3259-3269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Barranco-Medina S, Krell T, Finkemeier I, Sevilla F, Lázaro JJ, Dietz KJ. Biochemical and molecular characterization of the mitochondrial peroxiredoxin PsPrxII F from Pisum sativum. Plant Physiol Biochem. 2007;45:729-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Barranco-Medina S, Lázaro JJ, Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 179] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 31. | Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 32. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. NY: Cold Spring Harbor Laboratory 1989; . |
| 33. | Hoch JA. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 118] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029-38036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 372] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 35. | Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571-6578. [PubMed] |
| 36. | Bsat N, Chen L, Helmann JD. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579-6586. [PubMed] |
| 37. | Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis--a two-deimensional protein electrophoresis study. Microbiology. 1997;143:999-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 137] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 38. | Christman MF, Morgan RW, Jacobson FS, Ames BN. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 813] [Cited by in RCA: 765] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 39. | Jaeger T. Peroxiredoxin systems in mycobacteria. Subcell Biochem. 2007;44:207-217. [PubMed] |
| 40. | Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol. 2001;183:1961-1973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 154] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 41. | Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2011;15:795-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 261] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 42. | Chauhan R, Mande SC. Site-directed mutagenesis reveals a novel catalytic mechanism of Mycobacterium tuberculosis alkylhydroperoxidase C. Biochem J. 2002;367:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |