Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.218
Peer-review started: March 16, 2015
First decision: April 27, 2015
Revised: May 10, 2015
Accepted: June 15, 2015
Article in press: June 16, 2015
Published online: August 26, 2015
Processing time: 167 Days and 13.7 Hours
Proteinaceous infectious particles (prions) are unique pathogens as they are devoid of any coding nucleic acid. Whilst it is assumed that prion disease is transmitted by a misfolded isoform of the cellular prion protein, the structural insight of prions is still vague and research for high resolution structural information of prions is still ongoing. In this review, techniques that may contribute to the clarification of the conformation of prions are presented and discussed.
Core tip: Prions (proteinaceous infectious particles) are misfolded isoforms of cellular proteins that cause neurodegenerative diseases in mammals and humans. Several structural models are available for prions but a 3D-structure does still not exist. More structural information is demanded for the understanding of the conversion process and finally for the design of efficient therapeutic approaches. In this review, techniques that may contribute to the clarification of the conformation of prions are presented.
- Citation: Daus ML. Techniques to elucidate the conformation of prions. World J Biol Chem 2015; 6(3): 218-222
- URL: https://www.wjgnet.com/1949-8454/full/v6/i3/218.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.218
Prions are infectious proteins that cause fatal neurodegenerative diseases[1-5]. Rearrangements of the structure of the cellular prion protein (PrPC) are the key of infection. PrPC is characterized by a high α-helical content and conversion in a β-sheet rich conformer (PrPSC) transforms it into an infectious protein[6,7]. According to the “prion hypothesis” the protein itself is the causative agent of disease even though several cofactors that may influence the conversion process are discussed in the literature[2,8-12]. A 3D-structure of PrPSC does still not exist, albeit several structural models for PrPSC are described[13]. Definite structural information about prions is indispensable for the understanding of the conversion process and eventually for the design of new therapeutic strategies.
In the present review, several techniques to assess the conformation of prions are presented and discussed with a main focus on the application of Fourier-transform infrared spectroscopy. The intention of this mini-review article is to give an overview of current techniques used in prion research to elucidate the structure of prions. For details readers are referred to the literature cited in the text.
Fourier-transform infrared (FTIR) spectroscopy was one of the fundamental techniques that had demonstrated the high β-sheet content of PrPSC in contrast to PrPC[14-16]. This technique has several advantages compared to other analytical approaches. With FTIR spectroscopy protein samples can be analyzed (1) without labeling; (2) under native conditions; (3) with only minute amounts of material (in particular in case of micro-FTIR); and (4) in a dynamic process. Moreover results can be obtained within a very short time[16,17]. FTIR spectroscopy elucidates primarily the secondary structure of proteins and originates essentially from C=O stretching vibrations of the amide groups of the protein backbone. For the structural characterization of PrPSC the amide I band is the most useful infrared absorption band[18-20].
FTIR of proteins typically is performed in H2O, D2O or under dried conditions. For spectroscopic analyses the purity of sample material is of utmost importance[16,21]. This is particularly challenging when sample material has to be extracted from infected tissue and when the protein of interest is present in only small amounts. Improved purification protocols for the extraction of prions from tissue have been published and allowed us to analyze tiny amounts of sample material by micro-FTIR[16]. Micro-FTIR can be applied on dried protein samples using a FTIR spectrometer linked to an IR-microscope. This enables the performance of measurements on pure protein samples in the range of only a few nanograms.
Micro-FTIR has been shown to be an appropriate tool for the screening of different prion strains and in vitro generated prion protein (PrP)[16,17]. Particularly in case of in vitro generated PrP only very small amounts of prions are expected to be extractable. This could impede an analysis by other structure-sensitive techniques [e.g., circular dichroism (CD) spectroscopy]. We recently recorded spectra from PrPSC molecules generated by protein misfolding cyclic amplification (PMCA) using micro-FTIR[16]. PMCA mimics the conversion of PrPC to PrPSCin vitro[22,23]. Comparing spectra from native- and PMCA-derived PrPSC revealed structural differences of molecules before and after PMCA. This technical progress provides the possibility to analyze the influence of cofactors on the conversion activity and putative correlating structural rearrangements in future experiments.
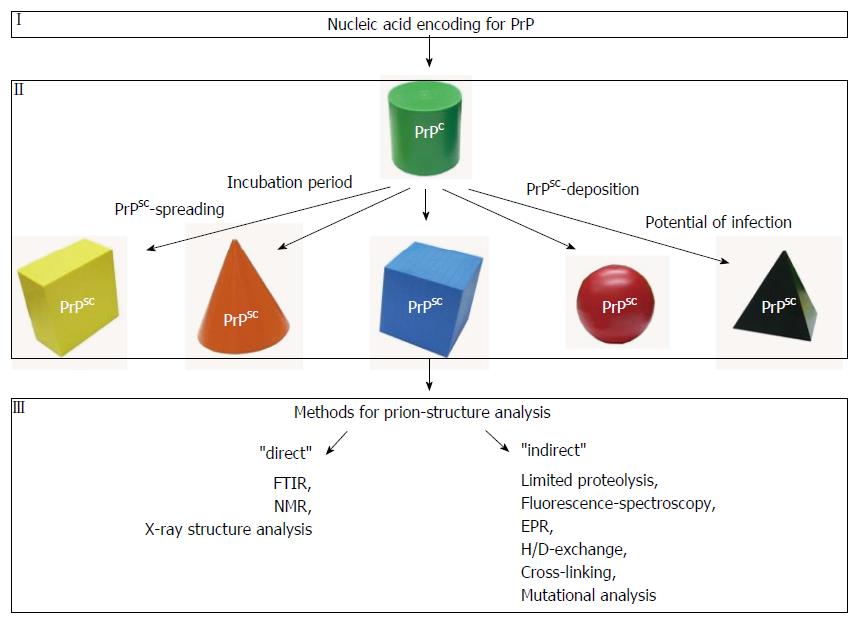
As mentioned above, micro-FTIR is also suitable to discriminate different prion strains[14,24,25]. In prion research, strains are defined as prion-isolates that, after inoculation into distinct hosts, cause disease with consistent characteristics, such as incubation period, distinct patterns of PrPSc distribution and spongiosis in the brain (Figure 1). A concrete example of use would be strain typing for chronic wasting disease (CWD)[17]. CWD is a prion disease occurring in cervids in North America and rapidly spreads among free ranging deer and elk[26,27]. To test whether and to which extent there are different strains for CWD in the environment or whether new strains emerge in the future, micro-FTIR (with only 100 mg of infected tissue needed to obtain a sufficient amount of protein extract for FTIR analysis) can be applied. The use of small amounts of sample material allows the preparation of multiple samples within a short time. Spectra from CWD field samples collected in the past, present or future could be compared. As different strains can correlate with different zoonotic potentials a rapid structural scan by FTIR may be suitable for a proper risk assessment.
Electron microscopy analyses directly reveal the morphological difference between PrPC and PrPSC as the latter appears as amyloid fibrils[28,29]. Moreover, it is possible to distinguish between some prion strains due to their distinct morphological shape. Electron crystallography had a limited scope to predict general conformational features because prions tend to aggregate[30,31]. Atomic force microscopy (AFM) is used as an alternative technique that needs no staining of samples[16,32]. AFM can be combined with infrared- and Raman-spectroscopy suitable for site-directed sample characterization. Tip enhanced infrared- or Raman-spectroscopy principally allow measurements on defined positions within a prion fibril[33,34]. These techniques could be useful for the analyses of mixtures of prion fibrils (“quasi species”) and would allow a precise biochemical characterization of prions[17,35].
The impact of specific amino acids on the prion conversion process can be determined by mutational analyses when subsequently tested in animal-, cell culture- or in vitro conversion assays[36-39]. By this means it could be demonstrated that specific amino acids accelerate or decelerate the conversion process and that the N-terminus is not essential for prion infectivity[40]. Limited proteolysis with Proteinase K is used to discriminate PrPC from PrPSC as the conversion in a β-sheet rich conformation (PrPSC) prevents the C-terminus from degradation while the less structured N-terminus is cleaved off[41-46]. Limited proteolysis at different pH values, with rising amounts of protease or in the presence of chaotropic salts can give further structural information and may help to discriminate prion strains with different conformations.
Recombinant PrP can (after site-directed mutagenesis) site-specifically be labelled with fluorophores or spin labels and then analyzed by fluorescence- or EPR spectroscopy, respectively. Such studies propose a parallel in-register β-sheet structure of PrP amyloids and enable the calculation of distances between specific residues within an amyloid fibril[11,47-49]. It has to be mentioned that misfolded recombinant PrP without the addition of cofactors usually lacks infectivity and may therefore not exactly represent the in vivo structure of PrPSC[11].
The exchange of hydrogen for deuterium ions and subsequent analysis by mass spectrometry allows the detection of unstructured and highly structured regions within PrP[50-52]. The C-terminal part of prions shows low exchange rates indicating a highly structured conformation[53].
While the above mentioned techniques reveal basic and to some extent indirect structural insights, high resolution structure information is needed to understand the conversion process of PrP. High resolution 3D-structural data from X-ray fiber diffraction and NMR are available for PrPC but not for the complete PrPSC molecule. Because of the insolubility and the propensity to aggregate only parts of PrPSC are structurally resolved until now[54-57].
Misfolding of a cellular protein into an infectious isoform sharing the same primary structure is unique for prions. This phenomenon is currently discussed to be a general principle for other neurodegenerative diseases[58-60]. Prions lack a coding nucleic acid thus diagnosis and the detection of misfolded isoforms cannot be done by genetics but e.g., by structural characterization.
A lot of structural models for PrPSC exist but as discussed recently by Requena and Wille “the results and/or their interpretation remain controversial”[61]. For a better understanding of the conversion of PrPC to PrPSC, more structural data from biochemical and biophysical experiments are required for PrPSC (Figure 1). Therapeutic strategies that aim at the prevention of this misfolding process would benefit from such a progress.
P- Reviewer: Bai G, Fujiwara N, Gurevich VV, Toth EA, Wang Y S- Editor: Tian YL L- Editor: A E- Editor: Wang CH
| 1. | Prusiner SB, DeArmond SJ. Prions causing nervous system degeneration. Lab Invest. 1987;56:349-363. [PubMed] |
| 2. | Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3695] [Cited by in RCA: 3504] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 3. | Johnson RT. Prion diseases. Lancet Neurol. 2005;4:635-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 128] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | Prusiner SB. The prion diseases. Brain Pathol. 1998;8:499-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 254] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1236] [Cited by in RCA: 1352] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 6. | Prusiner SB. Research on scrapie. Lancet. 1982;2:494-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Prusiner SB, Bolton DC, Groth DF, Bowman KA, Cochran SP, McKinley MP. Further purification and characterization of scrapie prions. Biochemistry. 1982;21:6942-6950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 318] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 663] [Cited by in RCA: 649] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 9. | Simoneau S, Thomzig A, Ruchoux MM, Vignier N, Daus ML, Poleggi A, Lebon P, Freire S, Durand V, Graziano S. Synthetic scrapie infectivity: interaction between recombinant PrP and scrapie brain-derived RNA. Virulence. 2015;6:132-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Supattapone S. Elucidating the role of cofactors in mammalian prion propagation. Prion. 2014;8:100-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Ma J, Wang F. Prion disease and the ‘protein-only hypothesis’. Essays Biochem. 2014;56:181-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Ma J. The role of cofactors in prion propagation and infectivity. PLoS Pathog. 2012;8:e1002589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Requena JR, Wille H. The structure of the infectious prion protein: experimental data and molecular models. Prion. 2014;8:60-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672-7680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 617] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962-10966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1667] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 16. | Daus ML, Wagenführ K, Thomzig A, Boerner S, Hermann P, Hermelink A, Beekes M, Lasch P. Infrared microspectroscopy detects protein misfolding cyclic amplification (PMCA)-induced conformational alterations in hamster scrapie progeny seeds. J Biol Chem. 2013;288:35068-35080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Daus ML, Beekes M. Chronic wasting disease: fingerprinting the culprit in risk assessments. Prion. 2012;6:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 18. | Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2200] [Cited by in RCA: 1991] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 19. | Bandekar J. Amide modes and protein conformation. Biochim Biophys Acta. 1992;1120:123-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 576] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 20. | Barth A, Zscherp C. What vibrations tell us about proteins. Q Rev Biophys. 2002;35:369-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1528] [Cited by in RCA: 1454] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 21. | Moore RA, Timmes AG, Wilmarth PA, Safronetz D, Priola SA. Identification and removal of proteins that co-purify with infectious prion protein improves the analysis of its secondary structure. Proteomics. 2011;11:3853-3865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Morales R, Duran-Aniotz C, Diaz-Espinoza R, Camacho MV, Soto C. Protein misfolding cyclic amplification of infectious prions. Nat Protoc. 2012;7:1397-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 23. | Pritzkow S, Wagenführ K, Daus ML, Boerner S, Lemmer K, Thomzig A, Mielke M, Beekes M. Quantitative detection and biological propagation of scrapie seeding activity in vitro facilitate use of prions as model pathogens for disinfection. PLoS One. 2011;6:e20384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Thomzig A, Spassov S, Friedrich M, Naumann D, Beekes M. Discriminating scrapie and bovine spongiform encephalopathy isolates by infrared spectroscopy of pathological prion protein. J Biol Chem. 2004;279:33847-33854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Spassov S, Beekes M, Naumann D. Structural differences between TSEs strains investigated by FT-IR spectroscopy. Biochim Biophys Acta. 2006;1760:1138-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Daus ML, Breyer J, Wagenfuehr K, Wemheuer WM, Thomzig A, Schulz-Schaeffer WJ, Beekes M. Presence and seeding activity of pathological prion protein (PrP(TSE)) in skeletal muscles of white-tailed deer infected with chronic wasting disease. PLoS One. 2011;6:e18345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Saunders SE, Bartelt-Hunt SL, Bartz JC. Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg Infect Dis. 2012;18:369-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Merz PA, Somerville RA, Wisniewski HM, Iqbal K. Abnormal fibrils from scrapie-infected brain. Acta Neuropathol. 1981;54:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 310] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Sim VL, Caughey B. Ultrastructures and strain comparison of under-glycosylated scrapie prion fibrils. Neurobiol Aging. 2009;30:2031-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci USA. 2002;99:3563-3568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 301] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 31. | Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342-8347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 421] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 32. | Ostapchenko V, Gasset M, Baskakov IV. Atomic force fluorescence microscopy in the characterization of amyloid fibril assembly and oligomeric intermediates. Methods Mol Biol. 2012;849:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Pozzi EA, Sonntag MD, Jiang N, Klingsporn JM, Hersam MC, Van Duyne RP. Tip-enhanced Raman imaging: an emergent tool for probing biology at the nanoscale. ACS Nano. 2013;7:885-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 34. | Harz M, Rösch P, Popp J. Vibrational spectroscopy--a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A. 2009;75:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 171] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 35. | Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep. 2011;12:1109-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 36. | Telling GC. Transgenic mouse models of prion diseases. Methods Mol Biol. 2008;459:249-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 37. | Muramoto T, Scott M, Cohen FE, Prusiner SB. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1996;93:15457-15462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 153] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 38. | Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255-1264. [PubMed] |
| 39. | Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 481] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 40. | Prigent S, Rezaei H. PrP assemblies: spotting the responsible regions in prion propagation. Prion. 2011;5:69-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim Biophys Acta. 1998;1382:191-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 329] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 42. | Kocisko DA, Lansbury PT, Caughey B. Partial unfolding and refolding of scrapie-associated prion protein: evidence for a critical 16-kDa C-terminal domain. Biochemistry. 1996;35:13434-13442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 84] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | Zou WQ, Capellari S, Parchi P, Sy MS, Gambetti P, Chen SG. Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J Biol Chem. 2003;278:40429-40436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 112] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 44. | Zanusso G, Farinazzo A, Prelli F, Fiorini M, Gelati M, Ferrari S, Righetti PG, Rizzuto N, Frangione B, Monaco S. Identification of distinct N-terminal truncated forms of prion protein in different Creutzfeldt-Jakob disease subtypes. J Biol Chem. 2004;279:38936-38942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 45. | Sajnani G, Pastrana MA, Dynin I, Onisko B, Requena JR. Scrapie prion protein structural constraints obtained by limited proteolysis and mass spectrometry. J Mol Biol. 2008;382:88-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Vázquez-Fernández E, Alonso J, Pastrana MA, Ramos A, Stitz L, Vidal E, Dynin I, Petsch B, Silva CJ, Requena JR. Structural organization of mammalian prions as probed by limited proteolysis. PLoS One. 2012;7:e50111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci USA. 2007;104:18946-18951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 263] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 48. | Seidel R, Engelhard M. Chemical biology of prion protein: tools to bridge the in vitro/vivo interface. Top Curr Chem. 2011;305:199-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 49. | Ngo S, Chiang V, Guo Z. Quantitative analysis of spin exchange interactions to identify β strand and turn regions in Ure2 prion domain fibrils with site-directed spin labeling. J Struct Biol. 2012;180:374-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 50. | Smirnovas V, Kim JI, Lu X, Atarashi R, Caughey B, Surewicz WK. Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by hydrogen/deuterium exchange. J Biol Chem. 2009;284:24233-24241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 51. | Nazabal A, Hornemann S, Aguzzi A, Zenobi R. Hydrogen/deuterium exchange mass spectrometry identifies two highly protected regions in recombinant full-length prion protein amyloid fibrils. J Mass Spectrom. 2009;44:965-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Lu X, Wintrode PL, Surewicz WK. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci USA. 2007;104:1510-1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 195] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 53. | Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2011;18:504-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 185] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 54. | Meier BH, Böckmann A. The structure of fibrils from ‘misfolded’ proteins. Curr Opin Struct Biol. 2015;30:43-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 55. | Nguyen JT, Inouye H, Baldwin MA, Fletterick RJ, Cohen FE, Prusiner SB, Kirschner DA. X-ray diffraction of scrapie prion rods and PrP peptides. J Mol Biol. 1995;252:412-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 128] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USA. 2009;106:16990-16995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 57. | Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 831] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 58. | Morales R, Callegari K, Soto C. Prion-like features of misfolded Aβ and tau aggregates. Virus Res. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 59. | Beekes M, Thomzig A, Schulz-Schaeffer WJ, Burger R. Is there a risk of prion-like disease transmission by Alzheimer- or Parkinson-associated protein particles? Acta Neuropathol. 2014;128:463-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 60. | Ashe KH, Aguzzi A. Prions, prionoids and pathogenic proteins in Alzheimer disease. Prion. 2013;7:55-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 61. | Acevedo-Morantes CY, Wille H. The structure of human prions: from biology to structural models-considerations and pitfalls. Viruses. 2014;6:3875-3892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |