Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.162
Peer-review started: April 12, 2014
First decision: May 14, 2014
Revised: May 8, 2015
Accepted: May 27, 2015
Article in press: May 28, 2015
Published online: August 26, 2015
Processing time: 501 Days and 17.7 Hours
MicroRNAs are small non-coding RNAs that participate in different biological processes, providing subtle combinational regulation of cellular pathways, often by regulating components of signalling pathways. Aberrant expression of miRNAs is an important factor in the development and progression of disease. The canonical myomiRs (miR-1, -133 and -206) are central to the development and health of mammalian skeletal and cardiac muscles, but new findings show they have regulatory roles in the development of other mammalian non-muscle tissues, including nerve, brain structures, adipose and some specialised immunological cells. Moreover, the deregulation of myomiR expression is associated with a variety of different cancers, where typically they have tumor suppressor functions, although examples of an oncogenic role illustrate their diverse function in different cell environments. This review examines the involvement of the related myomiRs at the crossroads between cell development/tissue regeneration/tissue inflammation responses, and cancer development.
Core tip: The roles of the canonical muscle-associated microRNAs are reviewed, including microRNA families miR-1 and miR-133, and single miR-206, which are collectively known as the “myomiRs”. The myomiRs act at the crossroads of the molecular regulation of muscle cells, linking between pathways for cell differentiation, development and maintenance, but also potentiate aberrant cell growth in numerous non-muscle cancers. Typically myomiRs are downregulated in cancers, but some myomiRs are upregulated in a few cancers, yet each dysregulation event advances tumor progression. The review examines normal and disease-linked molecular changes associated with the myomiRs, and collates the extensive literature into accessible tables.
- Citation: Mitchelson KR, Qin WY. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem 2015; 6(3): 162-208
- URL: https://www.wjgnet.com/1949-8454/full/v6/i3/162.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.162
MicroRNAs (miRs) are short single strand RNA molecules (typically 22 nt) which interact in a semi-complementary manner with numerous target gene mRNAs, directed by a short “seed sequence”, destining the targeted mRNA for degradation or for translational inhibition, and thus an miR can downregulate the functional expression of the target gene. In this manner a single miR can influence the abundance of numerous independent gene targets, and aid in the co-ordinate regulation of members of diverse cell signalling pathways, as well as metabolic pathways and basic cell proliferation or developmental processes. Three miR families, miR-1, miR-133 and miR-206 constitute the original (canonical) myomiRs and were considered muscle specific because of their prevalence in skeletal and cardiac muscle[1-5] and for their central roles in the regulation of myogenesis, muscle development and muscle remodelling[6-8]. Although other muscle enriched miRs such as miR-499 and -208, and others with key roles in cardiac muscle development have been identified, and although the term “myomiR” is now often used to denote several miRs encoded within myosin genes, for brevity this review is restricted to discussion of the three canonical myomiRs.
In man the genes encoding the canonical myomiR are organized into three cistrons encoding partners (miR-1-2, miR-133a-1), (miR-1-1, miR-133a-2) and (miR-133b, miR-206) and are located on chromosomes 18q11.2, 20q13.33 and 6p12.2, respectively. In this review we examine the roles of the myomiRs in normal tissue development and their emerging functions in various non-muscle tissues and their influence on the progression of cancers. The dysregulation of expression of the myomiRs in cancers is often related to a significant worsening patient prognosis, via the deregulation of a variety of validated gene targets.
The two mature miR-1 isomers have identical sequence, as have the two miR-133a isomers. The mature miR-133 isomers are also highly similar, differing only at the 3’-terminal base, with miR-133a1/2 terminating G-3’ and miR-133b with A-3’, respectively. Independent upstream enhancers have been identified for the cistronic miR-1-2 -133a-1 genes, as well as for the cistronic miR-1-1/-133a-2 genes which are intronic to the C20orf166 gene[9]. These independent enhancers allow the different isomer genes to be independently expressed under cell specific regulation.
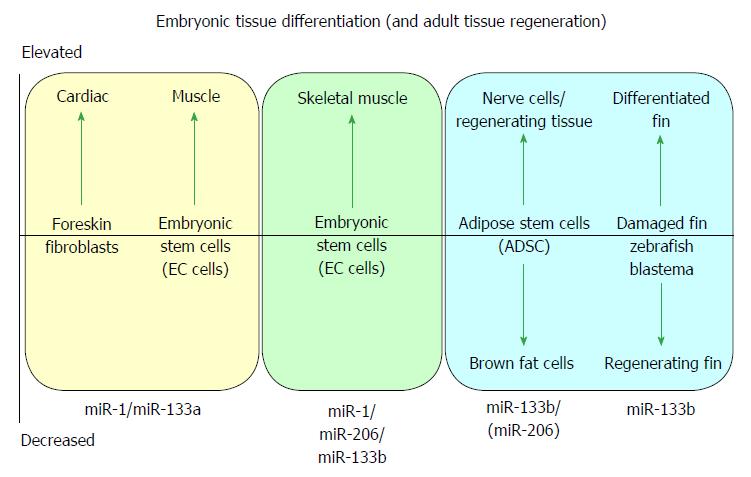
MicroRNA-1 and -133 were initially identified during the development and differentiation of skeletal muscle[7] and cardiac muscle[2,6]. Both miR-1/-133a gene cistrons are canonically expressed in skeletal and cardiac muscle[5,9], whilst the miR-133b/-206 gene cluster is expressed in developing skeletal muscle[5] but not (significantly) in cardiac muscle, defining seminal roles of miR-1 and miR-133a in muscle biogenesis, and specifically in cardiac biogenesis[2,6]. A cartoon illustrating some of the major effects of myomiRs during differentiation of embryonic tissue and during tissue regeneration is shown in Figure 1.
MiR-133a has a regulatory role from the earliest differentiation of myogenic stem cells into myoblasts[7,10] continuing throughout the growth of structurally complex muscle tissues[7,11], and has homeostatic functions for muscle maintenance and protection in mature muscle, or in muscle regeneration from muscle progenitor cells after skeletal muscle stress or injury[5]. Key studies show miR-1, -133b and -206 acting during early development of skeletal myocytes through to the homeostatic maintenance of skeletal muscle[3,4,8], with miR-133b/-206 also having functions in neuromuscular synapse development and maintenance[12], as detailed in Tables 1 and 2.
| Factor(s) | Regulation | Regulator | Tissue/cell | Ref. |
| Fish and lower vertebrates: Development and regeneration | ||||
| Ttk protein kinase (mps1) | Upregulated mps1: a target of miR-133 | Downregulation of miR-133 by Fgf | Regeneration of Zebrafish caudal fin (appendage) | [68] |
| RhoA | Downregulation of RhoA mRNA | Upregulation of miR-133b expression | Regenerating adult zebrafish spinal cord, axon outgrowth | [69] |
| RhoA | Downregulation of RhoA protein | Upregulation of miR-1 and miR-133 expression | Zebrafish muscle gene expression and regulation of sarcomeric actin organization | [166] |
| Cell cycle factors mps1, cdc37 and PA2G4, and cell junction components cx43 and cldn5 | Upregulated mps1, cdc37, PA2G4, cx43, cldn5 | Downregulated miR-133(a1) stimulates cardiac cell regeneration | Regenerating zebrafish cardiac muscle | [167] |
| miR-133b | MiR-133b found in developing somites, little in CNS tissues | Whole zebrafish embryos - normal development | [168] | |
| SRF activates muscle specific genes and miRs; | MiR-1 targets HDAC4, promoting myogenesis | In contrast, miR-133a represses SRF, enhancing myoblast proliferation | X. laevis embryos: skeletal muscle proliferation and differentiation in cultured myoblasts in vitro and in embryos in vivo | [7] |
| HDAC4 represses muscle gene expression | ||||
| nAChR subunits UNC-29, UCR-63; MEF2 | Subunits UNC-29, UCR-63, and MEF2 downregulated | miR-1 upregulated | C. elegans muscle at the neuromuscular junction | [34] |
| Mammalian pluripotent cells | ||||
| Muscle-specific microRNAs: miR-1 and miR-133a | MiR-1 and miR-133a have opposing functions during differentiation of progenitor cardiac muscles | Muscle-specific | Promotion of mesoderm formation from mouse ES cells | [13] |
| microRNAs, miR-1 and miR-133(a) upregulated | ||||
| Notch signalling, promotes neural differentiation and inhibits muscle differentiation; opposes miR-1 effects | Dll-1 translationally repressed | miR-1 upregulation, promotes cardiomycete differentiation | Mouse and human ES cell differentiation into muscle | [13] |
| SRF-/- EBs reflecting the loss of hematopoietic lineages in the absence of SRF | Early endoderm markers, Afp and Hnf4α: strongly down regulated | Increased miR-1 and miR-133a relieve the block on mesodermal differentiation | Mouse endoderm | [13] |
| Blood cell -specific genes, such as Cd53, CxCl4, and Thbs1, dramatically down regulated | Cd53, CxCl4, and Thbs1 expression was reinitiated by reintroduction of miR-1 or miR-133 | |||
| mES(miR-1)- and mES(miR-133a)- EBs compared to in control EBs | Nodal stimulated expression of endoderm markers Afp and Hnf4α in control EBs. Dramatically lower levels in mES(miR-1)- and mES(miR-133a)- EBs | miR-1 or miR-133 can each function as potent repressors of endoderm gene expression | mES cells, that lack either miR-1 or miR-133(a) during differentiation into EBs | [13] |
| IGF-1 | IGF-1 signalling and miR-133 co-regulate myoblast differentiation via a feedback loop | IGF-1 upregulates miR-133; | Myogenic differentiation of C2C12 myoblasts; Mouse during development from embryonic to mature skeletal muscle | [24] |
| IGF-1R | miR-133 downregulates IGF-1R | |||
| IGF-1 | IGF-1 signalling and miR-1 coregulate differentiation of myoblasts via a feedback loop | IGF-1 signalling downregulates miR-1 by repression of FoxO3a; | Differentiating C2C12 myoblasts | [25] |
| miR-1 down-regulates IGF-1 | ||||
| Reversine [2-(4-morpholinoanilino)-N6-cyclohexyladenine] | Decrease in active histone modifications; including trimethylation of histone H3K4/ H3K36, phosphorylation of H3S10; | miR-133a expression strongly inhibited by reversine; reduced acetylation of H3K14 at miR-133a promoter | Reversine dedifferentiates murine C2C12 myoblasts back into multipotent progenitor cells, via extensive epigenetic modification of histones resulting in chromatin remodelling, and altered gene expression | [20-23] |
| Stimulates expression of polycomb genes Phc1 and Ezh2 | Reduced expression of myogenin, MyoD, Myf5 and Aurora A and B kinases | |||
| FZD7 and FRS2 | miR-1 promotes cardiac differentiation; miR-1 targets FZD7 and FRS2 | Activitation of WNT and signalling cause MCPs differentiation into cardiomyocytes | Mouse and human ES cells | [169] |
| miR-206/133b cluster | PAX7 gene expression unchanged; | miR-206/133b cistron knock-out mice cells | Muscle satellite cell differentiation in vitro | [170] |
| miR-206/133b cluster is not required for development, and survival of skeletal muscle cells | ||||
| Differentiating skeletal muscle | ||||
| DNA polymerase alpha | Repression of Idl-3 protein expression | miR-206 up-regulated | Mouse skeletal muscle differentiation | [42] |
| Repression of p180 subunit of DNA polymerase alpha | ||||
| MEF2 transcription factor | MEF2 activates of miR-1-2 and 133a-1 transcription; binds muscle-specific enhancer | Bicistronic primary transcript of miR-1-2 and 133a-1 | Development of mammalian skeletal muscle | [9] |
| MRFs, Myf5, MyoD, Myogenin and MRF4 | Myf5 essential for miR-1 and miR-206 expression during skeletal muscle myogenesis | Forced expression of MRFs in neural tube induces miR-1 and miR-206 expression | Chicken and mouse embryonic muscle | [171] |
| PTB and neuronal homolog nPTB, exon splicing factors | Downregulation of PTB protein by miR-133 (and miR-206) | Concurrent upregulation of miR-133 and induction of splicing of several PTB-repressed exons | During myoblast differentiation, microRNAs control a developmental exon splicing program | [172] |
| BDNF | BDNF downregulated | miR-206 upregulated | Differentiation of C2C12 myoblasts into myotubes | [48] |
| Fstl1 and Utrn | Fstl1 and Utrn downregulated | miR-206 upregulated | Skeletal muscle differentiation | [40] |
| Utrophin A (muscle) | Utrophin A down-regulated by both miRs | Upregulated miR-133b, miR-206 | C2C12 mouse myoblasts, mouse soleus muscle | [173] |
| CNN3 gene | Negative correlation between miR-1 expression and CNN3 mRNA expression | Normal skeletal muscle | Tongcheng (Chinese) and Landrace (Danish) pigs | [174] |
| FGFR1 and PP2AC, members of ERK1/2 signalling pathway | miR-133 (a and b) activities increase during myogenesis | miR-133 directly downregulates expression of FGFR1 and PP2AC | Mouse C2C12 myoblast cells | [31] |
| ERK1/2 signalling pathway activity | ERK1/2 signalling activity suppresses miR-133 expression | Downregulation of expression of miR-133 | A reciprocal mechanism for regulating myogenesis | |
| BAF chromatin remodelling complex (BAF60a, BAF60b and BAF60c) | Positive inclusion of BAF60c in the BAF chromatin remodeling complex | Expression of miR-133 and miR-1/206 | Progression of developing somites in chick embryos | [63] |
| BAF chromatin remodelling complex | Negative regulation of BAF60a and BAF60b; exclusion from BAF chromatin remodelling complex | Expression of miR-133 | Progression of developing somites in chick embryos | [63] |
| BAF chromatin remodelling complex | Exogenous upregulation of BAF60a and BAF60b | Delay in developing somites in chick embryos | [63] | |
| Mitochondrial UCP2 and UCP3 | MyoD activates miR-133a expression which in turn directly downregulates UCP2 mRNA | Feedback network involving MyoD-miR-133a-UCP2 | Mouse skeletal and cardiac muscles; UCP2 imposes developmental repression | [56] |
| Mitochondrial UCP2 and UCP3 | Exogenous overexpression of myogenin and MyoD transcription factors | Strong increase in UCP3 promoter, expression, weak effect at the UCP2 promoter | Mouse C2C12 myoblasts | [57] |
| Proliferating myogenic skeletal muscle cells | ||||
| MiR-206/133b cluster | MiR-206/133b cluster is not required for survival and regeneration of skeletal muscle | Muscle regeneration proceeds in Mdx mice in vivo | miR-206/133b cistron knock-out mice | [170] |
| Enhanced translation of specific mitochondrial genome-encoded transcripts | miR-1 enters muscle mitochondria and binds mtRNA targets along with Ago factor | Increased expression of mtRNA targets | Proliferating myogenic skeletal muscle cells after muscle injury | [53] |
| mTOR (serine/threonine kinase) | MyoD stability regulated by mTOR | Regulates miR-1 expression via MyoD availability | Regenerating mouse skeletal muscle and differentiating myoblast cells | [32] |
| AMPK-CRTC2-CREB and Raptor-mTORC-4EBP1 pathways | mTORC regulates timing of satellite cell proliferation during myogenesis | Knockdown of mTORC reduces miR-1 expression | Myogenenic satellite SCs proliferating and differentiating into myogenic precursors following rat skeletal muscle injury | [58] |
| HDAC4 regulates Pax7-dependent muscle regeneration | Pax7 stimulates SCs differentiation toward the muscle lineage, and limits adipogenic differentiation | HDAC4 upregulated in SCs differentiating into muscle cells | Myogenenic satellite SCs | [175] |
| pcRNA encoded by the H strand of the rat mitochondrial genome | Introduction of mt pcRNAs into injured muscle restoring mitochondrial mRNA levels; Intramuscular ATP levels were elevated after pcRNA treatment of injured muscle | Enhanced organellar translation and respiration; similarly reactive oxygen species were reduced; Resulted in accelerated rate of wound resolution | Injured rat skeletal muscle is associated with general downregulation of mitochondrial function; reduced ATP, and increased ROS | [176] |
| Cardiac muscle precursor cells | ||||
| GATA binding protein 4, Hand2, T-box5, myocardin, and microRNAs miR-1 and miR-133 | Reprogrammed human fibroblasts show sarcomere-like structures and calcium transients; Some cells have spontaneous contractility | Forced over-expression of GATA binding protein 4, Hand2, T-box5, myocardin, and microRNAs miR-1 and miR-133 | Human embryonic and adult fibroblasts activated to express cardiac markers | [15] |
| SRF, MyoD and Mef2 transcription factors | miR-1-1 and miR-1-2 | miR-1 genes upregulated; | Cardiac muscle precursor cells | [30] |
| During cardiogenesis miR-1 genes titrate critical cardiac regulatory proteins, control ratio of differentiation to proliferation | Elevated miR-1 targets downregulation of Hand2 | |||
| Histone deacetylase inhibitor, trichostatin A forces differentiation, yet reduced miR-1 and miR-133a | miR-1 and miR-133a reduce cardiac specific Nkx2.5 protein and Cdk9 | miR-1 and miR-133a increase during spontaneous differentiation of cardiac myoblasts | Mouse cardiac stem cells (ES cells) | [10] |
| Specific inhibition of HDAC4 modulates CSCs to facilitate myocardial repair | Positively proliferative myocytes increased in MI hearts receiving HDAC4 downregulated CSCs | CSCs with downregulated HDAC4 expression improved ventricular function, attenuated ventricular remodeling, promoted regeneration and neovascularization in MI hearts | Mouse CSCs transplanted into MI mouse hearts | [177] |
| Snai1 | Overexpression of miR-133a (miR-133), Gata4, Mef2c, and Tbx5 (GMT) or GMT plus Mesp1 and MyocD improved cardiac cell reprogramming from mouse or human fibroblasts | miR-133a directly represses Snai1 expression, which silences fibroblast signatures; a key molecular process during cardiac reprogramming | Mouse/human fibroblasts more efficiently reprogrammed into cardiomycete-like cells | [16] |
| β1AR signal transduction cascade | Adenylate cyclase VI and the catalytic subunit of the cAMP-dependent PKA are components of β1AR transduction cascade | miR-133 directly targets β1AR, Adenylate cyclase VI and PKA | TetON-miR-133 inducible transgenic mice, subjected to transaortic constriction, maintained cardiac performance with attenuated apoptosis and reduced fibrosis via elevated miR-133 expression | [17] |
| ROS, MDA, SOD and GPx | miR-133 produced a reduction of ROS and MDA levels, and an increase in SOD activity and GPx levels | Overexpression of miR-133, a recognized anti-apoptotic miRNA | In vitro rat cardiomyocytes | [18] |
| Caspase-9 | miR-133 directly suppresses caspase-9 expression resulting in downregulation of downstream apoptotic pathways | Overexpression of miR-133 | In vitro rat cardiomyocytes | [18] |
| Spred1 | miR-1 directly targets Spred1 | miR-1 is upregulated in hCMPCs during angiogenic differentiation | hCMPCs | [178] |
| miRNA-1 and miRNA-133a | miRNA-1 and miRNA-133a have antagonistic roles in the regulation of cardiac differentiation | Forced overexpression of miR-1 alone enhanced cardiac differentiation, in contrast overexpression of miR-133a reduced cardiac differentiation, compared to control cells | Pluripotent P19.CL6 stem cells | [179] |
| Overexpression of both miRNAs promoted mesodermal commitment and decreased expression of neural differentiation markers | ||||
| Cardiac muscle | ||||
| Induction of GATA6, Irx4/5, and Hand2 | Cardiac myocytes show defective heart development, altered cardiac morphogenesis, channel activity, and cell cycling | miR-1-2-/- gene knockout | Cardiac myocytes with knockout of both miR-1-2 genes | [180] |
| mt-COX1 mRNA | 3’-UTR of mt-COX1 mRNA bound by miR-181c and Ago1 factor | Overexpression of miR-181c significantly decreased mt-COX1 protein, but not mt-COX1 mRNA level | Overexpression of miR-181c increased mitochondrial respiration and reactive oxygen species in neonatal rat ventricular myocytes | [54] |
| mt-COX1 mRNA | In vivo elevation of miR-181c in rat heart, reduces levels of mt-COX1 protein | Results in reduced capacity for strenuous exercise and evidence of heart failure | Rat cardiac muscle | [55] |
| Carvedilol, a β-adrenergic blocker | Induces upregulation of miR-133 | Cytoprotective effects against cardiomyocyte apoptosis | Rat cardiac tissue, in vivo | [18] |
| GLUT4, and SRF | Both miRs downregulate SRF and KLF15 | Both miR-133a and miR-133b target KLF15 | Mouse cardiac myocytes | [181] |
| GLUT4 expression | Both basal and insulin-stimulated glucose uptake are increased | KLF15 | Mouse muscle cell lines | [182] |
| MEF2 transcription factor | MEF2 directly activates transcription of miR-1-2 and 133a-1 binding muscle-specific enhancer between the genes | Bicistronic primary transcript of miR-1-2 and 133a-1 | Development of mammalian cardiac muscle | [9] |
| Myocardium tissue | Enriched in miR-1, miR-133b, miR-133a | Heart structures of rat, Beagle dog and cynomolgus monkey | [183] | |
| Gelsolin | One common miR-133a isomiR targets gelsolin gene more efficiently than standard isomer; New second rat miR-1 gene | Many isomiRs were detected by deep sequencing at higher frequency than the canonical sequence in miRBase | miRNA/isomiR expression profiles in the left ventricular wall of rat heart | [184] |
| CTGF | CTGF downregulated by both miRs | Exogenous upregulation of miR-133b (and miR-30c) | Cultured cardiomyocytes and ventricular fibroblasts | [185] |
| MT1-MMP | miR-133a upregulated | miR-133a targets MT1-MMP | Human left ventricular fibroblasts | [186] |
| Injured and regenerating cardiac muscle | ||||
| SERCA2a | Akt/FoxO3A-dependent pathway | Downregulation of miR-1 expression in failing heart muscle | Failing mouse heart muscle | [187] |
| Activated SERC2a reduces phosphorylation of FoxO3a, allowing entry to nucleus and activation of miR-1 expression | ||||
| IGF-1 | IGF-1 signalling and miR-1 co-regulate differentiation of myoblasts via a feedback loop | IGF-1 signalling down-regulates miR-1 by repression of FoxO3a; | Mouse heart muscle during cardiac failure states | [25] |
| miR-1 down-regulates IGF-1 | ||||
| Bim and Bmf | Only miR-133a expression enhanced under in vitro oxidative stress | miR-133a targets proapoptotic genes Bim and Bmf | Rat adult CPCs | [188] |
| miR-1 favors differentiation of CPCs, whereas | ||||
| Bim and Bmf | CPCs overexpressing miR-133a improved cardiac function by reducing Bim and Bmf | CPCs overexpressing miR-133a improved cardiac function, increasing vascularization and cardiomyocyte proliferation, reduced fibrosis and hypertrophy | CPCs overexpressing miR-133a in rat myocardial infarction model | [188] |
| MT1-MMP activity increased in both. Ischemia and reperfusion regions | Interstitial miR-133a decreased with ischemia in vitro and in vivo; reperfusion returned to steady-state | Phosphorylated Smad2 increased within the ischemia-reperfusion region | Ischemia-reperfusion Yorkshire pigs (90 min ischemia/120 min reperfusion) | [186] |
| Cardiovascular disease | ||||
| CNN2 | Strong upregulation of CNN2 expression | miR-133b downregulated; miR-133b directly targets CNN2 | Pre-inflammatory events in diseased cardiac tissues | [65] |
| Circulating platelet derived microparticles | Elevated miR-133 | Patients with stable and unstable coronary artery disease | [189] | |
| Acute MI causes upregulation of circulating serum miRs | miR-1, -133a, -133b, and -499-5p were about 15- to 140-fold elevated over control | Acute STEMI patients and experimental mouse MI model | [190] | |
| Circulating miRNAs in serum of cardiovascular disease patients | Released miR-1 and miR-133a are localized in exosomes, and are released by Ca(2+) stimulation | Levels of miR-1, miR-133a, reduced in infarcted mouse myocardium model heart | miR release indicates myocardial damage | [191] |
| LVM after valve replacement in aortic stenosis | microRNA-133a is a significant positive predictor of LVM normalisation | miR-133 is a key element of the reverse remodelling process | Patients following valve replacement | [192] |
| Circulating levels of miR-133a | Elevated miR-133a (11-fold) | Troponin-positive acute coronary syndrome patients | [193] | |
| Circulating levels of miR-133a | Elevated miR-133a | Improved potential regression of Left Ventricular Hypertrophy after valve replacement | Patients with aortic stenosis surgery | [194] |
| Apelin treatment reduces elevated circulating miRs | Elevated miR-133a, miR-208 and miR-1 reduced | High-fat diet elevated miRs and increased left ventricular diastolic and systolic diameters, and wall thickness | Obesity-associated cardiac dysfunction in mouse model | [195] |
| NAC treatment | Expressed miR-1, miR-499, miR-133a, and miR-133b were strongly depressed in the diabetic cardiomyocytes | NAC restored expression of miR-499, miR-1, miR-133a, and miR-133b significantly in the myocardium | Diabetic rat hearts | [196] |
| Myocardial junctin elevated | miR-1 targets junctin | NAC reduces junction levels | Development of diabetic cardiomyopathy in rat hearts | [196] |
| CAD associated ischemic heart failure | miR-133 expression decreased with increased severity of heart failure | Patients with CAD | [197] | |
| Runx2 | miR-133a targets Runx2 | Transition of VSMCs to osteoblast-like cells | [198] | |
| Increased alkaline phosphatase activity, osteocalcin secretion and Runx2 expression | miR-133a was decreased during osteogenic differentiation | Transition of VSMCs to osteoblast-like cells | [198] | |
| Circulating miR-133a and 208a levels | Cardiac muscle-enriched microRNAs (miR-133a, miR-208a) elevated | Patients with coronary artery disease | [199] | |
| Hypertrophic cardiac muscle | ||||
| Cx43 increased | miR-1 targets Cx43 | Downregulation of miR-1 mediates induction of pathologic cardiac hypertrophy | Hypertrophic rat cardiomyocytes in vitro and in vivo | [200] |
| Cx43 downregulated | miR-1 targets Cx43 | Cx43 protein downregulated in miR-1 Tg mice compared to WT mice | Cardiac-specific miR-1 transgenic (Tg) mouse model | [201] |
| Twf1 upregulated | miR-1 targets Twf1 | Strong downregulation of miR-1 in pathologic hypertrophic cardiac cells compared to normal, induces Twf1 expression | In vivo in hypertrophic mouse left ventricle; and in vitro in phenylephrine-induced hypertrophic cardiomyocytes | [202] |
| RhoA, Cdc42, Nelf-A/WHSC2 | Increased levels of RhoA, Cdc42, Nelf-A/WHSC2 | Reduction miR-133a | Hypertrophic cardiac muscle | [6] |
| Calcineurin, agonist of cardiac hypertrophy | Increased Calcineurin activity; | Reduced miR-133a; | Hypertrophic cardiac muscle; | [203] |
| Cyclosporin A inhibits calcineurin | Prevents miR-133 down-regulation | Cardiac hypertrophy reduced | ||
| NFATc4 | NFAFc4 targetted by miR-133a | miR-133a | Cardiomyocyte hypertrophic repression | [204] |
| Interdependent Calcineurin-NFAT and MEK1-ERK1/2 signalling pathways in cardiomyocytes | MEK1-ERK1/2 signalling augments NFAT and NFAF gene expression; Activated calcineurin activates NFAT, inducing cardiac hypertrophy | MEK1 is part of mitogen-activated protein kinase (MAPK) cascade; MEK1 activates ERK directly | Hypertrophic growth response of mouse cardiomyocytes | [205] |
| Innervating skeletal muscle | ||||
| Innervated skeletal muscle | MyoD, Myf5, Mrt4, nAChRα | Myogenin expression | Mouse skeletal muscle | [50,51] |
| Each is strongly repressed | ||||
| Denervated muscle (unstimulated) | Myogenin expression up-regulated MyoD, Myf5, Mrt4, nAChRα | Mouse skeletal muscle | [51] | |
| All strongly stimulated | ||||
| Electrically stimulated - Denervated muscle | Myogenin, MyoD, Myf5, Mrt4, partly stimulated; nAChRα inhibited | Mouse skeletal muscle | [51] | |
| HDAC4 | miR-1 promotes myogenesis by targetting HDAC4 | miR-133 enhances myoblast proliferation by targetting SRF | Skeletal muscle proliferation and differentiation in myoblast cultures | [7] |
| SRF | ||||
| Neural activity effect on muscle (HDAC4 - MEF2 Axis) | Loss of neural input leads to concomitant nuclear accumulation of HDAC4 | HDAC4 inhibits activation of muscle transcription factor MEF2; results in progressive muscle dysfunction | MEF-2 activity strongly inhibited in denervated mouse skeletal muscle and in ALS muscle | [49] |
| Innervation and formation of airway smooth muscle | Sonic hedgehog (Shh) /miR-206/ BDNF | Shh signalling blocks miR-206 expression, which in turn increases BDNF protein | Shh coordinates innervation and formation of airway smooth muscle | [206] |
| nAChR subunits (UNC-29 and UNC-63); retrograde signalling | Subunits UNC-29, UCR-63 and MEF2 downregulated | miR-1 upregulated | C. elegans muscle at the neuromuscular junction | [34] |
| MEF2 | Hnrpu, Lsamp, MGC108776, MEF2, Npy, and Ppfibp2 downregulated | miR-206 upregulated | Rat skeletal muscle/re-innervating muscle | [43] |
| HDAC4 | HDAC4 (miR-206 target, prospective miR-133b target) downregulated | miR-206/-133b upregulated (and miR-1/-133a downregulated) | Mouse fast twitch skeletal muscle/re-innervating muscle | [12] |
| Regenerating injured muscle | ||||
| Hnrpu and Npy downregulated | miR-1 upregulated | miR-1, -133a, downregulated 1 mo after denervation, then increased 2 × at 4 mo after re-innervation | Rat skeletal soleus muscle after sciatic nerve injury and subsequent re-innervation | [43] |
| Ptprd downregulated | miR-133a upregulated | |||
| Hnrpu, Lsamp, MGC108776, MEF2, Npy, and Ppfibp2 downregulated | 3 × increase in miR-206 1 mo later, after reinnervation; elevated at least 4 mo | Predominant type II fiber at 4 mo, after nerve re-innervation | Rat skeletal soleus muscle after sciatic nerve injury and subsequent re-innervation | [43] |
| PP2A B56a | PP2A B56a downregulated | 133a upregulated | Canine heart failure model: myocytes | [207] |
| CaMKII-dependent hyperphosphorylation of RyR2 | VF myocytes had increased reactive oxygen species and increased RyR oxidation | miR-1 upregulated | Canine post-myocardial infarction model | [208] |
| Collagen upregulated | TGF-b1 and TGFbRII: upregulated | miR-133a or miR-590: downregulated | Canine model of acute nicotine exposure. Atrial fibrosis in vivo; cultured canine atrial fibroblasts in vitro | [209] |
| miR-208 upregulated | miR-1 and miR-133a downregulated | Human MI compared to healthy adult hearts | [210] | |
| Myogenic proteins, MyoD1, myogenin and Pax7 | Induced expression of MyoD1, myogenin and Pax7 several days after miR injection | Exogenous injection of miR-1, -133 and -206 promotes myotube differentiation | Regenerating injured mouse skeletal muscle | [211] |
| Cyclin D1/ Sp1 | Cyclin D1/ Sp1 downregulated | miR-1/133 upregulated | Regenerating rat skeletal muscle | [212] |
| PRP, source of pro-inflammatory cytokines | Stong upregulation of the mRNA of pro-inflammatory cytokines IL-1β and TGF-β1; stimulation of both inflammatory and myogenic pathways; elevated heat shock proteins and increased phosphorylation of αB-cristallin | Stimulated tissue recovery via increased myogenic regulators MyoD1, Myf5, Pax7, and IGF-1Eb (muscle isoform) together with SRF; acts via increased expression of miR-133a with reduced levels of apoptotic factors (NF-κB-p65 and caspase 3) | Regenerating flexor sublimis muscle of rats, 5 d after injury and treated with PRP | [66] |
| Muscle degeneration | ||||
| Pro-inflammatory cytokine TWEAK | TWEAK upregulated | miR-1-1, miR-1-2, miR-133a, miR-133b and miR-206 downregulated | Degenerating/wasting mouse skeletal muscle | [59] |
| HMOX1 mediated by codependent inhibition of c/EBPδ binding to myoD promoter | HMOX1 inhibits differentiation of myoblasts and modulates miRNA processing | Downregulation of miR-1, miR-133a, miR-133b, and miR-206. | Degenerating/wasting mouse skeletal muscle | [60] |
| HMOX1 effects partially reversed by enforced expression of miR-133b and miR-206 | Downregulation of MyoD, myogenin and myosin, and disturbed formation of myotubes. Upregulation of SDF-1 and miR-146a | |||
| Dystrophic muscular disease | ||||
| Circulating serum microRNAs | miR-1, miR-133a, and miR-206 highly abundant in Mdx serum | miR-1, miR-133a, and miR-206 downregulated or modestly upregulated in muscle | Muscle tissue from patients with Duchenne muscular dystrophy (Mdx) | [213] |
| Laminin α2 chain deficiency | miR-1, miR-133a, and miR-206 are deregulated in laminin α2 chain-deficient muscle | Laminin α2 chain-deficient mouse | Congenital muscular dystrophy type 1A tissue | [214] |
| Dystrophic process advances from prominent inflammation with necrosis and regeneration to prominent fibrosis | Deficiency in calpain leads initially to accelerated myofiber formation followed by depletion of satellite cells | Pax7-positive SCs highest in the fibrotic patient group; correlated with down-regulation of miR-1, miR-133a, and miR-206 | Muscle from Limb-girdle muscular dystrophy 2 type I patients | [215] |
| Transgenic overexpression of miR-133a1 (in dystrophin point mutation Mdx mice) | Extensive overexpression in skeletal muscle, lesser increase in heart | Normal skeletal muscle and heart development | Mdx mice (model for human muscular dystrophy), extensor digitorum longus muscle | [216] |
| miR-206 located in nuclear in both normal and DM1 tissues by in situ hybridization | Only miR-206 showed an over-expression in majority of DM1 patients | No change in expression of profiled miRs, miR-1, miR-133 (miR-133a/-133b), miR-181 (miR-181a/-181b/-181c) | Skeletal muscle (vastus lateralis) of from patients with myotonic dystrophy type 1 (DM1) | [217] |
| FAPs facilitate myofiber regeneration | HDAC inhibitors can activate FAPs towards muscle regeneration | Inhibition of HDAC induces MyoD and BAF60C expression, which causes up-regulation of miR-1-2, miR-133, and miR-206 expression | Early stage disease dystrophic mouse muscles, regeneration of myofibres | [62] |
| TDP-43 | TDP-43 interacts with miR-1/-206 isomers, but not miR-133 isomers | Depleted miR-1/-206 allow targets IGF-1 and HDAC4 to accumulate in ALS muscle | Mouse ALS model injured motor neurons and muscle | [33] |
| Inflammation response in muscle | ||||
| Inflammatory myopathies | Increased expression of TNFα | Associated with decreased expression of miR-1, miR-133a, and miR-133b | Inflammatory myopathies including dermatomyositis, polymyositis, and inclusion body myositis | [64] |
| hBSMCs sensitized with IL-13 | Increased muscle RhoA | Reduction of muscle miR-133a | Sensitized human bronchial smooth muscle cells (hBSMCs) | [218] |
| Factor(s) | Regulation | Regulator | Tissue/cell | Ref. |
| Nerve tissues | ||||
| Pitx3 | Pitx3 downregulated | miR-133b | Mammalian midbrain DNs | [73] |
| Exosome-mediated transfer of miR-133b from MSC to brain astrocytes | miR-133b transfer from multipotent mesenchymal stromal cells to neural cells | miR-133b upregulated | Mouse MSCs to neural cells | [47] |
| Ctgf and RhoA | Ctgf and RhoA downregulated | miR-133b upregulated | Multipotent MSCs/Rat brain parenchymal cells | [72] |
| miR-133b null mice: Striatum dopamine levels unchanged, Pitx3 expression unaffected; motor coordination unaltered | miR-133b has no significant role on mDA neuron development and maintenance in vivo | Normal numbers of mDA neurons during development and aging of miR-133b null mice | Mouse mDA neuron development in -/-miR-133b mutant mice | [45] |
| Acute or chronic morphine administration, or morphine withdrawal | miR-133b levels not affected | Rat VTA/ nucleus accumbens shell | [219] | |
| GPM6A, a neuronal glycoprotein | microRNA-133b upregulation | Reduction in gmp6a at mRNA and protein level. Cell filopodium density was reduced | Hippocampus and prefrontal cortex of neonatal male rats stressed when in utero | [220] |
| Tac1 gene (neurotransmitter substance P) | Tac1 downregulated | miR-206 upregulated | MSCs-derived neural cells | [221] |
| Ketamine (antidepressive) administration | BDNF, a direct target gene of miR-206, was upregulated | miR-206 was downregulated by ketamine | Rat hippocampus tissue | [222] |
| Adipogenic tissues | ||||
| IGF-1 and IGF-1R | IGF-1 signalling and miR-133b co-regulate ADSC differentiation via a feedback loop | miR-133b downregulation of Pitx3; | Adipose tissue-derived stem cell differentiation into neuron-like cells | [71] |
| IGF-1 upregulates miR-133b; | ||||
| miR-133b downregulates IGF-1R | ||||
| Pdrm16 | miR-133a directly targets Prdm16. | Downregulation of miR-133 resultsin differentiation of pre-adipocyte precursors into BAT | Mouse adipocyte differentiation to BAT | [74] |
| Pdrm16 | miR-133 directly targets Prdm16 | Downregulation of miR-133 resulted in differentiation of pre-adipocyte precursors into BAT | Mouse primary brown adipocyte (and myogenic) progenitor cells - differentiate into BAT or SAT | [75] |
| Pdrm16 | miR-133 targets Prdm16 controlling brown adipose determination in skeletal muscle satellite cells | miR-133 downregulates Prdm16 | Adult mouse skeletal muscle stem cells (satellite cells) differentiate into BAT | [76] |
| HDAC4 downregulation directs SCs towards adipocyte differentiation | Brown adipose master regulator Prdm16 is upregulated, while its inhibitor miR-133 is also downregulated | HDAC4 downregulated in SCs differentiating into adipocyte progenitor cells | Myogenenic satellite SCs | [175] |
| GLUT4 expression | Both basal and insulin-stimulated glucose uptake are increased | KLF15 | Mouse 3T3-L1 preadipocytes differentiating into adipocytes | [182] |
| Intrinsic insulin resistance | Elevated miR-133b | Undefined role | Adipose tissue of women with PCOS | [223] |
| Upregulation of LIM homeobox 8 and Zic family member 1 and downregulation of Homeobox C8 and Homeobox C9 | Undefined relation of upregulated miR-206, miR-133b | Undefined relation with parallel upregulation of brite/beige markers, TBX1 and TMEM26 | Human BAT from the supraclavicular region | [224] |
| Obesity development | Downregulation of miR-133b, miR-1 | Undefined role | Adipose tissue from obese male C57BLJ6 mice | [225] |
| LXRα regulation of lipogenic genes | miR-1/miR-206 represses LXRα expression at both mRNA and protein levels | miR-1/miR-206-induces a decrease in lipogenic gene levels and lipid droplet accumulation | Mouse hepatocytes | [226] |
| Osteogenic tissues | ||||
| Development of bone on organic or inorganic substrates | miR-133 differentially expressed in osteoblasts grown on different substrates | Osteoblast | [227] | |
| Runx2 | miR-133 directly down-regulates Runx2 | miR-133 up-regulated | Osteogenic differentiation from C2C12 mesenchymal cells | [228] |
| HDAC4 | HDAC4 downregulates Runx2 | miR-1 targets HDAC4, increasing Runx2 activity | Chondrocyte proliferation in cartilage growth plate | [77] |
| Aggrecan | miR-1 promotes late-stage differentiation of growing cartilage cells | miR-1 targets Aggrecan gene expression | Chicken chondrocytes and human HCS-2/8 cells | [78] |
| Alveolar cells | ||||
| VAMP2/ lung surfactant secretion | miR-206 targets VAMP-2 | miR-206 overexpression decreased lung surfactant secretion | Lung alveolar type II cells | [229] |
| Hormonal regulation | ||||
| L-thyroxine | miR-206/miR-133b downregulated | L-thyroxine treatment | L-thyroxine treated hypothroidic skeletal muscle from thyroidectomized patients | [230] |
| miR-206/miR-133b upregulated | - | Hypothroidic human skeletal muscle | ||
| Thyroid hormone/TEAD1 | Thyroid hormone inhibits the slow muscle phenotype by upregulation of miR-133a1 which downregulates TEAD1 | miR-133a1 is enriched in fast-twitch muscle and regulates slow-to-fast muscle fiber type conversion | Mouse muscle | [231] |
| Thyroid hormone/miR-133a1 TEAD1 | myosin heavy chain I expression downregulated | TH indirectly downregulates myosin heavy chain I via miR-133a/TEAD1 | Mouse muscle | [232] |
| L-thyroxine | pre-miR-206 and pre-mir-133b downregulated | L-thyroxine | L-thyroxine treated hypothyroidic mouse liver; | [232] |
| 50-500x increase expression of miR-1/-133a and miR-206/-133b | - | Hypothyroidic mouse liver | ||
| Reduced insulin-mediated glucose uptake in cardiomycetes | Downregulation KLF15, which downregulates GLUT4 | Forced overexpression of miR-133a and miR-133b | Rat cardiac myocytes | [181] |
| Cardiac myocyte glucose metabolism | Upregulation KLF15, which upregulates GLUT4 | Silencing endogenous miR-133 | Rat cardiac myocytes | [181] |
| Metabolic control of glucose uptake by GLUT4 transporter | Downregulates KLF15, which results in downregulation of GLUT4 levels | Chronic heart failure has depressed miR-133a and -133b levels | Rat cardiac myocytes during chronic heart failure and cardiac hyperthrophy | [181] |
| Atrial natriuretic factor expression upregulation | Enhanced at LVH and dramatically increased at CHF stage | Both miR-133a and miR-133b downregulated at CHF stage | LVH and CHF in salt-sensitive Dahl rats | [181] |
| Estrogen | Estrogen replacement strongly decreased IGF-1 protein level in muscles at 1 wk | Ovariectomized rat skeletal muscle | [233] | |
| Multiple targets | miR-133a upregulated in BTBR mice | Pancreatic islets, adipose tissue, and liver from diabetes-resistant (B6) and diabetes-susceptible (BTBR) mice | [234] | |
| Augmentation of adipocyte differentiation by norepinephrine does not alter myomiR levels | miRNAs miR-1, miR-133a and miR-206 specifically expressed both in brown pre- and mature adipocytes | miRNAs miR-1, miR-133a and miR-206 were absent from white adipocytes | Mouse brown adipocytes | [235] |
| Foxl2 | miR-133b targets Foxl2; | Foxl2 regulates StAR and CYP19A1 transcriptionally | Estradiol production in ovarian granulosa cells | [236] |
| miR-133b inhibits Foxl2 binding to StAR and CYP19A1 promoter sequences | ||||
| Exosome release and cell to cell transfer | ||||
| Exosome-mediated transfer of miR-133b from MSCs to brain astrocytes | miR-133b transfer from multipotent mesenchymal stromal cells to neural cells | miR-133b upregulated | Mouse multipotent MSCs to neural cells | [47] |
| Cell to cell transfer of exosome-enriched extracellular particles | mir-133b promotes neural plasticity and recovery of function after stroke induced damage | miR-133b upregulated | Rat multipotent MSCs via transfer of exosome-enriched extracellular particles | [72] |
| Transplanted stem cells | ||||
| MSCs expressing miR-1 | Upregulated miR-1 | Increased rate of recovery, enhanced survival of transplanted MSCs and cardiomyogenic differentiation | Experimental ligation of the mouse left coronary artery to model myocardial infarction | [237] |
| Knockdown of Hes-1, member of Notch pathway | Upregulated miR-1 promotes the differentiation of MSCs into cardiac lineage | Role in survival of transplanted MSCs and cardiomyogenic differentiation | Mouse MSCs | [238] |
| Notch signalling and cardiomyocyte markers, Nkx2.5, GATA-4, cTnT, and Cx43 | MSCs expressing exogenous miR-1 | Mouse MSCs | [238] | |
| Tissue inflammation | ||||
| Selective release of miRs during inflammation into serum | miR-133 selectively released | Review | [239] | |
| Inflammation and cancer | MicroRNA, free radical, cytokine and p53 pathways | Review | [240] | |
| Immunological switch which shapes tissue responses | TWEAK/Fn14 pathway | Review | [241] | |
| Tumor biology | HMOX1 | Review | [242] | |
| GM-CSF | Direct supression of GM-CSF expression by miR-133 | Elevated expression of miR-133a/-133b during oxidative stress | Mouse alveolar epithelial cells during oxidative stress | [82] |
| PI3K/Akt and IGF-1 pathways | Activation of PI3K/Akt and IGF-1 pathway activities | Downregulation of miR-133a (and other miRs) by AOM/DSS induced chronic inflammation | Mouse model: AOM/DSS-induced colitis-associated gastro-intestinal cancer | [83] |
| CTGF, SMA, and COL1A1 | Increased expression of CTGF, SMA and COL1A1, which are miR-133b targets | Strong downregulation of miR-133b (and other miRs) | TGF-β treated rabbit corneal fibroblasts; Recovering mouse cornea after laser ablation, | [70] |
| IL-10 and TGF-β | Exogenous IL-10 and TGF-β induces miR-133b expression | Upregulation of miR-133b | Human tolerogenic dendritic cells during maturation | [79] |
| IL-17-producing T-cells | Upregulation of Il17a/f gene expression | miR-133b/-206 cistron transcription occurs along with nearby Il17a/f gene expression | Immunocompetent mouse Th17 cells | [80] |
| NLRP3 inflammasome which processes IL-1β by caspase-1 cleavage | miR-133a-1 suppresses activation of inflammasomes via suppression of expression of mitochondrial UCP2 | miR-133a-1 overexpression in cells increases caspase-1 p10 and IL-1β p17 cleavage, | Differentiated mouse THP1 cells | [81] |
| Concanavalin A-induced fulminant hepatitis | miR-133a is the most strongly differentially upregulated miR | Mouse liver following ConA injection | [243] | |
| Infection/immune response to influenza virus (H1N2) | miR-206 expression | Experimental influenza infection in pig lung | [244] | |
| HIF-1α, and its regulator Four-and-a-half LIM (Lin-11, Isl-1 and Mec-3) domain 1 (Fhl-1) | Downregulation of miR-206 and upregulated HIF-1α and Fhl-1 in hypoxic lung tissue and PASMCs | miR-206 targets HIF-1α directly. Hypoxia-induced down-regulation of miR-206 promotes PH in PASMCs | Hypoxia-induced PH in hypoxic rat model in cultured hypoxic PASMCs | [245] |
| miR-206/NR4A2/NFKB1; | NFKB1 stimulates inflammatory cytokines (IL6, IL1B, CCL5) | Liposaccharides induce miR-206 expression which targets NR4A2 downregulation, which in turn allows upregulation of NFKB1 activity | Astrocyte-associated inflammation during recovery from chronic central nervous system injury | [246] |
| Indirectly: inflammatory cytokines (IL6, IL1B, CCL5) | ||||
| Cellular factors influencing myomir expression/activity | ||||
| miR-1/miR-133a | ||||
| Skeletal muscle | ||||
| Positive regulator | Negative regulator | Regulated target miR | Tissue/cell | Ref. |
| Myogenin, MyoD | Upregulates miR-1-1 and miR-133a-2 | Primary human myoblasts; C2C12 cells | [11] | |
| Upregulates miR-1-2 and miR-133a-1 | ||||
| SRF, MyoD and MEF2 | Upregulates miR-1-2 | Muscle somites | [30] | |
| MEF2 | Upregulates miR-1 and miR-133a | Skeletal muscle | [9] | |
| KSRP (part of Drosha and Dicer complexes) | miR-206 binds 3’-UTR of KSRP and inhibits its expression | KSRP upregulates miR-1 expression | Skeletal muscle | [35,37] |
| RNA-binding protein LIN28 | LIN28 upregulates miR-1 expression; LIN28 promotes pre-miR-1 uridylation by ZCCHC11 (TUT4) | Cardiac muscle of patients with muscular dystrophy | [36] | |
| MBNL1 | MBNL1 downregulates miR-1 expression; MBNL1 binds to UGC motif in the loop of pre-miR-1 and competes for the binding of LIN28; MBNL1 blocks DICER processing of pre-miR-1 | Cardiac muscle of patients with muscular dystrophy | [36] | |
| CX43 and CACNA1C calcium channel | CX43 and CACNA1C both increased in both DM1-/DM2-affected hearts, contributing to the cardiac dysfunctions | CX43 and CACNA1C are direct targets of miR-1 repression | Cardiac muscle of patients with muscular dystrophy; | [36] |
| CACNA1C and CX43 encode the main calcium- and gap-junction channels in heart | ||||
| Utrophin A | miR-206 and KSRP are negative regulators of utrophin A | Overexpression of miR-206 promotes the upregulation of utrophin A, via the downregulation of KSRP | Normal and dystrophic muscle cells; | [37] |
| miR-206 can switch between (1) direct repression of utrophin A expression, and (2) activation of its expression by decreasing KSRP, allowing close regulation | ||||
| Myostatin | Downregulates miR-1, miR-133a, miR-133b, miR-206 | Mouse (35 d) pectoralis skeletal muscle | [29] | |
| SRF | Downregulates miRs-133a | Skeletal muscle | [1,3] | |
| Prmt5 and Prmt4 | Upregulates myomiR expression during differentiation | Mouse skeletal muscle | [247] | |
| Smooth muscle | ||||
| Sp-1 transcription factor | pERK1/2 | Upregulates miR-133(a) | VSMCs | [248] |
| Brg1 | Upregulates miR-133 (ChIP complex with SRF) | Smooth muscle | [249] | |
| Cardiac muscle | ||||
| GATA4, Nkx2.5, Myocardin, SRF | Upregulates miR-1 and miR-133a | Differentiating cardiac muscle | [5] | |
| SRF plus Myocardin | Upregulates miR-1-1 and miR-1-2 | Cardiomycetes | [30] | |
| Calcineurin | Downregulates miR-133a | Hypertrophic cardiac muscle | [203] | |
| miR-206/ miR-133b | ||||
| Skeletal muscle | ||||
| Mrf5 | Upregulates miR-1, miR-206 | Skeletal muscle | [171] | |
| Myogenin, MyoD | Upregulates miR-206 | Primary human myoblasts; C2C12 cells | [11] | |
| MyoD | Upregulates linc MD1 (encodes miR-133b) | Differentiating myoblasts | [11, 38] | |
| Binds to (E-box) enhancer of miR-206, miR-133b | skeletal muscle (mouse) | [12,40] | ||
| Upregulates miR-206/miR-133b | Differentiated human foetal skeletal muscle cells | [250] | ||
| FGF2 allows upregulation of Sp1/Cyclin D1 | Downregulates p38-mediated miR-1/133 expression | Regenerating rat skeletal muscle | [212] | |
| Myostatin | Downregulates miR-133a, mir-133b, miR-1, and miR-206 | Mouse (35 d) pectoralis skeletal muscle | [29] | |
| TWEAK downregulates myoD and MEF2c | Downregulates miR-1-1 and miR-133 | Degenerating/wasting skeletal muscle | [59] | |
| HMOX1 downregulates MyoD and myogenin | Downregulates all myomiRs | Inflamed skeletal muscle | [60] | |
| L-Thyroxine treatment | Downregulation of pri-miR-206 and pri-miR-133b | Human skeletal muscle | [230] | |
| No effect on miR-1/miR-133a pairs | ||||
| Smooth muscle | ||||
| p-ERK | Activated extracellular signal-regulated kinase p-ERK inversely correlated with VSMC growth | Downregulates miR-133 expression | VSMCs | [248] |
| Other tissues | ||||
| Myogenin | Binds miR-206 enhancer (ChIP) | Fibroblast cell line: | [40] | |
| IGF-I signalling | Upregulates miR-133b | Mouse Adipose derived stem cells | [71] | |
| L-Thyroxine deficiency | Upregulated Col5a3 | Strong upregulation of miR-133a and -133b | Hypothyroid mouse liver | [232] |
| Downregulated Slc17a8, Gp2, Phlda1, Klk1d3, Klk1 and Dmbt1 | Strong upregulation of miRs -1, -206 | |||
| Upregulated Vldlr and Akr1c19, and downregulated Upp2, Gdp2, Mup1, Nrp1, and Serpini2 | ||||
| L-Thyroxine treatment | Pre-miR-206 and Pre-miR-133b down-regulated | Upregulation of Gdp2 andMup1 | Hypothyroid mouse liver in vivo, and in vitro mouse hepatocyte AML12 cells | [232] |
| PA2G4, mps1, cdc37, cx43, cldn5; cx43 is a miR-133 target | Upregulation of cell cycle factors mps1, cdc37, and PA2G4, and cell junction components cx43 and cldn5 | Suppression of miR-133a1 stimulates cardiac cell proliferation | Regeneration of damaged Zebrafish cardiac muscle, associated with reduced miR-133a1 | [167] |
| Fgf | Upregulated Fgf | Downregulates miR-133 | Zebrafish regenerating fin blastema | [67] |
| SHP (nuclear receptor) | Downregulation of miR-206 in nuclear receptor SHP(-/-) mice | SHP(-/-) mice strain, mouse liver | [251] | |
| AP1 transcription factor complex | AP1 induced miR-206 promoter transactivity and expression; this is repressed by YY1 | ChIP analysis shows physical association of AP1 (c-Jun) and YY1 with miR-206 promoter | SHP(-/-) nuclear receptor mice strain, mouse liver | [251] |
| NR3B3 | YY1 promoter transactivated by ERRgamma; this inhibited by SHP (NROB2) | Nuclear receptor ERRgamma (NR3B3) binding site on the YY1 promoter | Mouse liver | [251] |
| Novel cascade "dual inhibitory" mechanism governing miR-206 gene transcription by SHP | (1) SHP inhibition of ERRgamma leads to decreased YY1 expression | (2) Derepression of YY1 on AP1 activity, leads to activation of miR-206 | Mouse liver | [251] |
| Il17a/f locus | miR-133b and miR-206 expression | Coregulated with IL-17 production | αβ and γδ T cells | [80] |
Others have noted that the canonical myomiRs act as balanced regulators, often specifying broadly opposing functions. The miRs-1 and -206 are semi-homologous with closely similar mature sequences (and identical seed sequences), and target some genes in common, as well as independent targets. The identical mature seed sequences of miRs-133a and -133b implies they would share many targets in common, yet each of these miRs appear to have distinct cellular functions, with miR-133a expression common to all muscle and miR-133b abundant in all muscle types, except cardiac muscle. Loosely, the cell signalling pathways targeted by miR-1/-206 tend to have opposing functions to the regulatory pathways targeted by miR-133a/-133b. Both miR-1/ -206 act to promote myogenic differentiation, while the miR-133 isomers maintain the undifferentiated state and promote cell growth; hence co-expression of the myomiRs likely aids maintenance of homeostasis under normal cellular conditions.
This difference in expression of the related myomiR members in cardiac muscle compared to skeletal muscle may be associated with the physiological specialization of cardiac muscle, or its greater constancy of fibre type and function. In contrast, skeletal muscles constitute a variety of differentiated fibre types and are more plastic, capable of undergoing marked changes in myofibre content and physiology related to the level of use and workload[1,3]. As understanding of the molecular regulation of muscle types have deepened, it is clear that the physiological and functional specializations are also reflected in the functions of the myomiRs.
Studies with mammalian stem cells reveal broad functions for the myomiRs in the definition of primary differentiation pathways. Both miR-133 and miR-1 have roles in early cell programs leading to differentiation of muscle[2,10,13]. Pluripotent mammalian embryonic stem (ES) cells undertake cell fate decisions controlled by activation and repression of lineage-specific gene sets. These decisions are dictated by signalling networks which progressively narrow and specify the potential of ES cells as differentiation progresses. Muscle specific miR-133(a) and miR-1 both promote mesoderm formation from ES cells and suppress ectoderm and endoderm fates[2], but later during further differentiation into cardiac muscle progenitors, these miRs appear to have opposing regulatory functions[11,13]. Many non-muscle cell genes are repressed by miR-1 and miR-133 during this early ES cell differentiation program, suggesting that these two miRNAs may have general roles to regulate early ES cell-fate decisions from pluripotent cells[13], with miR-1 specifically targeting the translational repression of Dll-1 and Cdk9[10].
In vivo, the deletion of both miR-133a1/2 genes causes lethal cardiac (ventricular-septal) abnormalities in about half of the mouse embryos or neonates, while mice deficient in only one of either miR-133a-1 or -133a-2 have phenotypically normal hearts[14]. Skeletal muscles are normal in both double and single mutant miR-133a mice (dead and surviving), implying that miR-133b can replace the absent miR-133a species in skeletal muscle and continue the regulation of normal development. In double mutant mice lacking all miR-133a, smooth muscle gene expression was activated 2-4 × and cardiomyocytes (but not cardio-fibroblasts) proliferated 2.5 × faster than normal, accompanied by increased expression of miR-133a targets, including PTBP2, CDC42, cell cycle control factors and cyclins D1, D2 and B1[14]. Recently, both adult and neonatal human foreskin fibroblasts were found capable of being reprogrammed towards cardiac muscle by exogenous expression of only several factors, myocardin, HAND2, T-box-5, GATA4, and miR-1, miR-133a and miR-499[15]. These stimulated cells expressed cardiac specific proteins and showed spontaneous contractility, emphasizing the role of these miRs in the control of specific cell development programs via the modulation of specific factor targets. Further, both human and mouse fibroblasts can be reprogrammed to form cardiomyocyte-like cells by overexpression of cardiac transcription factors (Gata4, Mef2c, and Tbx5 (GMT) or GMT plus Mesp1 and Myocd) along with miR-133a, which directly represses Snai1 which normally regulates EMT processes[16]. Interestingly, exogenous miR-133b can also downregulate Snai1 expression, suppressing fibroblast genes and upregulate the expression of a number of characteristic cardiac cell genes in vitro, yet it cannot replace miR-133a during normal heart development in vivo.
Heart contractility and heart rate are stimulated during chronic pressure overload by activation of the sympathetic nervous system causing catecholamine release. The catecholamines activate β-adrenergic receptors and overstimulation is a component of heart disease. MiR-133 directly targets adenylate cyclase VI and the catalytic subunit of PKA, both elements of the β1AR signal transduction cascade, reducing signalling[17]. Similarly carvedilol, an in vivoβ-adrenergic blocker, improves the cardiac function in infarcted rats by restoring miR-133 expression, resulting in reduced cardiomyocyte apoptosis[18]. In vitro overexpression of miR-133a in cardiac cells has similar effects to carvedilol by downregulating caspase-9 and caspase-3 expression in the presence of H2O2. Overexpression of miR-133a also reduces ROS and malondialdehyde content, and increases SOD activity and GPx levels, protecting cardiomyocytes from apoptosis. Studies in mouse by Caré et al[6] also demonstrated that downregulation of both miR-133 and miR-1 are involved in cardiac hypertrophy. Specific targets of miR-133 such as RhoA, Cdc42 and Nelf-A/ WHSC2 can accumulate and contribute to the hypertrophy of cardiac myocytes during infarction.
Liu et al[2,9] (2007, 2010) also established a fundamental model of differential expression of cistronic miR-1 and miR-133a genes during myogenesis and differentiation of skeletal muscle, smooth muscle and cardiac muscle. The factor MEF2 controls expression of the miR-1-2/-133a-1 cistron via at least two MEF2 enhancer loci: one MEF2 enhancer located 3’ upstream of the miR-1-2 gene, a second intragenic MEF2 enhancer located upstream of the miR-133a-1 gene and a third (less defined) locus far upstream that requires MyoD for expression[19]. Transcripts of pri-miR-1-2/-133-a-1 (bi-cistron), pri-miR-1 and pri-miR-133a-1 genes indicate that both proximal enhancers are functional[1]. Others have emphasized the distribution of these regulatory enhancers[2] drawing attention to the differential expression of these cistronic miRs which the regulatory factor binding sites provide. The regulation of expression of the cistronic miRs by these key muscle development regulatory factors, which are themselves targets of repression by these self-same miRs, also emphasizes the precise inter-regulatory control of each of the various developmental program factors.
Notably, the 2,6-disubstituted purine reversine can induce differentiation reversal (de-differentiation) of C2C12 murine myoblast cells back into multipotent progenitor cells[20]. This occurs by inhibition of Aurora A and B protein kinases, reducing histone H3 phosphorylation, which in turn induces chromatin remodelling and restores cell multipotentency. Reversine treatment also stimulates expression of polycomb genes, Phc1 and Ezh2, leading to inhibition of expression of the muscle-specific transcription factors, myogenin, MyoD, and Myf5[21]. Concomitantly, reversine strongly inhibits miR-133a expression in C2C12 cells through the reduced expression of SRF transcription factor and by reduction of its binding to the miR-133a enhancer and by reduced epigenetic histone modifications on the miR-133a promoter, including reduced trimethylation, phosphorylation, and acetylation[22]. The co-overexpression of a miR-133a mimic along with reversine treatment prevents C2C12 myoblast de-differentiation, indicating the central role of the inhibition of miR-133a expression to the de-differentiation process. Significantly, reversine induced de-differentiation of committed cells is not limited to myoblasts, and reversine treatment can transform primary murine dermal fibroblasts into myogenic-competent cells within regenerating muscle in vivo[23].
Skeletal muscle myogenesis also involves the IGF signalling pathway[24], which activates muscle proliferation and differentiation via the PI3K/AKT pathway. The IGF pathway is regulated by miR-133-a1 which directly inhibits translation of IGF-1R protein, resulting in repression of PI3K/AKT pathway activity. IGF-1, which increases and activates IGF-1R during myogenesis by binding and inducing its phosphorylation, also indirectly activates myogenin, which in turn activates miR-133 activity. Thus miR-133 provides a negative regulation loop to monitor and control PI3K/AKT pathway activity. Similarly, miR-1 targets and reduces the activity of IGF-1 in differentiating C2C12 skeletal muscle cells and in heart muscle during cardiac failure states[25], meanwhile active IGF-1 signalling pathway downregulates miR-1 via repression of FoxO3a transcription factor. Thus, miR-1 also mediates the activity of the IGF-1 signal pathway and is itself feed-back regulated by the IGF-1 signal transduction cascade. Significantly, IGF-1 signalling (IGF-1 and IGF-1R) has key roles in the growth and development of many tissues[26], and also in the progression of many cancers (see later).
Skeletal muscles are plastic tissues in which the ratios of muscle fibre type (slow or fast twitch, smooth muscle, etc.) are to some degree responsive to remodelling through environmental input and nerve control. The muscle fibre type is maintained by the type of nerve signals received by the muscle, and transition between fast and slow twitch fibres can occur over time if nerve signals are changed from slow to fast type, and vice versa[27]. Similarly, prolonged workload or exercise can alter muscle fibre type and its metabolism to allow it to better respond before exhaustion. Muscle development programs regulated by miR-1 and miR-133a play important roles in muscle remodelling[4], and in hypertrophic skeletal muscle miR-1 and -133 levels are decreased[28], indicating that functional overload of muscle induces regulatory alterations which are in part influenced via altered miR activities.
Myostatin is a repressor of myogenesis, and its downregulation allows increase of miR-1, -133a, -133b and -206 expression, activating muscle cell proliferation[29]. In contrast, myogenic factors myogenin and MyoD are well known positive regulators of myomiR expression in muscle that bind upstream of miR-1/-133a genes at defined enhancer regions[2,11]. SRF, MyoD and MEF2 are also direct transcriptional activators of myogenesis-related miR-1 expression in cardiac muscle[2,30]. Since downregulation of myostatin permits expression of miR-133b/-206, and MyoD and myogenin also binds the miR-206 promoter[11], suggesting that miR-206/-133b expression in muscle may also be in part controlled by MyoD/ myogenin.
The ERK1/2 signalling pathway also regulates expression of miR-133 during myogenesis in the C2C12 cell model[31], and its activity is also feedback influenced by miR-133. During myogenesis both miR-133a and -133b are upregulated, and both FGFR1 and PP2AC which function in the ERK1/2 pathway signal transduction are negatively regulated post-transcriptionally by both miRs. Inhibition of ERK1/2 pathway signalling inhibits C2C12 cell proliferation, stimulating initiation of differentiation and forming small truncated myotubes. Importantly, ERK1/2 signalling pathway activity negatively regulates expression of miR-133, providing a feedback loop between miR-133 levels and ERK1/2 signalling activity, forming an additional reciprocal mechanism for regulating myogenesis.
Recently other cellular factors have been identified that influence post-transcriptional maturation or bio-availability of myomiRs in muscle. mTOR regulates miR-1 indirectly in regenerating mouse skeletal muscle and differentiating myoblasts[32]. mTOR most likely affects MyoD protein stability, which then alters miR-1 expression through the availability of MyoD to bind its upstream enhancer. A pathway downstream of mTOR also operates in which miR-1 suppression of HDAC4 results in production of follistatin, which subsequently activates myocyte fusion. This suggests that an mTOR-miR-1-HDAC4-follistatin pathway regulates myocyte fusion during myoblast differentiation and in regenerating skeletal muscle.
King et al[33] (2014) demonstrated that the RNA-binding TDP-43 protein interacts with miR-1/-206 family (but not the miR-133 family) in skeletal myoblast cells, limiting their bioavailability by preventing interaction with the RISC silencing complex, noting this is the first observation of a mechanism differentiating between mature bicistronically encoded miRs, which selectively modulates the bioactivity of downstream targets of the sequestered miRs. TDP-43 accumulates in motor neurons during ALS, a neuromuscular wasting disease. Two miR-1/-206 targets, IGF-1 and HDAC-4 are elevated in both ALS-model transgenic mouse muscle and in cells modified to overexpress TDP-43. The authors suggest the decreased miR-1 (-206) activity in ALS affected muscle could alter retrograde signalling at the NMJ through the dysregulation of both HDAC-4 and MEF-2, whereby miR-1 refines synaptic function by coupling changes in muscle activity to changes in presynaptic function[34].
Factors KSRP[35], MBNL1 and RNA binding protein LIN28[36] also positively and negatively regulate miR-1 biogenesis respectively. Additionally, miR-206 binds to 3’-UTR sites of KSRP transcript to inhibit KSRP expression in skeletal muscle[37]. Independently, miR-206 and KSRP are negative regulators of utrophin A, but unexpectedly, overexpression of miR-206 in both normal and dystrophic muscle cells promotes upregulation of utrophin A, via the downregulation of KSRP. Thus, miR-206 appears capable of switching between direct repression of utrophin A expression and the activation of its expression through decreased KSRP, the two molecular mechanisms providing close counter-regulation of utrophin A expression.
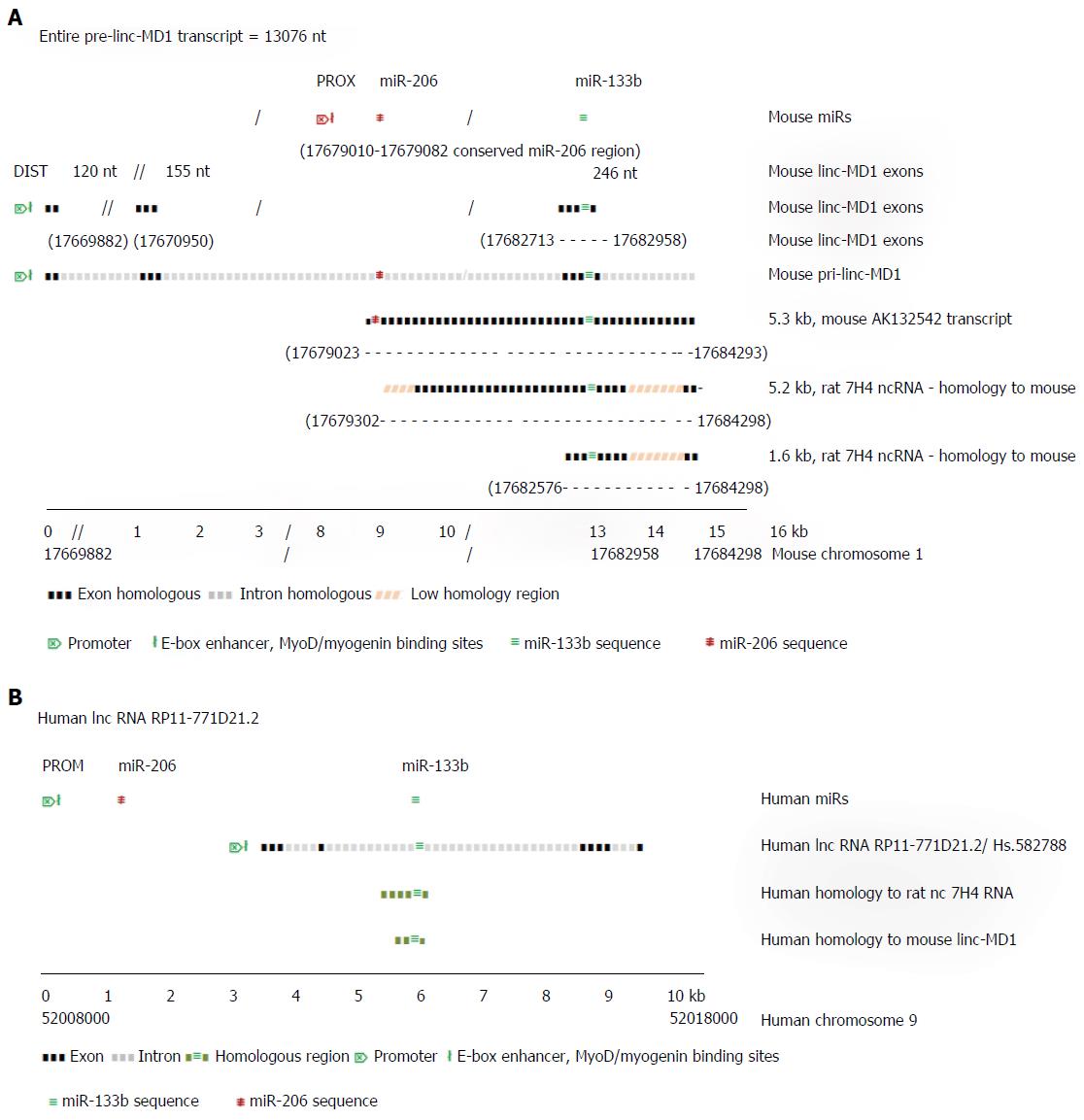
Although the microRNA-206 and -133b are thought typical of muscle specific miRs, little is known explicitly of their functions in skeletal muscle. Cesana et al[38] (2011) showed that miR-133b gene transcript in mouse is located within the precursor of the long (spliced) non-coding RNA linc-MD1 which is expressed under the control of an upstream distal (DIST) cistronic promoter[12] (Figure 2A), while the miR-206 gene, which is located within the intron of linc-MD1, is transcribed autonomously under control of its own proximal (PROX) promoter. In proliferating myoblasts, only primary miR-206 transcript is expressed strongly, initiated from the PROX promoter. During mouse muscle differentiation, long distance interactions bring the DIST promoter into conjunction with PROX and the polyA addition regions of linc-MD1, facilitating the co-expression of linc-MD1 RNA, as well as the primary miR-206 transcript, with the PROX promoter activated by both MyoD and myogenin binding[11]. Notably, mature linc-MD1 RNA contains binding sites for miR-133 (and miR-135) acting as a binding “sponge” to downregulate their free abundance, in turn contributing to the expression regulation of the targets of these miRs, which include key muscle transcription factors[38]. In mouse muscle, the expression of mature linc-MD1 RNA mutually excludes the expression of miR-133b, which must be excised from the linc-MD1 pre-transcript. In the rat genome, a distinct ncRNA 7H4, covering less than half of the mouse linc-MD1 precursor transcript, but closely similar to a mouse RNA AK132542 transcript, also encodes the miR-133b gene, suggesting a similar function to linc-MD1 may occur in rat[39].
Figure 2A uses information from Cesana et al[38] (2011) and Rosenberg et al[40] (2006) to illustrate the aligned transcript regions of the mouse pri-linc-MD-1, the 5.3 kb random cloned mouse transcript cDNA (GenBank sequence) AK132542 and the 5.2 kb rat 7H4 ncRNA transcript. The independent pre-miR-206 transcript overlaps almost completely with the AK132542 transcript which is likely the mouse homolog of the expressed rat ncRNA 7H4. The 7H4 RNA gene is almost fully conserved in the mouse genome, with the 7H4 RNA overlapping the 3’ exon of linc-MD1 gene (containing miR-133b gene) almost to the 3’ terminus of the miR-206 gene. Velleca et al[39] (1994) found two major transcripts of 7H4 RNA, a long 5.2 kb molecule, and a short 1.6 kb molecule coincident with the 3’-terminal region of the 5.2 kb transcript. The 1.6 kb fragment is much more abundant than the full length molecule, suggesting it is a product from the excision of miR-133b from the full length primary transcript. Notably, both long and short ncRNA 7H4 transcripts contain the entire miR-133b gene locus, suggesting they may function in rat similar to mature linc-MD1 in mouse, to bind complementary miRNAs, including miR-133. Similarly in man, ncRNA RP11-771D21.2 may represent the functional human homologue of mouse linc-MD1 RNA (Figure 2B).
Recently Legnini et al[41] (2014) reported that the mutually alternative synthesis of linc-MD1 and miR-133b is controlled by the pleiotropic mRNA regulator protein, HuR. In developing skeletal muscle, HuR favors accumulation of mature linc-MD1 by binding to it and repressing cleavage that would release pre-miR-133b. The level of HuR protein expression is also under the repressive negative control by miR-133 targetting, yet the sponging-up of miR-133 by the linc-MD1 helps consolidate HuR expression by forward positive control. Muscle developmental progression to later differentiation stages may involve overcoming this HuR-linMD1 repression of miR-133b expression by the independent miR-133a1/2 isomers which could downregulate HuR expression, allowing miR-133b excision, permitting developing muscle to exit from the control circuit. The level of linc-MD1 correlates inversely with the level of miR-135/-133b, which in turn control the expression of transcription factors MAML1 and MEF2C which are necessary for specific muscle gene expression. Thus, linc-MD1 activity provides another mechanism for pleiotropic regulation, slowing or activating muscle differentiation. Other evidence suggests the influence of HuR on many gene mRNA transcripts depends on the interplay of HuR with particular regulatory miRs that target and control the expression of the self-same mRNAs.
In sum, because the myomiRs target components of key signalling pathways and processes that control muscle cell development and maintenance, expression of the myomiRs is tightly regulated, often via feedback and feedforward circuits that provide both tight regulatory control and the ability to amplify myomiR expression. In muscle the myomiRs display high interconnectedness in terms of the regulation of their expression and the complementary processes that their regulatory functions influence. MyomiR genes are apparently cistronically encoded, yet expression of each of myomiR genes can be individually controlled by various transcription regulatory factors and other interactions such as RNA-RNA binding, such that expression of particular myomiR genes can be selectively enhanced under cellular conditions in which particular transcriptional regulatory factors are available.
MicroRNA miR-206 is expressed (virtually) exclusively in (developing) skeletal muscle[8], contributing to muscle differentiation programs through repression of Idl-3 protein expression, a downregulator of MyoD activity, as well as repressing the p180 subunit of DNA polymerase alpha, essential for DNA synthesis which occurs during differentiation[42]. MyoD itself promotes the expression of the miR-133 cistrons[11]. Further, in fast twitch muscle of mouse[12] and rat[43] miR-206 has been found to promote formation of new neuromuscular junctions following peripheral nerve denervation (scission). The expression of miR-206 and miR-133b are both upregulated strongly in muscle after denervation (as is 7H4 ncRNA), whereas miR-1 and miR-133a are downregulated. Four months after nerve scission, the re-innervated muscle was predominantly type II glycolytic fibres, suggesting that miR-206 may aid the determination of fibre type via down-regulation of MEF2 transcription factor activity[43]. Valdez et al[44] (2014) also examined the role of miR-133b and miR-206 on neuromuscular junction repair in injured mice. In miR-206 null mice, re-innervation was impaired following nerve injury, and in mice null for -133b and -206 genes the same impaired neuromuscular repair was seen as in single gene miR-206 null mice, whilst in single gene miR-133b null mice development and re-innervation proceeds normally following nerve injury. Together, these findings imply that miR-206 is the major regulator of nerve repair and reconnection to muscle following injury. In support, in miR-133b null mice Pitx3 levels were normal and impairment of locomotion was not detectable, controversially implying that miR-133b has no significant roles in neuron development, neuron maintenance and function in vivo[45]. In contrast, other studies with miR-206 null mice show no obvious phenotypic effects, muscles develop normally and mouse physiology appears normal, suggesting that other factors (including miR-133b) can replace miR-206 during development[46]. However, if the miR-206 null mice are then denervated, about 90% of both wt and miR-206 null mice recover and re-innervate after about 8 wk. This strongly suggests that other factors (including miR-133b) can provide redundant functions for the absent miR-206, including promoting compensatory peripheral nerve regeneration. Furthermore, miR-133b directly stimulates neurite outgrowth following nerve damage in rat brain after treatment with multipotent MSC cells[47], suggesting that elevation of levels of muscle miR-133b after muscle denervation is related to nerve regeneration, and that miR-133b may suffice in miR-206 null mice to replace absent functions. These various observations imply the likelihood that both miR-206 and -133b have functions in the recovery and maintenance of nerve-muscle signalling.
Additionally, miR-206 targets BDNF which promotes efficient skeletal muscle regeneration following damage[48]. BDNF also controls the initiation and maintenance of the differentiated state of muscle cells, potentially via the regulation of retrograde signalling at the neuromuscular junction. The loss of neural input to muscle also causes HDAC4 to accumulate, reducing MEF2-regulated gene expression. Importantly, miR-206 targets HDAC4 and fibroblast growth factor signalling pathways in muscle. HDAC4 regulates neuromuscular-related gene expression and acts in the regulation of muscle remodelling, influencing the formation of appropriate nerve types which connect to the muscle[49]. Significantly, it has been shown that expression of miR-1/miR-133a is also regulated by an intragenic MEF2-enhancer[9], and miR-1 also regulates a MEF-2 dependent retrograde signal at the neuromuscular junction, suggesting that members of both myomiR cistons act to maintain neuromuscular homeostasis[34].
In mouse, members of the MyoD muscle transcription factor family, myf-4 and myogenin, are progressively downregulated during maturation from embryonic day 15 to the first postnatal weeks (weeks 1-3), coinciding with induction of muscle innervation[50,51]. In contrast, muscle denervation results in strong expression of MyoD and myogenin, preceding the accumulation of nAChR, α-subunit[39]. Additionally during myogenic differentiation, acetylcholinesterase transcript levels increase dramatically (5 ×), principally due to its stabilization by binding with HuR protein[52], consistent with a regulatory role of HuR in neuron excitability. Normally the expression of MyoD and myogenin is suppressed by activated nerve signal pathways, including by electrical conduction per se, and sets of muscle genes regulated by the MyoD family and myogenin are downregulated by increasing electrical activity and other nerve-derived signals. Thus again, a pronounced neuromuscular maintenance function for miR-206/-133b can be implied from interplay of signalling control between skeletal muscle and nerve. Both myogenin and MyoD induce the expression of miR-133b and -206, while repression of these factors inhibits their expression. On balance it appears that cistronic miR-206 and -133b and linc-MD1 homologues may contribute to programs of regulatory developmental gene expression in growing muscle and peripheral nerve, facilitating programs to interregulate the developing nerve connections with muscle, and speculatively aid in coordinating appropriate nerve and muscle gene expression programs, establishing interactions between skeletal muscles and their appropriate innervating nerves to maintain muscle fibre type and their correct neuromuscular junction associations.
Zhang et al[53] (2014) reported that miR-1 enters skeletal muscle mitochondria efficiently during muscle development whereby it stimulates the translation of specific mitochondrial genome-encoded transcripts, contributing positive regulation to muscle development. This stimulation of translation requires specific base-pairing between miR and its target mtRNA as well as interactions with mt-located Ago2 protein. These observations contrast earlier findings of Das et al[54] (2012) who showed that the mature miR-181c translocates into rat cardiac muscle mitochondria, reducing mitochondrial cytochome oxygenase 1 (mt-COX1) compared to mt-COX2 and mt-COX3 proteins. The reduced mt-COX1 causes mitochondrial complex IV remodelling, resulting in increased mt respiration and increased ROS generation. Recently, Das et al[55] (2014) used cationic nanoparticles to deliver miR-181c into rat cardiac mitochondria in vivo, causing cardiac dysfunction and a tendency to develop heart failure. Taken together, these studies reveal important new miR-mediated regulatory pathways in muscle mitochondria involving direct manipulation of mitochondrial gene expression by cytosolic miRNAs, including by a myomiR.
Importantly in both cardiac and skeletal muscle, mitochondrial UPC2/UCP3 uncoupling proteins regulate energy homeostasis and the rate of development and differentiation, with UPC2 repressing differentiation and promoting cell proliferation. However, MyoD activates miR-133a expression which in turn directly downregulates UCP2 mRNA to alleviate the developmental repression, suggesting a feedback network involving MyoD-miR-133a-UCP2[56]. Additionally, overexpression of myogenin and MyoD in mouse C2C12 myoblasts[57] increase expression strongly from the UCP3 promoter, but act weakly at the UCP2 promoter. Together these observations suggest UCPs helps maintain balance between muscle differentiation and proliferation during myogenesis, regulated by a MyoD-miR-133a-UCP2 feedback network and by differential responsiveness of UCP2 and UCP3 promoters to activation by myogenin and MyoD.
Furthermore, a downregulation of mitochondrial function is associated with skeletal muscle injury, including increased ROS and reduced cellular ATP generation. However, the recovery and regeneration of post-injury skeletal muscle involves the activation and proliferation of resident stem cells, including satellite cells and endothelial precursor cells followed by their differentiation into myocytes. Jash et al[58] (2014) showed during recovery from muscle injury that the AMPK-CRTC2-CREB and Raptor-mTORC-4EBP1 pathways are activated in satellite cells. mTORC1 positively regulated Ccnd1 translation, yet destabilized Ccnd1 mRNA. These opposing effects of mTORC1 were mediated by two miRs which target the 3’-UTR of Ccnd1 mRNA: one being miR-1 which in mTORC-knockdown muscle was downregulated, allowing Ccnd1 mRNA to accumulate. The authors suggest that mTORC may act to coordinate satellite cell proliferation during the activation of myogenesis.
During conditions of skeletal muscle atrophy and wasting, the cytokine TWEAK and its binding receptor Fn14 are elevated, activating catabolic and pro-inflammatory processes[59]. TWEAK inhibits expression of myoD, MEF2C and myogenin which in turn inhibits expression of miR-1, -133, and -206, suppressing differentiation of progenitor cells into myocytes. HMOX1, another factor associated with inflammation[60], also inhibits myoblast differentiation and myotube formation, by inhibition of expression of each of the myomiRs, again by limiting their transcription factors, MyoD and myogenin. Thus, both HMOX1 and TWEAK may be potentially involved in the regulation of broadly common inflammation associated pathways in skeletal muscle.
Interestingly, TWEAK[61] has a role in stimulating the proliferation of normal neonatal rat cardiomyocytes, increasing cell numbers accompanied by expression of cell proliferation markers Cyclin D2 and Ki67, and other cell cycle factors. In contrast, adult rat cardiomyocytes cannot be stimulated by TWEAK because of the developmental downregulation of its receptor Fn14 in adult cells, coincident with the loss of proliferation capacity in mammalian cardiomycetes several weeks after birth. Fn14 is present in neonatal cardiomyocytes, interacting with TWEAK to activate downstream signalling via ERK and PI3K signalling pathways, as well as via inhibiting glycogen synthase kinase-3β.
FAPs are quiescent progenitor cells resident in normal muscle that can facilitate myofibre regeneration after muscle damage by providing factors which stimulate proliferating myogenic progenitor cells. In dystrophic muscle disease, FAPs typically proliferate and give rise to their differentiated progeny, fibroblasts and adipocytes which replace muscle tissue. However, Saccone et al[62] (2014) found in early-stage disease dystrophic (mdx) mouse muscles that HDAC inhibitors can activate and commit FAPs themselves towards regeneration of muscle tissue, by derepressing a “latent” myogenic program. The inhibition of HDAC induces two core components of the myogenic transcriptional machinery, MyoD and BAF60C, which upregulate expression of miR-1-2, miR-133, and miR-206. The structural subunits of the BAF chromatin remodelling complex (BAF60a, BAF60b and BAF60c) bind to Brg1 (the core complex ATPase) and provide functional specificity[63]. BAF60c is a specific member of the complex during myogenesis, and is essential for the myogenesis process. Interestingly, BAF60a and BAF60b are targets for downregulation by miR-133 and miR-1/206, suggesting that such negative regulation increases the availability of (non-target) BAF60c to join the muscle remodelling complex. Furthermore, a recent study of inflammatory myopathies[64] including dermatomyositis, polymyositis, and inclusion body myositis found increased expression of the inflammatory cytokine TNFα was associated with decreased expression of miR-1, miR-133a, and miR-133b in all subtypes, plus the decreased expression of miR-206 in dermatomyositis. TNFα inhibited the expression of myogenic miRNAs in cells in an NF-κB-dependent manner, while the overexpression of miR-1, miR-206, or miR-133a/b could relieve the TNFα blockage of myogenic cell differentiation. Overall, the dysregulation of myomiR expression in muscle degenerative diseases was demonstrated to be intrinsic to the disease progression.
In diseased cardiac tissues, pre-inflammatory reactions involve upregulation of CNN genes, and in vitro the downregulation of CNN2 blocks multiple proinflammatory and profibrotic pathways in mouse activated primary cardiac fibroblasts (PCFBs)[65]. Immune cell chemotaxis towards CCN2-depleted PCFBs is also reduced strongly. CCN2 is a direct regulation target of miR-133b, and silencing of CCN2 expression by siRNA strongly decreases the expression of stretch-induced chemokines, matrix metalloproteinases, extracellular matrix and a cell-to-cell contact protein, indicating that CCN2 is involved in control of multiple signal pathways for muscle regeneration. Exogenous factors also influence muscle recovery after injury. PRP plasma is an enriched source of autologous platelet α-granule-derived growth factors and cytokines which can stimulate tissue healing[66]. When PRP is applied to injured rat soleus muscle, the recruitment, proliferation and differentiation of cells for muscle recovery is stimulated. Molecular analysis showed that 5 d after PRP treatment the expression of proinflammatory cytokines IL-1β, and TGF-β1 was increased strongly, which in turn induced expression of myogenic factors MyoD1, Myf5 and Pax7, and muscle IGF-1Eb isoform, and muscle recovery was strongly accelerated. Concomitantly miR-133a and miR-1 were downregulated (miR-133a markedly), while SRF was upregulated, phosphoryled αB-cristallin was increased, as were apoptotic factors (NF-κB-p65 and caspase 3) which together indicate enhanced cell survival. Overall, PRP contributes to repair of injured skeletal muscle by via the control of secondary pathways (regulated by myomiRs and heat shock proteins) that modulate both inflammatory and myogenic pathways, with each contributing to the regulated tissue regeneration.
Whilst miR-133 plays a central role is the repair of damaged muscle and nerves in mammals, in lower vertebrates such as amphibians and teleost fish which retain the capacity for regeneration of entire limbs after damage or loss, here the downregulation of miR-133 plays a central role in organizing the reactivation of cells for the repair of complex tissues. Yin and Poss[67] (2008) found that miR-133 controls complex biological processes involving formation of the regeneration blastema, a proliferative mesenchymal cell mass which is the progenitor for regeneration of the lost structures, ultimately developing into organized organs, including connective tissues, muscle, blood vessels and nerve tissues[67]. When zebrafish fins are excised, the depletion of miR-133 is controlled by increased Fgf signalling (Tables 1 and 2). In normal developed fins, high levels of miR-133 are maintained, accompanied by a cessation of Fgf signalling, indicating that high miR-133 levels normally suppresses tissue proliferation factors and signalling pathways, maintaining developed tissue homeostasis[68]. Increased miR-133b also influences spinal cord regeneration in adult zebrafish, reducing the level of RhoA protein, an inhibitor of axonal growth, and stimulating spinal cord regeneration of axons from neurons in particular brain structures[69].
In mouse cornea after laser ablation injury[70], miR-133b is the most strongly reduced miR (amongst others) during wound recovery, allowing increased expression of its targets which include CTGF growth factor, SMA, and COL1A1. Transforming growth factor β1-treated rabbit corneal fibroblasts also produced significant decrease in miR-133b, associated with significantly increased expression of CTGF, SMA, and COL1A1, and helped to minimise scar development during corneal recovery. See Table 2 for other tissues.
The miR-133b has roles in early stem cell differentiation leading to nerve development. ADSC stem cells can be induced to differentiate into neuron-like cells by IGF-I signalling, which increases miR-133b expression via the downregulation of beta-III-tubulin, Pitx3 and IGF-IR by translational repression of their proteins[71]. Neural differentiation from ADSC involves a feedback control mechanism in which IGF-I upregulates miR-133b, while miR-133b in turn downregulates the signal receptor activity, IGF-IR. Xin et al[47,72] found multipotent MSC cells regulate the growth of neurites via direct exosomal transfer of miR-133b to neural cells. Middle cerebral artery occluded rat brains elevate miR-133b in MSC exosomes, which stimulates neural regeneration. Exosomes transfer to adjacent astrocytes and neurons, reducing the expression of selected miR-133b targets, including CTGF and RhoA[72]. The first identified molecular role for miR-133b was in neural tissue[73] where it regulated the maturation of mammalian midbrain dopaminergic neurons (DNs), along with the paired-like homeodomain transcription factor Pitx3, which itself then regulated the transcription of miR-133b in a feedback control loop.
Several studies found that miR-133 a/b isomers play key roles in the differentiation of brown fat tissues from precursor cells after cold exposure[74-76]. Strong reduction of the transcription regulator MEF2 caused reduction of miR-133a, allowing an increase in the adipocyte progenitor specific Prdm16 (a miR-133 target), which promotes the differentiation of both myogenic precursor cells and white fat precursor cells into brown adipocytes. Adult mouse skeletal muscle satellite cells can differentiate into brown adipose via miRNA-133 targeting of Prdm16, leading researchers to suggest that the presence of these miRs may indicate energy dissipating cell lineages (muscle, nerve), compared to energy storing cells, such as white adipose tissue[76]. Mice with knockdown of miR-133a1/a2 genes respond to cold exposure more strongly than wild-type animals and have increased insulin sensitivity and glucose tolerance associated with activation of the brown fat and thermogenic gene programs in subcutaneous white adipose tissue.
MiR-1 is highly expressed in the hypertrophic zone of growth plate cartilage, some 8-fold higher than in the proliferation zone[77]. MiR-1 strongly promotes chondrocyte proliferation and differentiation, including induction of the expression of chondrocyte markers Indian hedgehog and Col X, and acts by targetting HDAC4. Additionally HDAC4 negatively regulates chondrocyte hypertrophy by inhibiting Runx2, a critical transcription factor for chondrocyte hypertrophy. In contrast, miR-1 is repressed strongly during hypertrophic (late-stage) differentiation of chondrocytes in growth cartilage[78]. This differentiation could be reversed by transfection and overexpression of miR-1 which repressed the expression of aggrecan, the major cartilaginous proteoglycan gene in human chondrocytic HCS-2/8 cells and normal chicken chondrocytes. Thus miR-1 is a major effector in early growth and cell proliferation, and its repression at late differentiation stages is important for maintaining cartilage integrity.
Several reports of myomiR involvement in specialized immunological processes have recently emerged. Stumpova et al[79] (2014) found that several miRNAs, including miR-133b, were strongly expressed during in vitro maturation of human tolerogenic dendritic cells induced by exogenous IL-10 and TGF-β in comparison to miRs expressed in IL-4-induced and IFN-γ activated dendritic cells. MiR-133b and -206 have been previously reported to be expressed during differentiation of immunocompetent mouse Th17 cells, with miR-133b/-206 cistron transcription occurring along with expression at the nearby Il17a/f gene locus[80]. This feature of T cell differentiation towards an IL-17-producing phenotype is shared between αβ and γδ T cells, where the specific co-regulation of miR-133b and miR-206 with the Il17a/f locus extends to human Th17 cells, suggesting presence of miR-133b/-206 in T cells may be biomarkers for Th17-type immuno-reactions.
Bandyopadhyay et al[81] (2014) examined the effect of overexpression or suppression of miR-133a-1 in differentiated mouse THP1 cells which express the NLRP3 inflammasome, finding that miR-133a-1 overexpression suppresses expression of mitochondrial UCP2, resulting in increased caspase-1 p10 activity which subsequently causes IL-1β p17 cleavage. SiRNA silencing of UCP2 expression enhanced H2O2 stimulated inflammasome activity, and conversely, overexpression of UCP2 decreased the inflammasome activation, suggesting that the mechanism by which miR-133a-1 suppresses inflammasome activation involves the direct targetting of UCP2.
Further, the pulmonary cytokine GM-CSF is normally produced in lung alveolar epithelial cells (AECs) during a defense response. GM-CSF expression is suppressed by oxidative stress via altered mRNA turnover[82], in which miR-133a and -133b are upregulated in primary AECs. In vitro cell experiments confirmed that miR-133a and miR-133b bind the 3’-UTR of GM-CSF and suppress its expression, and that their inhibition can reverse oxygen-induced suppression of GM-CSF expression, suggesting that miR-133 isomers are important regulators of GM-CSF expression in AECs during oxidative stress. Studies of mouse colorectal epithelial cells involving the azoxymethane/dextran sulfate-induced mouse model of colitis-associated cancer found that the induced chronic colorectal epithelial cell inflammation downregulates miR-133a and other miRs associated with the modulation of PI3K/Akt and IGF-1 associated pathways, particularly during the earlier inflammatory stages[83].
Overall, whilst myomiRs miR-1-2, miR-133 and miR-206 have central roles muscle regeneration and (neuromuscular) repair, and in the regulation of aspects of inflammation and immunological responses during regeneration of injured and diseased muscles, miR-133a and miR-133b also appear to be involved in other regulatory processes affecting, repair, inflammation and immunological responses a number of other non-muscle cells/tissues, suggesting that these may involve common signalling pathways in a range of tissues in which myomiR play a regulatory role on the levels of signalling pathway components.
This review also examines the numerous cancers with prominent dysregulation of one or more of the canonical myomiRs, and the molecular events influenced by them. We focus on the deregulated myomiRs and their key target genes (up- or down-regulated) which have demonstrable effects in vitro on cell migration, proliferation or apoptosis, or those which are in vivo risk factors for cancer progression or patient survival. Although cancers characteristically have changed expression of large numbers of different protein genes and numerous different miRs which may contribute to the cancer pathology, these other deregulated genes and factors are not discussed except in relation with the myomiRs or their known targets. Table 3 lists the deregulated myomiRs reported in many different cancers. In addition, Tables 4 and 5 list targets and pathways influenced by downregulated and upregulated myomiRs respectively, and Table 6 lists validated myomiR targets related to enhanced cancer progression.
| MiR-1 | MiR-206 | MiR-133a | MiR-133b | Cancer type | Ref. |
| + | + | Progressive bladder cancer (TCC) | [122] | ||
| - | - | Bladder cancer (TCC) | [93,156] | ||
| - | - | - | Bladder cancer (TCC) | [92,144] | |
| - | - | - | - | Bladder cancer | [127]1 |
| - | - | - | - | Bladder cancer | [128]1 |
| - | - | - | Muscle-invasive bladder cancer | [92] | |
| - | Proliferating breast cancer | [108] | |||
| - | ERα-positive breast cancer | [109] | |||
| - | - | Breast cancer | [95] | ||
| + | Progressive cervical carcinoma | [117,121] | |||
| - | - | Chordoma | [90] | ||
| + | Colon cancer | [116] | |||
| - | Colon cancer | [101] | |||
| - | CRC | [104,136] | |||
| - | CRC | [86,87] | |||
| +/- | Liver metastasis compared to primary CRC | [118] | |||
| - | - | Colon cancer | [95] | ||
| - | - | Progressive GIST | [281] | ||
| - | HNSCC | [143] | |||
| - | HNSCC | [114] | |||
| - | EEC | [110] | |||
| - | ESCC | [132] | |||
| - | - | - | ESCC | [94] | |
| - | Laryngeal SCC cells | [113] | |||
| - | - | MSSCC | [137] | ||
| - | TSCC | [111,112] | |||
| - | HCC | [98] | |||
| + | + | Liver cancer | [95] | ||
| - | Lung cancers: (NSCLC, adenocarcinomas, lung SCC, large cell carcinoma, and bronchoalveolar cell carcinoma) | [100] | |||
| - | High metastasis lung tumors | [107] | |||
| - | Lung SCC tissue; lung-SCC cell lines | [115] | |||
| - | NSCLC | [85,105] | |||
| - | - | Lung adenocarcinomas; NSCLC cells | [99] | ||
| - | - | Lung carcinomas | [85,115,131] | ||
| - | - | Lung cancer | [95] | ||
| - | - | Lymphoma | [95] | ||
| + | AML | [120,151] | |||
| + | Multiple myeloma | [152] | |||
| + | Progressive prostate cancer | [123] | |||
| - | (-) | - | Prostate cancer | [102] | |
| - | - | Prostate cancer | [95] | ||
| - | - | Recurrent prostate cancer compared to non-recurrent cancer | [96] | ||
| - | - | Hormone-insensitive prostate cancer cells | [102] | ||
| - | - | - | Osteosarcoma | [91] | |
| - | - | Ovarian cancer | [95] | ||
| - | PDAC | [103] | |||
| - | RCC | [138] | |||
| - | - | Rhabdomyosarcoma | [88,89] | ||
| - | - | Testicular cancer | [95] |
| Downregulated miR-1 | ||||
| miR-1 downregulation influences multiple cancer-related pathway processes, and promotes cell proliferation and motility | Epigenetic promoter hypermethylation reduces miR-1/-133a expression in (a subset of) human prostate tumors | Reduced miR-1, miR-133a (and miR-206) | Human prostate tumors | [102] |
| Actin filament network-associated genes: FN1, LASP1, XPO6, CLCN3 and G6PD; Cell cycle and DNA damage control genes: BRCA1, CHK1, MCM7; Histone acetylation: HDAC4; Oncogenes: NOTCH3 and PTMA | miR-1 downregulation associated with upregulation of multiple cancer-related pathway processes | Reduced miR-1; | Human prostate cell lines, LNCaP, 22Rv1, PC-3 and RWPE-1 | [102] |
| Exogenous introduction of miR-1 or miR-206 caused similar inhibition of various cancer-related pathway genes | ||||
| HSPB1 | HSPB1 restores oncogenic pathways in prostate cancer cells | Downregulates miR-1 expression | Progressive prostate cancer PCa cells | [252] |
| XPO6 and TWF1 (PTK9) | Inverse expression between miR-1, XPO6 and TWF1 proteins in prostate cancer cell lines | Downregulated miR-1 expression | Prostate cancer cell cultures | [253] |
| CCND2, CXCR4, and SDF-1α | Inverse expression between miR-1 and CXCR4 and SDF-1α protein levels in thyroid carcinomas | Strongly downregulated miR-1 expression in thyroid adenomas and carcinomas | Thyroid adenomas and carcinomas | [254] |
| MET | MET upregulated | Reduced miR-1 | Colon cancer | [101] |
| Reduced miR-1, -133b | Colon cancer | [87] | ||
| MET, Pim-1 (Ser/Thr kinase), FoxP1 and HDAC4 | miR-1 downregulated, | MET, Pim-1, FoxP1 and HDAC4 are often upregulated in lung cancer | NSCLC tissue and A549 cell line | [100] |
| miR-1 targets MET, Pim-1, and may regulate FoxP1 and HDAC4 | ||||
| Fibronectin1 | Fibronectin1 upregulated | miR-1 downregulated | Laryngeal SCC Hep2 cells | [255] |
| Met, Twf1 and Ets1 and Bag4 | Met, Twf1 and Ets1 and Bag4 activities upregulated | miR-1 downregulated | Mouse cutaneous squamous cell carcinomas | [256] |
| Mediator complex subunit 1 (Med1) and 31 (Med31) | Med1 and Med31 activation result in increased Met activity | Reduced miR-1; miR-1 targets Med 1 and Med 31 | Osteosarcoma | [257] |
| NOTCH3 upregulates Asef expression, activating the Asef promoter, enhancing cell migration | NOTCH3 upregulated | Reduced miR-1; miR-1 targets NOTCH3 | Colorectal tumor cells | [258] |
| Overexpression of PIK3CA correlates with low miR-1 expression in NSCLC tissues | 71% of NSCLC samples had high PIK3CA expression | 69% of NSCLC samples had low miR-1 expression | Predictors of lymph node metastasis in NSCLC tissues | [259] |
| SLUG expression downregulated by miR-1 | Transcriptional repressor of E-cadherin, or an inducer of epithelial-to-mesenchymal transition | Overexpression of miR-1 induces morphological change from a mesenchymal to an epithelial character | NSCLC A549 cell line | [260] |
| SLUG expression high in chordoma tissue | miR-1 inhibited cell proliferation both time- and dose-dependently in chordoma | Transfection of MiR-1 inhibited Slug expression | mR-1 transfected chordoma cells | [261] |
| Slug overexpressed in advanced chordoma tissues and chordoma cells | ||||
| MET expression high in chordoma tissue | miR-1 downregulated 97% of chordoma samples | MiR-1 directly targets MET | Decreasing miR-1 expression levels correlated with severity of clinical prognosis | [262] |
| SRSF9/SRp30c | Exogenous upregulation of miR-1 expression | Novel apoptosis pathway involving SRSF9/SRp30c mediates tumor suppression | Bladder cancer (TCC) cells | [263] |
| ANXA2 is essential for glioblastoma growth and invasion | ANXA2 is highly abundant protein in glioblastoma-derived extracellular vesicles | miR-1 directly targets ANXA2; | Human Glioblastoma cells; miR-1 orchestrates glioblastoma extracellular vesicle function | [264] |
| Reduced miR-1 in glioblastoma | ||||
| EDN1 | miR-1 downregulated in gastric cancer | miR-1 causes ET-1 silencing in gastric cancer cell lines | Gastric cancer tissue compared with adjacent normal tissue | [265] |
| EDN1 | Elevated expression of EDN1 and reduced miR-1 level | miR-1 directly targets EDN1 | Human liver cancer tissues | [266, 267] |
| Overexpressed EDN1 | Enhanced in vitro cell proliferation and cell migration. Upregulation of several cell cycle/proliferation- and migration-specific genes | Upregulated UPR pathway mediators, spliced XBP1, ATF6, IRE1, and PERK at both RNA and protein levels | 293T cells | [267] |
| AKT inhibitor diminished the unfolded protein response and eliminated EDN1-induced cell migration | EDN1 effects act via activation of the AKT pathway | Results to enhance the UPR and subsequently activate the expression of downstream genes | 293T cells | [267] |
| Edn1 | Induced steatosis, fibrosis, glycogen accumulation, bile duct dilation, hyperplasia, and HCC | Liver-specific edn1 expression | Transgenic Zebrafish liver | [267] |
| API5 | API5 expression upregulated thus inhibiting apoptosis | miR-1 expression downregulated | Human liver cancer tissues | [268] |
| Apoptosis activated, API5 reduced | Overexpression of miR-1 | HepG2 liver cancer cells | ||
| Phosphorylation of ERK and AKT; LASP1 | Overexpression of miR-1 inhibits phosphorylation of ERK and AKT and reverses EMT process via inhibition of MAPK and PI3K/AKT pathways | MAPK and PI3K/AKT pathways | Transgenic miR-1 expressing CRC cell lines | [269] |
| LASP1 expression upregulated | Upregulated LASP1 stimulates EMT resulting in cell proliferation and migration | miR-1 downregulated | Colorectal tumor tissue | [269] |
| PIK3CA | Increased expression of PIK3CA | Downregulated miR-1 expression in lung cancer | NSCLC tissue with poor patient prognosis | [259] |
| PIK3CA indirectly regulating pAKT and survivin proteins | Overexpressed miR-1 downregulated PIK3CA causing reduced pAKT and survivin proteins | Exogenously overexpressed miR-1 targets PIK3CA directly. | NSCLC A549 cell line | [270] |
| Signalling pathways such as TGF-β, ErbB3, WNT and VEGFA, and cell motility or adhesion | Ectopic expression of miR-1 and miR-145 downregulates VEGFA and AXL, respectively | Highly downregulated expression of miR-1, miR-133, miR-143 and miR-145 in gall bladder cancer | Gall bladder tumor samples and GBC NOZ cell line | [271] |
| lncRNA UCA1 | Lnc RNA UCA1 upregulated in bladder cancer (TCC); | Downregulated miR-1 expression in bladder cancer (TCC); miR-1 targets lnc RNA UCA1 for downregulation | Human bladder cancer (TCC) tissue | [156] |
| Inverse relationship between miR-1 and lnc UCA1 | ||||
| Downregulated miR-133a | ||||
| Moesin | Moesin upregulated | Reduced miR-133a | HNSCC | [143] |
| ARPC5 | ARPC5 upregulated | Reduced miR-133a | HNSCC | [272] |
| ARPC5 and GSTP1 | ARPC5 and GSTP1 upregulated | Reduced miR-133a (and miR-206) | Lung carcinoma | [115] |
| IGF-1R, TGFBR1, and EGFR are downregulated | Restoration of ectopic-expression of miR-133a in NSCLC suppresses metastatic capacity | miR-133a inhibits cell invasiveness and cell growth via suppression of IGF-1R, TGFBR1 and EGFR | NSCLCs | [131] |
| Low expression of miR-133a is characteristic of pancreas tissue | Reduced miR-133a | PDAC | [103] | |
| CDC42 | CDC42 upregulated causing downstream activation of PAKs | miR-133 downregulated | Gastric cancer tissues | [273] |
| GSTP1 | Upregulated GSTP1 | Downregulation of miR-133a in cancer | Bladder cancer (TCC) cell lines | [144] |
| Enforced downregulation of GSTP1 inhibits cell proliferation and growth; | Enforced upregulation of miR-133a and miR-133b induces cell apoptosis | |||
| GSTP1 in cancer specimens | GSTP1 upregulated | Reduced miR-133a | Bladder cancer (TCC) tissue | [144] |
| Actin-binding protein, FSCN1 | Upregulated FSCN1; | Downregulation of miR-133a; | Bladder cancer (TCC) tissue | [274] |
| Enforced downregulation of FSCN1 inhibits cell proliferation, migration and invasion | Forced UP exp of miR-133a inhibits cell proliferation, migration and invasion | |||
| EGFR/AKT signalling pathway | Upregulated EGFR; | Downregulated miR-133a; | Human MCF-7 and MDA-MB-231 breast cancer cell lines | [275] |
| Activated pAkt-1 | Enforced expression of miR-133a inhibits EGRF translation; causes inhibition of Akt protein phosphorylation and its nuclear translocation | |||
| Bcl-xL and Mcl-1 expression | Upregulated Bcl-xL and Mcl-1 | Downregulated miR-133a correlated with tumor progression and poor patient prognosis; | Primary human osteosarcoma tissues; | [276] |
| Osteosarcoma cell lines | ||||
| E3 ubiquitin protein ligase | Downregulation of p21 and p53 proteins | Downregulated miR-133a | Primary CRC tissues | [277] |
| Enhanced sensitivity to doxorubicin and oxaliplatin | Enhancing apoptosis and inhibited cell proliferation | Ectopic upregulation of miR-133a | CRC cell lines | [277] |
| LASP1 upregulated | miR-133a expression downregulated | miR-133a targets LASP1 | CRC tissues and cell lines | [278] |
| FTL protein upregulated | miR-133a expression downregulated | miR-133a targets downregulation of FTL protein | Patient breast cancer tissue | [279] |
| Increased sensitivity to chemotherapeutic drugs doxorubicin and cisplatin | Exogenous upregulation of miR-133a expression | Downregulation of FTL protein | Human MCF-7 breast cancer cells | [279] |
| Poor survival during breast cancer; upregulated FSCN1 | Loss of miR-133a expression | FSCN1 is a direct target gene of miR-133a | Breast cancer tissue | [280] |
| FSCN1 downregulated | Restoration of miR-133a expression | Inhibited breast cancer cell growth and invasion | Breast cancer cell line | [280] |
| lncRNA Malat1/Srf/miR-133 regulatory loop | Malat1 transcript has a functional miR-133 target site, miR-133 acts as a competing endogenous RNA, regulating Malat1 levels | In vitro depletion of Malat1 in C2C12 cells reduces Srf activity, Srf is an enhancer of miR-133 expression; feed-back regulation loop involving miR-133 | Mouse myoblast C2C12 cells | [164] |
| lncRNA MALAT1 | MALAT1 is overexpressed in 46% of ESCC tissues, primarily in high-stage tumors, high expression correlates with lymph node metastasis | In vitro depletion of MALAT1 suppresses tumor cell proliferation, cell migration and invasion; G2/M phase arrest was induced and the ratio of apoptotic cells increased | Human ESCC | [162] |
| WIF1/lncRNA MALAT1 | WIF1 (strong tumor suppressor) is systematically downregulated in glioblastoma | WIF1 down regulation correlates with strong upregulation of MALAT1. In vitro depletion of MALAT1 suppresses tumor cell proliferation | Glioblastoma | [163] |
| Downregulated miR-133b | ||||
| Fascin-1 mRNA | FSCN1 upregulated | Reduced miR-133b | High-grade GIST tissue | [281] |
| BCL-2 family (MCL-1 and BCL2L2) | MCL-1 and BCL2L2 upregulated | Reduced miR-133b | Lung cancer | [85] |
| FAIM antiapoptotic protein and GSTP1 | miR-133b directly targets FAIM and GSTP1 | Downregulated miR-133b | miR-133b expression significantly downregulated in 75% of prostate cancer tumor specimens | [282] |
| Gli1 | Gli1 upregulated | Gli1 inversely correlated with downregulated expression of miR-133b | Gastric cancer | [283] |
| Bcl-w and Akt1 | Bcl-w and Akt1 proteins overexpressed significantly | miR-133b significantly downregulated | Bladder cancer tissues | [284] |
| miR-133b downregulated in tumors compared to surrounding tissue | Gastric and esophageal adenocarcinomas | [285] | ||
| Endometrial sarcoma, leiomyosarcoma, and mixed epithelial-mesenchymal tumors | [286] | |||
| Downregulated miR-206 | ||||
| Notch3/ miR-206 | Downregulated Notch3, blocking of the anti-apoptotic activity of Notch3 | Forced expression of miR-206 strongly induced apoptotic cell death via; also inhibited cell migration and focus formation | HeLa cells | [287] |
| Met | Upregulated Met | miR-206 downregulated | Human rhabdomyosarcoma | [288] |
| HGFR | Upregulated HGFR | miR-206 downregulated | Human breast cancer cells | [289] |
| KLF4 | Upregulated KLF4 | miR-206 downregulated | RK3E breast epithelium cells | [108] |
| KLF4; RAS-ERK signalling | Upregulated KLF4 promotes RAS-ERK signalling | miR-206 downregulated | TNBC cells | [290] |
| Endogenous KLF4 binds the promoter regions stimulates expression of miR-206 | ||||
| RASA1 and SPRED1 | miR-206 inhibits translation of the RAS pathway suppressors RASA1 and SPRED1 | Suppression of RASA1 or SPRED1 increased levels of GTP-bound, wild-type RAS and activated ERK 1/2 | ||
| VEGF | VEGF upregulated in Laryngeal SCC tissues | MiR-206 strongly downregulated in LSCC tissues | Laryngeal SCC cancer tissue and cells | [113] |
| VEGF | VEGF upregulated in ccRCC tissues | MiR-206 strongly downregulated in ccRCC tissues | ccRCC tissues assayed by Deep Sequencing | [291] |
| Cdc42, MMP-2 and MMP-9 | Upregulated Cdc42, MMP-2 and MMP-9 | miR-206 downregulated | Human breast cancer tissues | [292] |
| ERα | miR-206 directly targets ERα 3'-untranslated region | MiR-206 inhibited by ERα agonists, indicating a mutually (double) inhibitory feedback loop; | Estrogen stimulated breast cancer cell lines | [293] |
| miR-206 downregulated | [109] | |||
| Upregulated ERα | MCF-7 breast cancer cells | [294] | ||
| ERα | Upregulated ERα | miR-206 downregulated | EEC tissue | [110] |
| K-Ras | K-Ras is direct target of miR-206; | Low miR-206 potentiates metastases, and shorter overall survival | OSCC tissue samples and cell lines | [295] |
| MiR-206 expression significantly downregulated and k-Ras upregulated on OSCC tissues | ||||
| MiR-206 | Enforced upregulated of miR-206 attenuated cell proliferation, increased apoptosis and inhibited cell migration and invasion | MiR-206 strongly downregulated in lung cancer tissues | Lung cancer - tissues and cell lines | [107] |
| EGFR/MAPK signalling switches MCF-7 breast cancer cells from ERα-positive, Luminal-A phenotype to ERα-negative, basal-like phenotype | EGFR signalling represses estrogenic responses in MCF-7 cells by enhancing miR-206 activity | miR-206 downregulates steroid receptor co-activators SRC-1 and SRC-3 and GATA-3 transcription factor, directly | MCF-7 breast cancer cells | [296] |
| Elevated miR-206 reduces cell proliferation, enhances apoptosis, and reduces numerous estrogen-responsive genes | ||||
| Greater lymph node metastasis, venous invasion, and at a more advanced stage | miR-206 expression strongly downregulated | Correlates with tumor progression | Human gastric cancer tissue | [297] |
| CCND2 | miR-206 expression strongly downregulated | Correlates with upregulation of CCND2 and cancer progression | Human breast cancer | [298] |
| Human gastric cancer | [299] | |||
| MET | miR-206 expression strongly downregulated | Upregulation of MET | Papillary thyroid carcinoma | [300] |
| Prognostic signature of metastatic colorectal cancer | miR-206 expression strongly downregulated | Prognostic signature of metastases: miRs 21, 135a, 335, 206 and let-7a | Metastatic CRC | [301] |
| Notch3, Hes1, Bcl-2 and MMP-9; | Exogenous upregulation of miR-206 expression; | Notch3, Hes1, Bcl-2 and MMP-9 downregulated at both mRNA and protein level; | Human HHC Hep2 cells. | [302] |
| p57, Bax and caspase-3 | miR-206 is a potent tumor supressor | p57 and Bax upregulated, and cleaved caspase-3 protein upregulated | Reduced apoptosis, and cell migration in HepG2 cells overexpressing miR-206 | |
| STC2, HDAC4, KLF4, IGF1R, FRS2, SFRP1, BCL2, BDNF and K-ras | Exogenous upregulation of miR-206 expression; | STC2, HDAC4, KLF4, IGF1R, FRS2, SFRP1, BCL2, BDNF, and K-ras downregulated strongly in SCG-7901 cells overexpressing miR-206 | Gastric carcinoma SCG-7901 cells | [303] |
| miR-206 is a potent tumor supressor | Reduced apoptosis, and cell migration in SCG-7901 cells overexpressing miR-206 | |||
| Cyclin C, CCND1 and CDK4 | Cyclin C, CCND1 and CDK4 upregulated in melanoma tissue; | hsa-miR-206 downregulated in melanoma tissue | Human melanoma cancer tissue, and cell lines | [304] |
| Exogenous upregulation of miR-206 expression reduced growth and migration/invasion of several melanoma cell lines; | Overexpression of miR-206 in melanoma cells strongly downregulated cyclin C, CCND1 and CDK4 | |||
| G1 arrest in melanoma cells | ||||
| Coronin, actin-binding protein | Silencing of coronin expression reduced tumor cell migration and altered the cellular actin skeleton and cell morphology, but did not effect cell proliferation | Downregulated miR-206 allowed upregulation of coronin, a direct target; | TNBC cell lines | [305] |
| Upregulated miR-206 reduced TNBC cell migration and cell proliferation | ||||
| RNA binding protein DEAD-END (DND1), DNA cytosine deaminase (AICDA), and APOBEC3 | DND1 blocks miRNA interaction with 3'-UTR of specific mRNAs, restores protein expression; APOBEC3G binds DND1 counteracts repression and restores miRNA activity | APOBEC3G blocks DND1 to restore miR-206 inhibition of CX43 translation | Mouse cells | [306] |
| Advanced clinical stage, T classification, metastasis and poor histological differentiation | Significant association with decreased miR-206 expression | Paired human osteosarcoma and normal adjacent tissues | [307] | |
| Ellagic acid inhibits E2-induced mammary tumorigenesis | Reverses the downregulation of miR-206 | ACI model rat mammary tissue | [308] | |
| Actin-like 6A (BAF53a), a subunit of the SWI/SNF chromatin remodeling complex | Elevated BAF53a | Downregulation of miR-206 | Primary rhabdomyosarcoma tumors | [309] |
| Actin-like 6A (BAF53a) | BAF53a transcript is significantly higher in primary rhabdomyosarcomas than in normal muscle | Restoration of miR-206 expression downregulated BAF53a, which inhibits proliferation and anchorage independent growth; | Primary rhabdomyosarcoma tumors | [309] |
| BAF53a and is a direct target of miR-206 | ||||
| Wnt and transcription factors Tbx3 and Lef1 | Exogenous upregulation of miR-206 expression | Inhibition of Wnt, Tbx3 and Lef1 activities | Estrogen receptor alpha (ER-α)-positive human breast cancer; developing mammary buds | [310] |
| ANXA2 and KRAS | Stimulation of KRAS activity then induces NFKB1 expression; | Downregulated miR-206 in PDAC | PDAC tissues and cell lines | [311] |
| Induces NFKB1 | Increased KRAS results in stimulation of cytokines CXCR2, CXCL1, CCL2, as well as CSF2 (GM-CSF) and VEGFC | Increased cell cycle progression, cell proliferation, migration and invasion | ||
| Downregulated miR-1 and miR-133a | ||||
| PNP | PNP upregulated | Reduced miR-1, -133a | Prostate cancer | [97] |
| TAGLN2 | TAGLN2 upregulated | Reduced miR-1, -133a | RCC | [136] |
| TAGLN2 and PNP | TAGLN2 and PNP upregulated | Reduced miR-1, -133a | MSSCC | [137] |
| PTMA and PNP | PTMA and PNP upregulated | Reduced miR-1, -133a | Bladder cancer (TCC) | [312] |
| LASP1 | LASP1 upregulated | Reduced miR-1, 133a, (and miR-218) | Bladder cancer (TCC) | [313] |
| Forced expression of each miR decreased LASP1 in cell lines | ||||
| DNA methylation regulates miR-1-1 and miR-133a-2 cistron expression | Inverse correlation with TAGLN2 levels | CpG islands upstream of miR-1-133a hypermethylated | Colorectal carcinoma tissue and liver cancer tissue | [314] |
| Downregulated miR-1 and miR-133b | ||||
| miR-1 and mir-133b have sufficient power to distinguish recurrent specimens from non-recurrent prostate cancer | miR-1 and mir-133b are significantly downregulated in recurrent prostate cancer tissue specimens | Recurrent prostate cancer tissue | [96] | |
| Downregulated miR-1 and miR-206 | ||||
| NRF2 upregulated | Downregulated miR-1 and miR-206 expression | Upregulated expression of NRF2 induces increased expression HDAC4 | Primary lung adenocarcinoma; DU145 human prostate cancer cell line | [99] |
| Loss of NRF2 | Decreased expression histone deacetylase (HDAC4) | Results in increased expression of miR-1 and miR-206; which inhibits PPP expression; Reduced PPP acts as a regulatory feedback loop stimulates HDAC4 expression | A549 human NSCLC cell line | [99] |
| c-Met | c-Met upregulated | miR-1 and -206 downregulated | Human rhabdomyosarcoma | [89] |
| ARPC5 and GSTP1 | ARPC5 and GSTP1 upregulated | Reduced miR-133a (and miR-206) | Lung SCC cell lines | [115] |
| Downregulated miR-133a and miR-133b | ||||
| PKM2 | PKM2 upregulated | Downregulated miR-133a, -133b | TSCC | [111] |
| FSCN1 | FSCN1 upregulated | Downregulated miR-133a, -133b, (miR-145) | ESCC | [132] |
| miR-133a, miR-133b downregulated | ESCC | [94] | ||
| KRT7 | KRT7 upregulated | Downregulated (miR-133a and miR-133b) | Bladder cancer (TCC) and in vitro in BC KK47 cells | [315] |
| Downregulated miR-1, miR-206 and miR-133 | ||||
| myomiRs | Patient to patient variation in the up or down regulation of miR expression in both tumor and matched normal tissues | In tumors strong down regulation of highly expressed miR-1/133a; (downregulation of weakly expressed miR-206/-133b) | Bladder cancer assayed by deep sequencing | [315] |
| Candidate tumor suppressor miRNAs in RCC | Each of miR-206, miR-1, miR-133b strongly downregulated | Restored expression strongly inhibited cancer cell proliferation, | RCC | [316] |
| Shorter overall survival and disease-free survival | Correlated with increased downregulated of miR-133b and/or miR-206 | Both miR-133b and miR-206 significantly downregulated | Osteosarcoma tissues | [317] |
| Cell invasion and metastasis | miR-1, miR-133a, miR-133b downregulated | miR-133a, miR-133b involved in invasion and metastasis | ESCC | [94] |
| Factor(s) | Regulation | Regulator | Tissue/cell | Ref. |
| Upregulated miR-133b | ||||
| Activated p-ERK, pAKT1 cause in vitro proliferation of cervical cancer cell lines, and promote in vivo tumorigenesis and metastasis | Downregulation of MST2, CDC42, RHOA | Upregulated miR-133b | Human cervical carcinoma tissue compared to surrounding normal cervical tissue | [121] |
| Decreased patient survival | Upregulated miR-133b | Progression bladder cancer | [122] | |
| Androgen receptor | miR-133b directly represses CDC2L5, PTPRK, RB1CC1, CPNE3 | miR-133b directly upregulated by AR | Hormone-sensitive human prostate cancer (LNCaP) cells stimulated by androgen | [123] |
| Activativated neuroendocrine neoplasia proliferation | Mutation in von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase gene (VHL) | Upregulated miR-133b expression in VHL- deficient pheochromocytoma | Human pheochromocytoma (PCCs) and paraganglioma (PGLs) neuroendocrine neoplasias | [318] |
| Upregulated miR-206 | ||||
| Cell reprogramming factor KLF4 | KLF4 downregulated in colon cancer tissue, associated with increased miR-206 | miR-206 strongly upregulated in colon cancer tissues | Human colon cancer tissue | [116] |
| Upregulated miR-1 and miR-133 | ||||
| Decreased survival of R172 IDH2-mutated subset of CN-AML patients, increases resistance to chemotherapy | Distinctive gene and microRNA expression profiles accurately predicted R172 IDH2 mutations | Upregulated expression of miR-1 and miR-133 | De novo CN-AML patient bone marrow and blood samples | [151] |
| EVI1 increases aggressive cancer growth | EVI1 expression upregulated in established patient samples | Upregulated expression of miR-1-2 and miR-133-a-1 | EVI1 expressing AML subset of patients | [120] |
| ChIP assays show EVI1 binds to miR-1-2 gene promoter directly | ||||
| CCND2 | miR-1 and miR-133a were specifically overexpressed in the cases with t(14;16) translocation, correlates with down-regulated CCND2 expression | Upregulated miR-1 and miR-133-a | Multiple myeloma | [152] |
| Secreted myomiRs | ||||
| miRs selectively released into serum (within exosome microparticles) | miR-1, miR-133a, and miR-133b selectively released | Human breast cancer | [319] | |
| Circulating microRNA | Tumor-derived exosomes | Human non-small-cell lung cancer | [320] | |
| Up regulated cell factor | Down regulated cell factor | Cancer type | Ref. | |
| Downregulated myomiRs | ||||
| Downregulated miR-1 | ||||
| Mediator complex subunit 1 (Med1) and 31 (Med31) upregulated | miR-1 downregulated | Osteosarcoma | [257] | |
| Slug expression upregulated; enhanced cell migratory and invasive activities | miR-1 downregulated | Chordoma | [261] | |
| Slug expression upregulated; stimulation of EMT process | miR-1 reduced increasingly with cancer progression | Prostate adenocarcinoma | [321] | |
| Downregulated miR-133a | ||||
| ARPC5 upregulated; | Downregulated miR-133a (> miR-206) | Lung SCC | [115] | |
| HNSCC | [272] | |||
| CAV1 upregulated; | miR-133a downregulated | HNSCC | [322] | |
| Moesin upregulated; | miR-133a downregulated | HNSCC | [143] | |
| FSCN1 upregulated; | miR-133a/ miR-133b downregulated | ESCC | [132] | |
| Bladder cancer (TCC) | [274] | |||
| GSTP1 upregulated; | miR-133a downregulated | HNSCC | [323] | |
| Bladder cancer (TCC) | [144] | |||
| Lung SCC | [115] | |||
| LASP1 upregulated; | miR-133a downregulated in 83% of colorectal tumors | Colorectal cancer | [136] | |
| CAV1 downregulated with miR-133a levels, and is lowest in metastatic cancers; | Contrastingly, higher levels of miR-133a correlate with poor prognosis and increased metastasis | |||
| FSCN1 upregulated in non-metastatic tumors | ||||
| LASP1 upregulated; | miR-133a downregulated | Bladder cancer (TCC) | [313] | |
| PKM2 upregulated; | miR-133a downregulated | TSCC | [111] | |
| Moesin upregulated; | miR-133a downregulated | HNSCC | [143] | |
| EGFR upregulated; | miR-133 downregulated | Hormone-sensitive prostate cancer cell lines | [324] | |
| Human TERT telomerase catalytic subunit upregulated; | miR-133a downregulated | [325] | ||
| TCF7 transcription factor upregulated; | miR-133a downregulated | [325] | ||
| FSCN1 and MMP14 upregulated; | miR-133a downregulated | ESCC | [326] | |
| Reduced miR-133a expression correlated significantly with advanced clinical stages, poor histological differentiation and lymph node metastasis | Marked downregulation of miR-133a in primary EOC tumors and OVCAR-3 cell line | Epithelial ovarian cancer (EOC), and in OVCAR-3 cell line | [327] | |
| Downregulated miR-133b | ||||
| FSCN1 upregulated; | miR-133a/-133b downregulated | ESCC | [132] | |
| FSCN1 mRNA upregulated; | miR-133b downregulated | Progressive GIST | [281] | |
| BCL2L2 upregulated; | miR-133b downregulated | Lung cancer | [85] | |
| MCL1 upregulated; | miR-133b downregulated | Lung cancer | [85] | |
| MET upregulated; | miR-133b downregulated | Colorectal cancer | [87] | |
| MET protein upregulated; | miR-133b downregulated | high grade osteosarcoma tumor samples and cell lines | [328] | |
| EGFR upregulated; | miR-133b downregulated | NSCLC | [105] | |
| Multiple cell factors elevated; | miR-133b downregulated | Prostate cancer | [282] | |
| FGFR1 downregulated; | miR-133b downregulated | Gastric cancer | [329] | |
| Gli1 protein downregulated by miR133b, Gli1 target genes, OPN and Zeb2, are indirectly regulated | miR-133b downregulated | Gastric cancer | [283] | |
| TAp63 supresses metastasis; downregulation target of miR-133b | miR-133b is a transcription target of TAp63, downregulated | Colon cancer cells | [330] | |
| Chemokine (C-X-C motif) receptor 4 protein downregulated by miR133b; upregulated in advanced cancer | miR-133b downregulated | CRC | [331] | |
| TBP-like 1 mRNA and protein are upregulated in CRC | miR-133b downregulated in CRC | CRC | [332] | |
| Strong additional down regulation of miR-133b aids liver metastatic niche for CRC cells | miR-133b downregulated 3 × (significant) in liver metastasis compared to primary CRC | miR-133b downregulated in primary CRC compared to surrounding tissue | Metastatic cancer arising from primary hCRC | [333] |
| Interestingly, miR-133b is not downregulated significantly in lung metastasis compared to primary CRC | ||||
| SP1 targeted directly by miR-133, causing reduced expression of MMP-9 and Cyclin D1 | miR-133a and -133b downregulated | Gastric cancer | [334] | |
| miR-133b target MMP-9 is upregulated | miR-133b downregulated | RCC | [335] | |
| Downregulated miR-206 | ||||
| ERα | ERα downregulates miR-206 | ERα-positive breast cancer; | [294] | |
| miR-206 downregulated | Double feedback loop | [109] | ||
| miR-206 downregulated | [293,336] | |||
| ERα | miR-206 downregulated | EEC tissue | [110] | |
| SRC-1, SRC-3 and GATA-3 proteins contribute to estrogenic signalling | miR-206 downregulated | ERα-positive breast cancer | [296] | |
| Signalling contributes to Luminal-A phenotype | ||||
| KLF4 over expressed in proliferating cells and cancers. | miR-206 levels are KLF4 dependent. KLF4 and miR-206 feedback pathway oppositely affect KLF4 protein translation | Breast cancer cells and normal cells | [108] | |
| FGBP1 | miR-206 gene double knockdown | miR-206-/- mouse skeletal muscle. | [12] | |
| VEGF upregulated | miR-206 downregulated | Laryngeal SCC cells | [113] | |
| VEGF upregulated | miR-206 downregulated | CRC tumors compared to matched normal tissue; (1DS assay) | [337] | |
| miR-206 correlates with negative ER status, negative PR status, and negative HER-2 status | Downregulated miR-206 | Breast cancer tumor tissue | [338] | |
| miR-206 was downregulated in clinical TNBC tumor samples, one of its targets, actin-binding protein coronin was upregulated | Downregulated miR-206 associates with increased metastasis potential in breast cancers | High metastatic capacity TNBC tumors | [305] | |
| Downregulated miR-1 and miR-133a | ||||
| PNP upregulated | miR-1/miR-133a downregulated | MSSCC | [137] | |
| Prostate cancer | [97] | |||
| Bladder cancer (TCC) | [312] | |||
| TAGLN2 upregulated; | miR-1/miR-133a downregulated | MSSCC; | [137] | |
| RCC | [138] | |||
| HNSCC | [339] | |||
| Bladder cancer (TCC) | [93] | |||
| PTMA upregulated | miR-1 and miR-133a downregulated | Bladder cancer (TCC) | [312] | |
| Downregulated miR-1 and miR-206 | ||||
| MET levels correlated inversely with miR-1/206 expression | miR-1/206 downregulated | Up-regulation of MET in rhabdomyosarcoma | [89,288] | |
| HGFR upregulated | miR-1/206 downregulated | Breast cancer cells | [289] | |
| G6PD; PGD; TKT; GPD2 upregulated | miR-1/206 downregulated | Primary lung adenocarcinoma | [99] | |
| Upregulated myomiRs | ||||
| Upregulated miR-133b | ||||
| miR-133b | miR-133b strongly upregulated | MST1, CDC42, RHOA, and DUSP1 downregulated | Cervical carcinoma | [121] |
| miR-133b | miR-133b is directly upregulated by AR | miR-133b represses CDC2L5, PTPRK, RB1CC1, and CPNE3 | PCa prostate cancer cell line | [123] |
| Upregulated miR-206 | ||||
| miR-206 | Strongly upregulated miR-206 | KLF4 downregulated | Human colon cancer tissue | [116] |
| Upregulated miR-1 and miR-133a | ||||
| miR-1-2 and miR-133-a-1 | Upregulated miR-1-2 and miR-133-a-1 | EVI1 (transcriptional activator of miR-1 and miR-133b) | AML | [120,151] |
| miR-1 and miR-133-a | Upregulated miR-1 and miR-133-a | Downregulated CCND2 | Multiple myeloma | [152] |
| Up-regulation of exogenous myomiR expression in cell lines | ||||
| Reduced cell proliferation | Estrogen receptor alpha | Overexpression of miR-206 has an inhibitory effect on cell proliferation | ERα-positive breast cancer cells over expressing mir-206 | [289] |
| miR-133b | GSTP1 downregulated | Transgenic miR-133b overexpression | HeLa cervical cancer cells | [282] |
| miR-133b | FAIM downregulated | Transgenic miR-133b overexpression | HeLa cervical cancer cells | [282] |
| Apoptosis increased | TNFα-induced cell death is activated | Transgenic miR-133b overexpression | HeLa cervical cancer cells | [282] |
| Increased cell proliferation and migration | Downregulation of MST2 | Transgenic miR-133b overexpression | CaSki cervical cancer cells | [121] |
| Downregulation of CDC42 | ||||
| Downregulation of RHOA | ||||
| Increased cell proliferation and migration | Indirect upregulation of p-AKT1 activity | Transgenic miR-133b overexpression | CaSki cervical cancer cells | [121] |
| Indirect upregulation of p-ERK activity | ||||
| RB1CC1 downregulated | Exogenous upregulation of miR-133b; | miR-133bm promotes cell apoptosis, but suppressed cell proliferation and cell-cycle progression in aggressive PC-3 cells | PC3 prostate cancer cell line | [106] |
| miR-133b directly targets RB1CC1 in LNCaP cells | In contrast in low-aggression LNCaP cells, miR-133b stimulate cell proliferation and cell-cycle progression, but inhibit apoptosis | Hormone sensitive prostate cancer LNCaP cell line | ||
| Cell proliferation decreased and apoptosis increased | Met, Twf1 and Ets1 and Bag4 activities downregulated | miR-1 expression is lower in mouse cSCCs compared to normal skin | Mouse cutaneous squamous cell carcinomas (cSCCs); A5 and B9 cSCCcell lines | [256] |
| Transgenic miR-1 overexpression | ||||
| Ets1 proto-oncogene | Repression of Ets1 expression inhibited HepG2 cell invasion and migration | Transgenic miR-1 overexpression | HCC HepG2 cells | [340] |
| lncRNA UCA1 | Knockdown of lnc UCA1 expression phenocopied the effects of upregulation of hsa-miR-1 | hsa-miR-1 decreased the expression of lnc UCA1 in bladder cancer cells in an Ago2-slicer-dependent manner | Human bladder cancer (TCC) cells | [156] |
| NOTCH3 signalling | miR-206 had a direct inhibition of NOTCH3 signalling and indirect interaction with other signalling pathways via CDH2 and MMP-9 | miR-206 upregulation blocks the cell cycle, inhibits cancer cell proliferation and migration and activates cell apoptosis | SW480 (plus its metastatic strain) and SW620 colon cancer cell lines | [341] |
| FSCN1 | miR-133b targets FSCN1 in GC cells; the direct knockdown of FSCN1 can also inhibit GC cell growth and invasion | Up regulation of miR-133b in GC cells inhibits cell proliferation, cell migration and invasion | miR-133b is significantly downregulated in GC tissues compared with adjacent normal tissues, as well as in GC cell lines | [342] |
| FSCN1 | miR-133a targets FSCN1 in CRC cells; | Up regulation of miR-133a expression and downregulation of FSCN1 protein expression both suppress colorectal cancer cell invasion | miR-133a is significantly downregulated in some colorectal cancer cell lines, as well as in colorectal cancer tissues compared with the normal adjacent tissues | [343] |
| Overexpression of FSCN1 can reverse the inhibitory effect of miR-133a upregulation, reactivating CRC cell invasion | ||||
An excellent review by Nohata et al[84] (2012) reported changes in the expression of the miR-1/-133a and miR-206/-133b cistron clusters in numerous cancers, typically finding reduced expression of different combinations of myomiRs, such as in lung cancer[85], colorectal cancer[86,87], rhabdomyosarcoma[88,89], chordoma[90] osteosarcoma[91], muscle-invasive bladder cancer[92], bladder cancer (transitional cell carcinoma) (TCC)[93], ESCC[94], breast cancer[95] and prostate cancer[95-97]. In different cancers (Figure 3) deregulation typically involves downregulation of one or more myomiR isomers, indicating they normally function as tumor suppressors in the tissues, limiting the abundance of factors involved in aspects of cell proliferation[84].
Some cancers however show that elevation of myomiR levels promotes cancer progression (Figure 3), indicating that in such cell environments the deregulated myomiR is oncogenic and their targets here are normal tumor-suppressor cell factors. These are both regulatory roles that myomiRs normally exert, either by influencing particular cell signalling pathways that maintain a differentiated non-proliferating state in mature cell development stages, or by influencing other signalling pathways involved in cell proliferation and tissue genesis stages. Notably, the reports of deregulated myomiRs in cancers often reveal significant relation between the myomiR concentration and tumor severity, with greatest disregulation of expression statistically associated with metastasis, or with tumors of high metastatic potential, or with decreasing prospects of patient survival[88,98-102]. Yet, double knockout mice lacking both miR-133a1/2 gene alleles[14] do not display an elevated incidence of tumors in any organs or tissues in vivo, similarly animals with knockout of miR-206 expression[46], or knockdown of miR-133b or double knockdown of -133b/-206[44] are not reported to be associated with tumors, indicating that the singular lack of myomiR(s) alone is not (usually) sufficient to initiate tumorigenesis. Hence the oncogenic role of the deregulated myomiRs is a significant potentiating one, dysregulating cell signalling pathways in concert with the multiplicity of other molecular and cell environmental changes occurring in the developing tumor.
It is also notable that expression of cistronic myomiR isomers is not co-ordinated in different carcinomas, emphasizing individual deregulation of control of expression of each myomiR gene. For example, at least one miR of either cistronic pair is expressed in 16/20 cancers having altered miR-133 expression, showing also that each miRs has independent expression in non-muscle cells and tissues. Since the myomiRs influence numerous developmental pathways in normal muscle and other tissues, the differential expression of the myomiR isomers may potentially influence the pathology of cancers differently.
Reduced expression of myomiRs was associated with numerous cancers, such as with PDAC[103] colorectal cancer[86,104], NSCLC[105], HCC[98], prostate cancer[102,106], metastatic lung tumors[107], proliferating breast cancer[108], ERα-positive breast cancer[109], EEC[110], NSCLC[85], TSCC[111,112], laryngeal SCC[113], and HNSCC[114] (Table 3). Many of the reports identify reduction in a single myomiR, but in case studies of particular cancers often the different myomiRs have been identified as downregulated, for example in NSCLC tumor samples, miR-1[100], -133a[85,115], -133b[85,115], -206[107] or miR-1/-206[99] have been reported, by implication dysregulation of all of the myomiRs. Similarly, for colorectal cancer, downregulated miR-1[101], -133a[104], or -133b[86], or upregulated miR-206[116], while in prostate cancer downregulated miR-1[101,102] and miR-206[102], miR-133a[97,102] and miR-133b[95,96] have been variously reported (Table 3). Either these studies have examined different subclasses of the particular cancer which have different myomiR profiles, or more likely the method of miR detection or the purpose of the study (e.g., relating a particular miR to a particular deregulated target gene) may have influenced the findings reported, leading to some potentially conflicting and inconsistent reports. Additionally, the mature miR isomers are difficult to distinguish by molecular assay and some reports note cross-identification of miR-133b by Taqman mature miR-133a specific probe[14], which is used in many studies. Others[114,117] also make this point, that the different miR assay platforms generate different profiles which may obscure or confuse the identity of observed molecular changes, and at minimum generate apparently different miR profiles of the same tissues or diseases[118,119].
It is essential to accurately determine the specific expression of particular myomiR isomers and their alleles to understand the control processes of particular pathological states. The expressed alleles of the myomiRs can be assessed accurately by employing pri- or pre-miR RT-PCR assays. For example, only cistron miR-1-2/-133a1 is found expressed in AML, presumably due to specific activation of that cistron[120] and amongst the myomiRs only pre-miR-133b is elevated in cervical cancer[121], in bladder cancer[122], and in progressive prostate cancer[123]. We suggest that initial microarray profiling be confirmed by pri- or pre-miRNA assay of each miR isomer independently.
Significantly, a deep sequencing profile of miR expression in mouse heart[124] found that miR-133b is expressed at about 1/6 of the level of miR-133a, reflecting that microarray-based studies may underestimate the relative levels of important miR isomers. Importantly, such deep sequencing analyses also question the canonical view that miR-133b is not expressed in cardiac tissue, reinforcing the need to employ sensitive and accurate analysis to extend our understanding of miR involvement in biological processes. Discrepancies in miR profiles detected between deep sequencing analysis of liver cancer[344] and microarray/Taqman expression profiling[95], and in comparison of level 3 expression data from the Cancer Genome Atlas (TCGA) with deep sequence data from ovarian cancer patients[119] which found only 1 out of 12 survival-associated miRs identified by sequencing correlated by the TCGA data, emphasize the need for robust analytical and computational methods for in-depth profiling of tumors. Expression profiling of prostate tumors from individual patients by deep sequencing revealed that the expression of numerous miRs changed according to tumor stage[125]; however qRT-PCR of individual miRs at each tumor stage could not consistently confirm these alterations. A detailed survey of miRs using qRT-PCR accompanied by in situ hybridization to confirm the identity of the changed miR expression in matched prostate tumor tissue found less than 50% identity between major altered species when compared with deep sequencing analysis of pooled tumors by the same authors[126]. Furthermore, a deep sequencing profile of expressed miRs in bladder cancer[127] also showed that individual patient profiles varied greatly, and remarkably found also that the majority of deregulated miRs are upregulated in tumor tissue compared to matched normal tissue, with up to 3-5 × more upregulated than downregulated species in some patient tumor samples. Generally however, the myomiRs expressed in normal tissue are found downregulated in tumors when analysed by both qRT-PCR and by sequencing platforms. Significantly, another deep sequencing profile of expressed miRs in bladder cancer[128] revealed a different myomiR expression profile to the above deep sequencing study[127], yet confirmed the pattern of myomiR expression detected previously by the same group using microarray and qRT-PCR techniques[93] (Table 3). Overall, these various platform comparisons suggest the need to re-evaluate the genome-wide miR expression profiles of different cancers by use of deep sequencing and then to confirm findings using independent molecular methods.
Teicher[129] (2012) and Frith et al[130] (2013) noted in sarcoma tumors, which arise in diverse tissues of mesenchymal origin, that the upregulation of cMET (MET), HIF-1α, IGFR-1 or EGFR, CDK4, MCL1 or mTOR is observed with some elevated frequency in (particular) sarcomas, related to increased cancer severity and enhanced tumor progression, with the significance of each altered pathway likely due to the differentiated tissue in which the sarcoma arises. Downregulated expression of myomiRs occurs in both some sarcomas and carcinomas, linked specifically to the upregulated expression of some of the above gene targets. The different deregulated myomiRs modulate the levels of numerous mRNA/gene targets, but often particular deregulated gene targets are seen in several different cancers. For example, MET is upregulated due to reduced miR-1 levels in colon cancer[87,101], NSCLC[100] and rhabdomyosarcoma[89]. MET binds specifically with HGF, resulting in activation of pathways causing malignant progression via increased cell mobility, tissue invasion, and reduction of apoptosis. Reduced miR-1 targetting is only one of several cell alterations which can contribute to MET activation, yet the downregulation of miR-1 relates significantly to increased cancer severity, indicating the likely importance of miR-1 as a pleiotropic suppressor of several pathways important for tumor development.
Oncogenic membrane receptors, such as IGFR-1 and EGFR are upregulated in association with reduced miR-133 expression in NSCLC[131], and a similar EGFR upregulation occurs in hormone-sensitive prostate cancer and in breast cancer (Tables 4-6). Furthermore, a marked upregulation of cyclin C, cyclin D1 and CDK4 is seen in skin melanoma tissues associated with the reduced expression miR-206; whilst the BCL2 family of pro-survival molecules (Mcl-1 and BCL2L2) are both strongly upregulated due to a marked decrease in miR-133b levels (> 20-fold) in lung carcinoma tissue[85]. In addition, a detailed investigation of the role of myomiRs in prostate cancer[102] found that miR-1, miR-133a (and miR-206) were epigenetically supressed through promotor modification in numerous tumor samples and in prostate cancer cell lines. In vitro studies with prostate cancer cells showed that the downregulation of miR-1 allowed overexpression of multiple target genes that regulate key pathways affecting cell proliferation and cell migration, such as the actin filament network (FN1, LASP1, XPO6, CLCN3, G6PD), cell cycle and DNA damage control (BRCA1, CHEK1, MCM7) and histone acetylation (HDAC4); and with further downregulation of miR-1 the activation of oncogenic genes such as NOTCH3 and PTMA. Cell studies also showed that miR-206 has similar effects on prostate cancer cell biology as miR-1, suggesting that the combined downregulation of the myomiRs contributes significantly to prostate cancer progression.
In addition, FSCN1 expression is elevated in progressive metastatic ESCC, in breast cancer and in high-grade GIST, in part due to the loss of miR-133 expression[132]. FSCN1 is an actin-binding protein critical to cell adhesion, cell motility, and cell-cell interactions. In normal prenatal pig skeletal muscle, FSCN1 expression increases during major muscle developmental spurts, but postnatally it is expressed highly only in brain tissues during accelerated neural cell development[133]. The CREB pathway is often activated in different metastatic human cancers, causing the upregulation of FSCN1 expression[134]. In HNSCC patients, expression of RSK2 and FSCN1 proteins correlate closely. RSK2 protein expression potentiates filopodia formation and cell bundling, increasing cell invasiveness. Overexpression of FSCN1 can rescue the invasion phenotypes in RSK2 knockdown cells, linking RSK2-CREB signalling to the upregulation of FSCN1. In the highly metastatic PDAC[135], relative FSCN1 expression correlates with expressed HIF-1α levels, suggesting the hypoxic tumor microenvironment might induce FSCN1 expression, contributing to invasion and metastasis. Taken together, the downregulation of the myomiRs can contribute to the deregulated overexpression of oncogenic cell factors such as FSCN1, TAGLN2, KLF4, MET (cMET), IGFR and others, each of which can potentiate dysregulation of other cell signalling pathways, enhancing oncogenesis and metastasis (Tables 4-6).
Interestingly, the oncogenic membrane receptors, IGF-1R, TGFBR1 and EGFR are upregulated in NSCLCs due to the downregulation of miR-133a, whilst NSCLC patients with highly expressed levels of miR-133a tend to survive longer[131] presumably because these target oncogenic proteins are less strongly expressed. In contrast, although downregulation of miR-133a occurs generally in colorectal tumors, tumors with higher miR-133a levels are associated with increased metastasis, adverse clinical characteristics and poor prognosis[136], suggesting that other cell factors contribute to these differences in outcome. Expression of other targets of miR-133a: LASP1, CAV1, and FSCN1 are also deregulated in a complex pattern. While LASP1 showed negative correlation with miR-133a levels, CAV1 instead had significant positive correlation, which increased in patients with distant metastases, while negative correlation of FSCN1 was only seen in non-metastatic patients. Again the relationship between the deregulated myomiR and its deregulated target(s) display a complexity which suggests the involvement of other factors in early developing and metastatic cancer stages.
The upregulation of yet other cell factors were seen with reduced myomiR expression, such as TAGLN2 with the downregulation of miR-1 and -133a in MSSCC[137], in RCC[138], and in CRC, liver cancer and in bladder cancer (TCC) (Table 6). Interestingly, TAGLN2 is a known tumor suppressor, yet its upregulated expression is associated with increased metastasis, and worse patient prognosis. This outcome may be related to findings in HCC where phosphorylation of TAGLN2 by PFTK1 (an oncogenic serine/threonine protein kinase) inactivates its actin-binding function, abrogating its suppression of cell motility[139]. Furthermore, in mouse metaphase I oocytes after IGF-1 treatment, miRNA-133b was upregulated more than 30-fold and target TAGLN2 was downregulated, stimulating growth and maturation of the oocytes[140]. These several reports indicate that both the miR-133 isomers may target TAGLN2.
The (semi-) homologous miRs-1 and -206 are downregulated in many cancers, acting as tumor suppressors (Table 3). The homologues share many of the same oncogenic targets, which may be released from negative regulation in tumors. Recently, Singh et al[99] (2013) demonstrated that downregulation of miR-1/-206 in lung adenocarcinomas (and NSCLC) have major effects on carbon flux and the activity of metabolic pathways associated with cell proliferation and growth. The inhibition of miR-1/-206 expression results (indirectly) from elevated NRF2 which in turn increases the activity of redox-sensitive HDAC4, affecting myomiR expression. Importantly, key targets of miR-1-2 and -206 include enzymes of the pentose phosphate pathway (including G6PD, PGD and TKT) and the tricarboxylic acid cycle enzyme GPD2, which increase their activities and potentiate cancer cell proliferation and compromise patient survival. Pleiotropic factors, such as KLF4, HDAC4 and IGFR-1 are also elevated in gastric carcinoma and in breast cancer cells in part due to the loss of miR-206[108], yet in contrast KLF4 is reduced in colon cancer tissue due to strong upregulation of -206[116]. KLF4 is a zinc-finger transcription factor that binds to the TGF-β1 promoter and is important during normal cellular differentiation and proliferation. KLF4 is highly expressed in normal cardiac fibroblasts where it promotes the formation of myofibroblasts[141].
Some common molecular targets of the isoforms of miR-133 are elevated in different cancers (Tables 4 and 5). Moesin, a member of the ezrin-radixin-moesin protein family which links functionally between the plasma membrane and the actin-based cytoskeleton of cells and is found in invadopodia of metastatic cells[142], is deregulated in HNSCC, breast cancer, CRC, and prostate cancer. In HNSCC cells both miR-133a/-133b target moesin specifically, and restoration of miR-133b levels in HNSCC cells, as well as in PC10 and H157 lung SCC cell lines, reduces moesin expression, inhibits cell proliferation, cell migration and invasion[143]. The phenotype is also associated with elevation of other miR-133 targets such as ARPC5 and GSTP1[115]. Both ARPC5 and GSTP1 levels are elevated in lung-SCC and elevated GSTP1 is seen in bladder cancer (TCC) associated with reduced levels of miR-133a[144].
Several large cohort studies have examined the changes in expressed miR networks in a variety of cancers. Navon et al[95], Hart et al[125], Han et al[127], Itesako et al[128] and Volinia et al[145] noted that the downregulated expression of the miR-133/-1/-206 genes is associated with a variety of solid tissue cancers, with numerous other miRs also having highly significant oncogenic roles in particular cancer types. In sum, the downregulation of the various myomiRs contributes significantly to the upregulation of oncogenic proteins linked to regulation of cell cycle progression in solid tumors, sarcomas and carcinomas.
In yet other cancers in which the upregulation of a myomiR is associated with worsening metastasis or potentiation of cancer severity, the downregulation of different tumor-suppressor factors appears to be critical. In cervical cancer the upregulation of miR-133b is associated with downregulation of Mst2 protein kinase (STE20), Cdc42 and RhoA, which in turn lead to increased p-ERK and pAKT1 signalling activity, resulting in tumorigenesis and metastatic cancer proliferation[121]. RhoA, Cdc42 and other small Rho GTPases are components the Rho-kinase pathway which is a key controller of fundamental cellular processes such as cell motility, cell proliferation, cell division, cell differentiation, cell apoptosis, as well as morphological structure development, epithelial and skin morphogenesis, nerve system and limb development[146-148]. The downregulation of these key factors restricts cell apoptosis, favouring tumorigenesis. Interestingly, elevated levels of RhoA, Cdc42, Nelf-A/WHSC2 are also observed in hypertrophic cardiac muscle, associated with reduced levels of miR-133a[6], suggesting that both isomers of miR-133 may normally regulate these factors (in muscle). Similarly, the disruption of the Mst2 pathway is also associated with the disruption of cell apoptosis, of abrogating cell cycle regulation and with increased tumorigenesis in mouse intestinal epithelium[149], specifically by derepressing the accumulation of Yes-associated protein[150] which results in strong activation of β-catenin and Notch signalling. Elevation of Notch has also been seen in colorectal tumors associated with reduced miR-1 levels.
In AML the levels of expression of both miR-133a and miR-1 were elevated significantly[120,151]; similar to multiple myeloma where upregulation of these myomiRs was associated with the downregulation of the cyclin CCND2[152]. Wang et al[153] (2007) also found in patients with AML that downregulated CCND2 and CCND3 results in dephosphorylation of phospho-retinoblastoma protein and induction of G(1) cell-cycle arrest. These findings suggest that CCND2 may also be downregulated in AML in association with the upregulation of miR-133a and miR-1 levels, similar to multiple myeloma. During development or regeneration of normal muscle, the downregulation of CCND2 strongly enhances the myogenic terminal differentiation of muscle progenitor cells[154]. Taken together, the upregulation of these myomiRs in particular cancers can contribute to the deregulated repression of cell factors such MST2, RHOA, CDC42, CCND2 and others, which then also contribute to dysregulation of other cell signalling pathways, potentiating oncogenesis and metastasis. In contrast, a study of the miR network associated with altered mRNA profiles in AML[155] found that miRs other than myomiRs had highly significant roles in key deregulated pathways. Overall, the several studies suggest that myomiRs also contribute to development of AML.
Long non-coding RNAs (lncRNAs) are emerging as important new regulators of oncogenic pathways in cancers, and miRs are emerging as important regulators of lncRNA activities. Recently Wang et al[156] (2014) reported that deregulated expression of lncRNA UCA1, an important new oncogene in human bladder cancer (TCC), can be downregulated by miR-1 in vitro. These two factors are inversely expressed in bladder cancer tissue in vivo. lncRNA UCA1 expression is induced by HIF-1α which stimulates bladder cancer cell proliferation, migration, and invasion under hypoxic growth conditions[157]. It is also induced by the transcription factor CCAAT/enhancer binding protein α[158], and in another route for potentiation of bladder cancer cell growth and reduction of cell apoptosis the transcription factor Ets-2 binds directly at the UCA1 promoter, stimulating UCA1 promoter activity[159]. In bladder cancer cell lines the transgenic alteration of lncRNA UCA1 levels positively influences AKT expression and activity, and cell cycle progression could be reduced by inhibition of the PI3-K pathway, indicating that lncRNA UCA1 affected cell cycle progression through CREB[160]. Taken together, lncRNA UCA1 regulates the cell cycle through CREB and via PI3K-AKT-dependent pathways in bladder cancer. Interestingly, several of the cellular factors which are induced by lncRNA UCA1 are also regulated at the expression level by myomiRs. miR-206 targets HIF-1α directly, hence hypoxia-induced downregulation of miR-206 promotes pulmonary hypertension via elevated HIF-1α in hypoxic rat model pulmonary artery smooth muscle cells (Table 6). Additionally, during regeneration of injured muscle, the AMPK-CRTC2-CREB and Raptor-mTORC-4EBP1 pathways are activated in satellite cells, which involve regulation by miR-1[58]. The involvement of myomiRs in the regulation of these cellular factors in muscle cells suggests a potential for involvement of lncRNA UCA1 in the regulation of normal cellular processes involving the above protein factors.
Recently, the lncRNA MALAT1 which is upregulated during the differentiation of myoblasts into myotubes in normal muscle biogenesis[161] was also reported to be upregulated in several non-muscle cancers associated with worsening patient outcomes[162,163]. In skeletal muscle MALAT1 expression is downregulated by myostatin[161], whilst the silencing of MALAT1 expression in the mouse myoblast C2C12 cells results in the reduction of SRF transcription factor at both RNA and protein levels as well as reduced myocyte differentiation[164]. The MALAT1 transcript has a functional miR-133 target site, thus miR-133 acts as a competing endogenous RNA, regulating MALAT1 levels, which in turn modulates SRF activity. SRF also regulates the expression of miR-133a in C2C12 cells by its binding to the miR-133a enhancer[22], indicating a complex regulatory loop involving SRF, miR-133 and MALAT1.
Considering the central role of the myomiRs in the cell biology of myogenesis and muscle cell differentiation, in muscle metabolism, as well as in muscle remodelling and recovery from injury, it may be expected that they regulate the expression of numerous other target genes, in addition to the well documented pathway genes associated with key processes. In non-muscle-tissue cancers in which myomiRs play critical roles, the myomiRs influence expression of a variety of intermediary regulatory pathway genes, causing the potentiation of tumour development and metastasis. Literature searches (PubMed) show that essentially all myomiR targetted genes detected in different cancers have identified roles in several aspects of muscle cell biology. Thus, the myomiRs likely influence the expression of these gene targets in muscle in a normal regulatory manner, yet in non-muscle cancers the deregulated expression of myomiRs contributes to the dysregulated expression of these various gene targets, to the advancement of cancer development.
This review examines the diverse and complex regulatory functions of the cistronic myomiRs miR-133, -1 and -206 in numerous tissues. The myomiRs are intimately involved in the regulation of many processes of muscle development, muscle cell metabolism and homeostasis. Indeed some of the myomiRs are critical cell factors that commit stem cells onto the path of muscle cell differentiation and development, and their removal can elicit de-differentiation of committed muscle cells to an undifferentiated state. This centrality of cell regulatory functions suggests that, by necessity, these miRs would also be involved in redeveloping and repairing tissue after damage or injury, and that dysfunctional expression of myomiRs would play important roles during disease states. Furthermore, individual myomiRs have functional roles in the development of numerous non-muscle cells and tissues, beyond their original classification as muscle-specific factors, and hence the observation that myomiRs have roles in an increasing number of different cancer types should perhaps not be surprising. Whilst the myomiRs can display either tumor suppressor or tumor stimulator roles in different cells and tissues, independent cellular function assays confirm that the altered expression of the myomiRs typically correlates with the potentiation of cancer severity. This apparently contradictory ability of miRs to cause tumor suppression or tumor stimulation actions in different tissues relates to the multiple regulatory pathway genes targeted by each miR and the specific regulatory functions targeted in each tissue type. Interestingly, this ambiguity parallels the alternate roles of key signalling pathway regulatory genes in cancers, for example the increased expression of members of the FOX family of genes can cause either tumor suppression or tumor stimulation in different cancer types[165]. Significantly, the number of validated targets of each of the myomiRs has increased greatly in recent years, yet the extent to which each myomiR, miR-133, miR-1 or miR-206, contributes to specific tumorigenesis or tumor progression must await fuller clarification and integration with complex cellular regulatory pathway processes which are not yet fully defined.
P- Reviewer: Hatzaras I, Hosoda T, Yang BF S- Editor: Ji FF L- Editor: A E- Editor: Wang CH
| 1. | McCarthy JJ. The MyomiR network in skeletal muscle plasticity. Exerc Sport Sci Rev. 2011;39:150-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 2. | Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 379] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 3. | Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19-37. [PubMed] |
| 4. | Potthoff MJ, Olson EN, Bassel-Duby R. Skeletal muscle remodeling. Curr Opin Rheumatol. 2007;19:542-549. [PubMed] |
| 5. | Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252-1255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 271] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 6. | Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613-618. [PubMed] |
| 7. | Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-233. [PubMed] |
| 8. | McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682-691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 315] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 9. | Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844-20849. [PubMed] |
| 10. | Takaya T, Ono K, Kawamura T, Takanabe R, Kaichi S, Morimoto T, Wada H, Kita T, Shimatsu A, Hasegawa K. MicroRNA-1 and MicroRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J. 2009;73:1492-1497. [PubMed] |
| 11. | Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721-8726. [PubMed] |
| 12. | Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 590] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 13. | Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 460] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 14. | Liu N, Bezprozvannaya S, Shelton JM, Frisard MI, Hulver MW, McMillan RP, Wu Y, Voelker KA, Grange RW, Richardson JA. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J Clin Invest. 2011;121:3258-3268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 15. | Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588-5593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 405] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 16. | Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565-1581. [PubMed] |
| 17. | Castaldi A, Zaglia T, Di Mauro V, Carullo P, Viggiani G, Borile G, Di Stefano B, Schiattarella GG, Gualazzi MG, Elia L. MicroRNA-133 modulates the β1-adrenergic receptor transduction cascade. Circ Res. 2014;115:273-283. [PubMed] |
| 18. | Xu C, Hu Y, Hou L, Ju J, Li X, Du N, Guan X, Liu Z, Zhang T, Qin W. β-Blocker carvedilol protects cardiomyocytes against oxidative stress-induced apoptosis by up-regulating miR-133 expression. J Mol Cell Cardiol. 2014;75:111-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 19. | Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303-317. [PubMed] |
| 20. | Amabile G, D’Alise AM, Iovino M, Jones P, Santaguida S, Musacchio A, Taylor S, Cortese R. The Aurora B kinase activity is required for the maintenance of the differentiated state of murine myoblasts. Cell Death Differ. 2009;16:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Shan SW, Tang MK, Chow PH, Maroto M, Cai DQ, Lee KK. Induction of growth arrest and polycomb gene expression by reversine allows C2C12 cells to be reprogrammed to various differentiated cell types. Proteomics. 2007;7:4303-4316. [PubMed] |
| 22. | Kim M, Yi SA, Lee H, Bang SY, Park EK, Lee MG, Nam KH, Yoo JH, Lee DH, Ryu HW. Reversine induces multipotency of lineage-committed cells through epigenetic silencing of miR-133a. Biochem Biophys Res Commun. 2014;445:255-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Anastasia L, Sampaolesi M, Papini N, Oleari D, Lamorte G, Tringali C, Monti E, Galli D, Tettamanti G, Cossu G. Reversine-treated fibroblasts acquire myogenic competence in vitro and in regenerating skeletal muscle. Cell Death Differ. 2006;13:2042-2051. [PubMed] |
| 24. | Huang MB, Xu H, Xie SJ, Zhou H, Qu LH. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS One. 2011;6:e29173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 25. | Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377-2385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 311] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 26. | Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell. 1993;75:59-72. [PubMed] |
| 27. | Pette D. Historical Perspectives: plasticity of mammalian skeletal muscle. J Appl Physiol (1985). 2001;90:1119-1124. [PubMed] |
| 28. | McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol (1985). 2007;102:306-313. [PubMed] |
| 29. | Rachagani S, Cheng Y, Reecy JM. Myostatin genotype regulates muscle-specific miRNA expression in mouse pectoralis muscle. BMC Res Notes. 2010;3:297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-220. [PubMed] |
| 31. | Feng Y, Niu LL, Wei W, Zhang WY, Li XY, Cao JH, Zhao SH. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 2013;4:e934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 32. | Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189:1157-1169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 33. | King IN, Yartseva V, Salas D, Kumar A, Heidersbach A, Ando DM, Stallings NR, Elliott JL, Srivastava D, Ivey KN. The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J Biol Chem. 2014;289:14263-14271. [PubMed] |
| 34. | Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 35. | Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010-1014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 565] [Cited by in RCA: 531] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 36. | Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 213] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 37. | Amirouche A, Tadesse H, Miura P, Bélanger G, Lunde JA, Côté J, Jasmin BJ. Converging pathways involving microRNA-206 and the RNA-binding protein KSRP control post-transcriptionally utrophin A expression in skeletal muscle. Nucleic Acids Res. 2014;42:3982-3997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1874] [Cited by in RCA: 2163] [Article Influence: 154.5] [Reference Citation Analysis (0)] |
| 39. | Velleca MA, Wallace MC, Merlie JP. A novel synapse-associated noncoding RNA. Mol Cell Biol. 1994;14:7095-7104. [PubMed] |
| 40. | Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77-85. [PubMed] |
| 41. | Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 42. | Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677-687. [PubMed] |
| 43. | Jeng SF, Rau CS, Liliang PC, Wu CJ, Lu TH, Chen YC, Lin CJ, Hsieh CH. Profiling muscle-specific microRNA expression after peripheral denervation and reinnervation in a rat model. J Neurotrauma. 2009;26:2345-2353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 44. | Valdez G, Heyer MP, Feng G, Sanes JR. The role of muscle microRNAs in repairing the neuromuscular junction. PLoS One. 2014;9:e93140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 45. | Heyer MP, Pani AK, Smeyne RJ, Kenny PJ, Feng G. Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice. J Neurosci. 2012;32:10887-10894. [PubMed] |
| 46. | Sokol NS. The role of microRNAs in muscle development. Curr Top Dev Biol. 2012;99:59-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 47. | Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Shang X, Zhang ZG, Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 613] [Cited by in RCA: 698] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 48. | Miura P, Amirouche A, Clow C, Bélanger G, Jasmin BJ. Brain-derived neurotrophic factor expression is repressed during myogenic differentiation by miR-206. J Neurochem. 2012;120:230-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 49. | Cohen TJ, Barrientos T, Hartman ZC, Garvey SM, Cox GA, Yao TP. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J. 2009;23:99-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 50. | Eftimie R, Brenner HR, Buonanno A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc Natl Acad Sci USA. 1991;88:1349-1353. [PubMed] |
| 51. | Buonanno A, Apone L, Morasso MI, Beers R, Brenner HR, Eftimie R. The MyoD family of myogenic factors is regulated by electrical activity: isolation and characterization of a mouse Myf-5 cDNA. Nucleic Acids Res. 1992;20:539-544. [PubMed] |
| 52. | Deschênes-Furry J, Bélanger G, Mwanjewe J, Lunde JA, Parks RJ, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuR binds to acetylcholinesterase transcripts and regulates their expression in differentiating skeletal muscle cells. J Biol Chem. 2005;280:25361-25368. [PubMed] |
| 53. | Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 377] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 54. | Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596-1603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 287] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 55. | Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One. 2014;9:e96820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 56. | Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, Guo X, Wang G, Yang R, Zhu L. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem. 2009;284:5362-5369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 57. | Kim D, Jitrapakdee S, Thompson M. Differential regulation of the promoter activity of the mouse UCP2 and UCP3 genes by MyoD and myogenin. J Biochem Mol Biol. 2007;40:921-927. [PubMed] |
| 58. | Jash S, Dhar G, Ghosh U, Adhya S. Role of the mTORC1 complex in satellite cell activation by RNA-induced mitochondrial restoration: dual control of cyclin D1 through microRNAs. Mol Cell Biol. 2014;34:3594-3606. [PubMed] |
| 59. | Panguluri SK, Bhatnagar S, Kumar A, McCarthy JJ, Srivastava AK, Cooper NG, Lundy RF, Kumar A. Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PLoS One. 2010;5:e8760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Kozakowska M, Ciesla M, Stefanska A, Skrzypek K, Was H, Jazwa A, Grochot-Przeczek A, Kotlinowski J, Szymula A, Bartelik A. Heme oxygenase-1 inhibits myoblast differentiation by targeting myomirs. Antioxid Redox Signal. 2012;16:113-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 61. | Novoyatleva T, Diehl F, van Amerongen MJ, Patra C, Ferrazzi F, Bellazzi R, Engel FB. TWEAK is a positive regulator of cardiomyocyte proliferation. Cardiovasc Res. 2010;85:681-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 62. | Saccone V, Consalvi S, Giordani L, Mozzetta C, Barozzi I, Sandoná M, Ryan T, Rojas-Muñoz A, Madaro L, Fasanaro P. HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev. 2014;28:841-857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 63. | Goljanek-Whysall K, Mok GF, Fahad Alrefaei A, Kennerley N, Wheeler GN, Münsterberg A. myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development. 2014;141:3378-3387. [PubMed] |
| 64. | Georgantas RW, Streicher K, Greenberg SA, Greenlees LM, Zhu W, Brohawn PZ, Higgs BW, Czapiga M, Morehouse CA, Amato A. Inhibition of myogenic microRNAs 1, 133, and 206 by inflammatory cytokines links inflammation and muscle degeneration in adult inflammatory myopathies. Arthritis Rheumatol. 2014;66:1022-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 65. | Tank J, Lindner D, Wang X, Stroux A, Gilke L, Gast M, Zietsch C, Skurk C, Scheibenbogen C, Klingel K. Single-target RNA interference for the blockade of multiple interacting proinflammatory and profibrotic pathways in cardiac fibroblasts. J Mol Cell Cardiol. 2014;66:141-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 66. | Dimauro I, Grasso L, Fittipaldi S, Fantini C, Mercatelli N, Racca S, Geuna S, Di Gianfrancesco A, Caporossi D, Pigozzi F. Platelet-rich plasma and skeletal muscle healing: a molecular analysis of the early phases of the regeneration process in an experimental animal model. PLoS One. 2014;9:e102993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 67. | Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008;18:381-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 68. | Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 69. | Yu YM, Gibbs KM, Davila J, Campbell N, Sung S, Todorova TI, Otsuka S, Sabaawy HE, Hart RP, Schachner M. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur J Neurosci. 2011;33:1587-1597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 70. | Robinson PM, Chuang TD, Sriram S, Pi L, Luo XP, Petersen BE, Schultz GS. MicroRNA signature in wound healing following excimer laser ablation: role of miR-133b on TGFβ1, CTGF, SMA, and COL1A1 expression levels in rabbit corneal fibroblasts. Invest Ophthalmol Vis Sci. 2013;54:6944-6951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 71. | Ning H, Huang YC, Banie L, Hung S, Lin G, Li LC, Lue TF, Lin CS. MicroRNA regulation of neuron-like differentiation of adipose tissue-derived stem cells. Differentiation. 2009;78:253-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 72. | Xin H, Li Y, Liu Z, Wang X, Shang X, Cui Y, Zhang ZG, Chopp M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells. 2013;31:2737-2746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 580] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 73. | Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220-1224. [PubMed] |
| 74. | Liu W, Bi P, Shan T, Yang X, Yin H, Wang YX, Liu N, Rudnicki MA, Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 75. | Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14:1330-1335. [PubMed] |
| 76. | Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 77. | Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C, Wei L. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28:3930-3941. [PubMed] |
| 78. | Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Nishida T, Shimo T, Yamashiro T, Takigawa M. Identification of miR-1 as a micro RNA that supports late-stage differentiation of growth cartilage cells. Biochem Biophys Res Commun. 2010;402:286-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 79. | Stumpfova Z, Hezova R, Meli AC, Slaby O, Michalek J. MicroRNA profiling of activated and tolerogenic human dendritic cells. Mediators Inflamm. 2014;2014:259689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 80. | Haas JD, Nistala K, Petermann F, Saran N, Chennupati V, Schmitz S, Korn T, Wedderburn LR, Förster R, Krueger A. Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γδ T cells. PLoS One. 2011;6:e20171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 81. | Bandyopadhyay S, Lane T, Venugopal R, Parthasarathy PT, Cho Y, Galam L, Lockey R, Kolliputi N. MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2. Biochem Biophys Res Commun. 2013;439:407-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 82. | Sturrock A, Mir-Kasimov M, Baker J, Rowley J, Paine R. Key role of microRNA in the regulation of granulocyte macrophage colony-stimulating factor expression in murine alveolar epithelial cells during oxidative stress. J Biol Chem. 2014;289:4095-4105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 83. | Josse C, Bouznad N, Geurts P, Irrthum A, Huynh-Thu VA, Servais L, Hego A, Delvenne P, Bours V, Oury C. Identification of a microRNA landscape targeting the PI3K/Akt signaling pathway in inflammation-induced colorectal carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G229-G243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 84. | Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9-21. [PubMed] |
| 85. | Crawford M, Batte K, Yu L, Wu X, Nuovo GJ, Marsh CB, Otterson GA, Nana-Sinkam SP. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun. 2009;388:483-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 86. | Akçakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 87. | Hu G, Chen D, Li X, Yang K, Wang H, Wu W. miR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;10:190-197. [PubMed] |
| 88. | Missiaglia E, Shepherd CJ, Patel S, Thway K, Pierron G, Pritchard-Jones K, Renard M, Sciot R, Rao P, Oberlin O. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769-1777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 89. | Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596-29604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 235] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 90. | Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, Schwab J, Mankin H, Xavier R, Hornicek FJ. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res. 2010;28:746-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 91. | Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 236] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 92. | Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana D. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479-2491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 93. | Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808-818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 232] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 94. | Fu HL, Wu de P, Wang XF, Wang JG, Jiao F, Song LL, Xie H, Wen XY, Shan HS, Du YX. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67:657-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 95. | Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 96. | Karatas OF, Guzel E, Suer I, Ekici ID, Caskurlu T, Creighton CJ, Ittmann M, Ozen M. miR-1 and miR-133b are differentially expressed in patients with recurrent prostate cancer. PLoS One. 2014;9:e98675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 97. | Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 98. | Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049-5058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 374] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 99. | Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123:2921-2934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 100. | Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394-33405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 293] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 101. | Migliore C, Martin V, Leoni VP, Restivo A, Atzori L, Petrelli A, Isella C, Zorcolo L, Sarotto I, Casula G. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res. 2012;18:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 102. | Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689-3703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 103. | Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452. [PubMed] |
| 104. | Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 260] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 105. | Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang R, Huang P, Yin Y, Shu Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279:3800-3812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 106. | Li X, Wan X, Chen H, Yang S, Liu Y, Mo W, Meng D, Du W, Huang Y, Wu H. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res. 2014;20:2312-2325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 107. | Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken). 2011;294:88-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 108. | Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol. 2011;31:2513-2527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 109. | Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 248] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 110. | Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, Liu X, Wan X. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314:41-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 111. | Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing Yuen A, Wai-Man Ng R, Ignace Wei W. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J Cancer. 2008;123:251-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 112. | Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588-2592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 602] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 113. | Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res. 2011;31:3859-3863. [PubMed] |
| 114. | Liu X, Chen Z, Yu J, Xia J, Zhou X. MicroRNA profiling and head and neck cancer. Comp Funct Genomics. 2009;837514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 115. | Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 2012;57:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 116. | Parasramka MA, Dashwood WM, Wang R, Saeed HH, Williams DE, Ho E, Dashwood RH. A role for low-abundance miRNAs in colon cancer: the miR-206/Krüppel-like factor 4 (KLF4) axis. Clin Epigenetics. 2012;4:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 117. | Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA. MicroRNA expression variability in human cervical tissues. PLoS One. 2010;5:e11780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 118. | Ellermeier C, Vang S, Cleveland K, Durand W, Resnick MB, Brodsky AS. Prognostic microRNA expression signature from examination of colorectal primary and metastatic tumors. Anticancer Res. 2014;34:3957-3967. [PubMed] |
| 119. | Wan YW, Mach CM, Allen GI, Anderson ML, Liu Z. On the reproducibility of TCGA ovarian cancer microRNA profiles. PLoS One. 2014;9:e87782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 120. | Gómez-Benito M, Conchillo A, García MA, Vázquez I, Maicas M, Vicente C, Cristobal I, Marcotegui N, García-Ortí L, Bandrés E. EVI1 controls proliferation in acute myeloid leukaemia through modulation of miR-1-2. Br J Cancer. 2010;103:1292-1296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 121. | Qin W, Dong P, Ma C, Mitchelson K, Deng T, Zhang L, Sun Y, Feng X, Ding Y, Lu X. MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways. Oncogene. 2012;31:4067-4075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 122. | Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851-4860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 123. | Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, Wan X, Zhou C, Guo F, Huang Y. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8:e56592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 124. | Hu Y, Matkovich SJ, Hecker PA, Zhang Y, Edwards JR, Dorn GW. Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc Natl Acad Sci USA. 2012;109:19864-19869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 125. | Hart M, Nolte E, Wach S, Szczyrba J, Taubert H, Rau TT, Hartmann A, Grässer FA, Wullich B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res. 2014;12:250-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 126. | Wach S, Nolte E, Szczyrba J, Stöhr R, Hartmann A, Ørntoft T, Dyrskjøt L, Eltze E, Wieland W, Keck B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 127. | Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, Jiang Z, Zhang Z, Yang R, Chen J. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 128. | Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014;9:e84311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 129. | Teicher BA. Searching for molecular targets in sarcoma. Biochem Pharmacol. 2012;84:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 130. | Frith AE, Hirbe AC, Van Tine BA. Novel pathways and molecular targets for the treatment of sarcoma. Curr Oncol Rep. 2013;15:378-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 131. | Wang LK, Hsiao TH, Hong TM, Chen HY, Kao SH, Wang WL, Yu SL, Lin CW, Yang PC. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS One. 2014;9:e96765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 132. | Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804-2814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 392] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 133. | Tang ZhL, Zhang XJ, Yang ShL, Mu YL, Cui WT, Ao H, Li K. The chromosomal localization, expression pattern and polymorphism analysis of porcine FSCN1 gene differently expressed from LongSAGE library. Mol Biol Rep. 2010;37:2361-2367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 134. | Li D, Jin L, Alesi GN, Kim YM, Fan J, Seo JH, Wang D, Tucker M, Gu TL, Lee BH. The prometastatic ribosomal S6 kinase 2-cAMP response element-binding protein (RSK2-CREB) signaling pathway up-regulates the actin-binding protein fascin-1 to promote tumor metastasis. J Biol Chem. 2013;288:32528-32538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 135. | Zhao X, Gao S, Ren H, Sun W, Zhang H, Sun J, Yang S, Hao J. Hypoxia-inducible factor-1 promotes pancreatic ductal adenocarcinoma invasion and metastasis by activating transcription of the actin-bundling protein fascin. Cancer Res. 2014;74:2455-2464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 136. | Wan TM, Lam CS, Ng L, Chow AK, Wong SK, Li HS, Man JH, Lo OS, Foo D, Cheung A. The clinicopathological significance of miR-133a in colorectal cancer. Dis Markers. 2014;2014:919283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 137. | Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Sasaki K, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol. 2011;39:1099-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 138. | Kawakami K, Enokida H, Chiyomaru T, Tatarano S, Yoshino H, Kagara I, Gotanda T, Tachiwada T, Nishiyama K, Nohata N. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer. 2012;48:827-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 139. | Leung WK, Ching AK, Chan AW, Poon TC, Mian H, Wong AS, To KF, Wong N. A novel interplay between oncogenic PFTK1 protein kinase and tumor suppressor TAGLN2 in the control of liver cancer cell motility. Oncogene. 2011;30:4464-4475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 140. | Xiao G, Xia C, Yang J, Liu J, Du H, Kang X, Lin Y, Guan R, Yan P, Tang S. MiR-133b regulates the expression of the Actin protein TAGLN2 during oocyte growth and maturation: a potential target for infertility therapy. PLoS One. 2014;9:e100751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 141. | Zhang Y, Wang Y, Liu Y, Wang N, Qi Y, Du J. Krüppel-like factor 4 transcriptionally regulates TGF-β1 and contributes to cardiac myofibroblast differentiation. PLoS One. 2013;8:e63424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 142. | Chakraborty PK, Zhang Y, Coomes AS, Kim WJ, Stupay R, Lynch LD, Atkinson T, Kim JI, Nie Z, Daaka Y. G protein-coupled receptor kinase GRK5 phosphorylates moesin and regulates metastasis in prostate cancer. Cancer Res. 2014;74:3489-3500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 143. | Kinoshita T, Nohata N, Fuse M, Hanazawa T, Kikkawa N, Fujimura L, Watanabe-Takano H, Yamada Y, Yoshino H, Enokida H. Tumor suppressive microRNA-133a regulates novel targets: moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2012;418:378-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 144. | Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013;31:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 145. | Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 146. | Donnelly SK, Bravo-Cordero JJ, Hodgson L. Rho GTPase isoforms in cell motility: Don’t fret, we have FRET. Cell Adh Migr. 2014;8:526-534. [PubMed] |
| 147. | Wojnacki J, Quassollo G, Marzolo MP, Cáceres A. Rho GTPases at the crossroad of signaling networks in mammals: impact of Rho-GTPases on microtubule organization and dynamics. Small GTPases. 2014;5:e28430. [PubMed] |
| 148. | Duquette PM, Lamarche-Vane N. Rho GTPases in embryonic development. Small GTPases. 2014;5:8. [PubMed] |
| 149. | Romano D, Matallanas D, Frederick DT, Flaherty KT, Kolch W. One Hippo and many masters: differential regulation of the Hippo pathway in cancer. Biochem Soc Trans. 2014;42:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 150. | Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci USA. 2011;108:E1312-E1320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 390] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 151. | Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrózek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 607] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 152. | Gutiérrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV, Fermiñán E, Martín-Jiménez P, Chillón C, Risueño A. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010;24:629-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 172] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 153. | Wang L, Wang J, Blaser BW, Duchemin AM, Kusewitt DF, Liu T, Caligiuri MA, Briesewitz R. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110:2075-2083. [PubMed] |
| 154. | Khanjyan MV, Yang J, Kayali R, Caldwell T, Bertoni C. A high-content, high-throughput siRNA screen identifies cyclin D2 as a potent regulator of muscle progenitor cell fusion and a target to enhance muscle regeneration. Hum Mol Genet. 2013;22:3283-3295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 155. | Havelange V, Stauffer N, Heaphy CC, Volinia S, Andreeff M, Marcucci G, Croce CM, Garzon R. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer. 2011;117:4696-4706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 156. | Wang T, Yuan J, Feng N, Li Y, Lin Z, Jiang Z, Gui Y. Hsa-miR-1 downregulates long non-coding RNA urothelial cancer associated 1 in bladder cancer. Tumour Biol. 2014;35:10075-10084. [PubMed] |
| 157. | Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol. 2014;35:6901-6912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 158. | Xue M, Li X, Wu W, Zhang S, Wu S, Li Z, Chen W. Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein α contributes to bladder cancer cell growth and reduced apoptosis. Oncol Rep. 2014;31:1993-2000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 159. | Wu W, Zhang S, Li X, Xue M, Cao S, Chen W. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One. 2013;8:e73920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 160. | Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 161. | Watts R, Johnsen VL, Shearer J, Hittel DS. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013;304:C995-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 162. | Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. [PubMed] |
| 163. | Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 164. | Han X, Yang F, Cao H, Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054-3064. [PubMed] |
| 165. | Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198-206 [. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 303] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 166. | Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 167. | Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012;365:319-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 168. | Barreto-Valer K, López-Bellido R, Macho Sánchez-Simón F, Rodríguez RE. Modulation by cocaine of dopamine receptors through miRNA-133b in zebrafish embryos. PLoS One. 2012;7:e52701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 169. | Lu TY, Lin B, Li Y, Arora A, Han L, Cui C, Coronnello C, Sheng Y, Benos PV, Yang L. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J Mol Cell Cardiol. 2013;63:146-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 170. | Boettger T, Wüst S, Nolte H, Braun T. The miR-206/133b cluster is dispensable for development, survival and regeneration of skeletal muscle. Skelet Muscle. 2014;4:23. [PubMed] |
| 171. | Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Münsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 209] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 172. | Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71-84. [PubMed] |
| 173. | Basu U, Lozynska O, Moorwood C, Patel G, Wilton SD, Khurana TS. Translational regulation of utrophin by miRNAs. PLoS One. 2011;6:e29376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 174. | Tang Z, Liang R, Zhao S, Wang R, Huang R, Li K. CNN3 is regulated by microRNA-1 during muscle development in pigs. Int J Biol Sci. 2014;10:377-385. [PubMed] |
| 175. | Choi MC, Ryu S, Hao R, Wang B, Kapur M, Fan CM, Yao TP. HDAC4 promotes Pax7-dependent satellite cell activation and muscle regeneration. EMBO Rep. 2014;15:1175-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 176. | Jash S, Adhya S. Induction of muscle regeneration by RNA-mediated mitochondrial restoration. FASEB J. 2012;26:4187-4197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 177. | Zhang LX, DeNicola M, Qin X, Du J, Ma J, Tina Zhao Y, Zhuang S, Liu PY, Wei L, Qin G. Specific inhibition of HDAC4 in cardiac progenitor cells enhances myocardial repairs. Am J Physiol Cell Physiol. 2014;307:C358-C372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 178. | van Mil A, Vrijsen KR, Goumans MJ, Metz CH, Doevendans PA, Sluijter JP. MicroRNA-1 enhances the angiogenic differentiation of human cardiomyocyte progenitor cells. J Mol Med (Berl). 2013;91:1001-1012. [PubMed] |
| 179. | Izarra A, Moscoso I, Cañón S, Carreiro C, Fondevila D, Martín-Caballero J, Blanca V, Valiente I, Díez-Juan A, Bernad A. miRNA-1 and miRNA-133a are involved in early commitment of pluripotent stem cells and demonstrate antagonistic roles in the regulation of cardiac differentiation. J Tissue Eng Regen Med. 2014;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 180. | Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618-627. [PubMed] |
| 181. | Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, Kuwabara Y, Takanabe R, Hasegawa K, Kita T. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389:315-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 148] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 182. | Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Krüppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322-34328. [PubMed] |
| 183. | Vacchi-Suzzi C, Hahne F, Scheubel P, Marcellin M, Dubost V, Westphal M, Boeglen C, Büchmann-Møller S, Cheung MS, Cordier A. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One. 2013;8:e52442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 184. | McGahon MK, Yarham JM, Daly A, Guduric-Fuchs J, Ferguson LJ, Simpson DA, Collins A. Distinctive profile of IsomiR expression and novel microRNAs in rat heart left ventricle. PLoS One. 2013;8:e65809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 185. | Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170-178, 6p following 178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 186. | Eckhouse SR, Akerman AW, Logdon CB, Oelsen JM, O’Quinn EC, Nadeau EK, Stroud RE, Mukherjee R, Jones JA, Spinale FG. Differential membrane type 1 matrix metalloproteinase substrate processing with ischemia-reperfusion: relationship to interstitial microRNA dynamics and myocardial function. J Thorac Cardiovasc Surg. 2013;145:267-275, 277.e1-4; discussion 275-277. [PubMed] |
| 187. | Kumarswamy R, Lyon AR, Volkmann I, Mills AM, Bretthauer J, Pahuja A, Geers-Knörr C, Kraft T, Hajjar RJ, Macleod KT. SERCA2a gene therapy restores microRNA-1 expression in heart failure via an Akt/FoxO3A-dependent pathway. Eur Heart J. 2012;33:1067-1075. [PubMed] |
| 188. | Izarra A, Moscoso I, Levent E, Cañón S, Cerrada I, Díez-Juan A, Blanca V, Núñez-Gil IJ, Valiente I, Ruíz-Sauri A. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Reports. 2014;3:1029-1042. [PubMed] |
| 189. | Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, Ziemann M, Helbing T, El-Osta A, Jowett JB. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633-644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 354] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 190. | D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765-2773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 625] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 191. | Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, Watanabe S, Baba O, Kojima Y, Shizuta S. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446-454. [PubMed] [DOI] [Full Text] |
| 192. | Villar AV, Merino D, Wenner M, Llano M, Cobo M, Montalvo C, García R, Martín-Durán R, Hurlé JM, Hurlé MA. Myocardial gene expression of microRNA-133a and myosin heavy and light chains, in conjunction with clinical parameters, predict regression of left ventricular hypertrophy after valve replacement in patients with aortic stenosis. Heart. 2011;97:1132-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 193. | De Rosa S, Fichtlscherer S, Lehmann R, Assmus B, Dimmeler S, Zeiher AM. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 194. | García R, Villar AV, Cobo M, Llano M, Martín-Durán R, Hurlé MA, Nistal JF. Circulating levels of miR-133a predict the regression potential of left ventricular hypertrophy after valve replacement surgery in patients with aortic stenosis. J Am Heart Assoc. 2013;2:e000211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 195. | Ceylan-Isik AF, Kandadi MR, Xu X, Hua Y, Chicco AJ, Ren J, Nair S. Apelin administration ameliorates high fat diet-induced cardiac hypertrophy and contractile dysfunction. J Mol Cell Cardiol. 2013;63:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 196. | Yildirim SS, Akman D, Catalucci D, Turan B. Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: junctin as a target protein of miR-1. Cell Biochem Biophys. 2013;67:1397-1408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 197. | Danowski N, Manthey I, Jakob HG, Siffert W, Peters J, Frey UH. Decreased expression of miR-133a but not of miR-1 is associated with signs of heart failure in patients undergoing coronary bypass surgery. Cardiology. 2013;125:125-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 198. | Liao XB, Zhang ZY, Yuan K, Liu Y, Feng X, Cui RR, Hu YR, Yuan ZS, Gu L, Li SJ. MiR-133a modulates osteogenic differentiation of vascular smooth muscle cells. Endocrinology. 2013;154:3344-3352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 199. | Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T, Müller-Ardogan M. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 200. | Curcio A, Torella D, Iaconetti C, Pasceri E, Sabatino J, Sorrentino S, Giampà S, Micieli M, Polimeni A, Henning BJ. MicroRNA-1 downregulation increases connexin 43 displacement and induces ventricular tachyarrhythmias in rodent hypertrophic hearts. PLoS One. 2013;8:e70158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 201. | Zhang Y, Sun L, Zhang Y, Liang H, Li X, Cai R, Wang L, Du W, Zhang R, Li J. Overexpression of microRNA-1 causes atrioventricular block in rodents. Int J Biol Sci. 2013;9:455-462. [PubMed] |
| 202. | Li Q, Song XW, Zou J, Wang GK, Kremneva E, Li XQ, Zhu N, Sun T, Lappalainen P, Yuan WJ. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci. 2010;123:2444-2452. [PubMed] |
| 203. | Dong DL, Chen C, Huo R, Wang N, Li Z, Tu YJ, Hu JT, Chu X, Huang W, Yang BF. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: a novel mechanism for progressive cardiac hypertrophy. Hypertension. 2010;55:946-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 204. | Li Q, Lin X, Yang X, Chang J. NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression. Am J Physiol Heart Circ Physiol. 2010;298:H1340-H1347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 205. | Sanna B, Bueno OF, Dai YS, Wilkins BJ, Molkentin JD. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol Cell Biol. 2005;25:865-878. [PubMed] |
| 206. | Radzikinas K, Aven L, Jiang Z, Tran T, Paez-Cortez J, Boppidi K, Lu J, Fine A, Ai X. A Shh/miR-206/BDNF cascade coordinates innervation and formation of airway smooth muscle. J Neurosci. 2011;31:15407-15415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 207. | Belevych AE, Sansom SE, Terentyeva R, Ho HT, Nishijima Y, Martin MM, Jindal HK, Rochira JA, Kunitomo Y, Abdellatif M. MicroRNA-1 and -133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS One. 2011;6:e28324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 208. | Terentyev D, Belevych AE, Terentyeva R, Martin MM, Malana GE, Kuhn DE, Abdellatif M, Feldman DS, Elton TS, Györke S. miR-1 overexpression enhances Ca(2+) release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 209. | Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 275] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 210. | Bostjancic E, Zidar N, Stajer D, Glavac D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010;115:163-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 236] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 211. | Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495-2505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 212. | Zhang D, Li X, Chen C, Li Y, Zhao L, Jing Y, Liu W, Wang X, Zhang Y, Xia H. Attenuation of p38-mediated miR-1/133 expression facilitates myoblast proliferation during the early stage of muscle regeneration. PLoS One. 2012;7:e41478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 213. | Roberts TC, Blomberg KE, McClorey G, El Andaloussi S, Godfrey C, Betts C, Coursindel T, Gait MJ, Smith CI, Wood MJ. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic Acids. 2012;1:e39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 214. | Holmberg J, Alajbegovic A, Gawlik KI, Elowsson L, Durbeej M. Laminin α2 Chain-Deficiency is Associated with microRNA Deregulation in Skeletal Muscle and Plasma. Front Aging Neurosci. 2014;6:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 215. | Rosales XQ, Malik V, Sneh A, Chen L, Lewis S, Kota J, Gastier-Foster JM, Astbury C, Pyatt R, Reshmi S. Impaired regeneration in LGMD2A supported by increased PAX7-positive satellite cell content and muscle-specific microrna dysregulation. Muscle Nerve. 2013;47:731-739. [PubMed] |
| 216. | Deng Z, Chen JF, Wang DZ. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet Disord. 2011;12:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 217. | Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C, Vergani L, Novelli G, Botta A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 218. | Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 219. | García-Pérez D, López-Bellido R, Hidalgo JM, Rodríguez RE, Laorden ML, Núñez C, Milanés MV. Morphine regulates Argonaute 2 and TH expression and activity but not miR-133b in midbrain dopaminergic neurons. Addict Biol. 2015;20:104-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 220. | Monteleone MC, Adrover E, Pallarés ME, Antonelli MC, Frasch AC, Brocco MA. Prenatal stress changes the glycoprotein GPM6A gene expression and induces epigenetic changes in rat offspring brain. Epigenetics. 2014;9:152-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 221. | Greco SJ, Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc Natl Acad Sci USA. 2007;104:15484-15489. [PubMed] |
| 222. | Yang X, Yang Q, Wang X, Luo C, Wan Y, Li J, Liu K, Zhou M, Zhang C. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. Neuromolecular Med. 2014;16:594-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 223. | Chen YH, Heneidi S, Lee JM, Layman LC, Stepp DW, Gamboa GM, Chen BS, Chazenbalk G, Azziz R. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278-2286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 200] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 224. | Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013;17:798-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 440] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 225. | Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE, Habeos IG. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS One. 2012;7:e34872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 226. | Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F. MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in hepatocytes. Cell Signal. 2013;25:1429-1437. [PubMed] |
| 227. | Palmieri A, Pezzetti F, Brunelli G, Zollino I, Scapoli L, Martinelli M, Arlotti M, Carinci F. Differences in osteoblast miRNA induced by cell binding domain of collagen and silicate-based synthetic bone. J Biomed Sci. 2007;14:777-782. [PubMed] |
| 228. | Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906-13911. [PubMed] |
| 229. | Zhang H, Guo Y, Mishra A, Gou D, Chintagari NR, Liu L. MicroRNA-206 regulates surfactant secretion by targeting VAMP-2. FEBS Lett. 2015;589:172-176. [PubMed] |
| 230. | Visser WE, Heemstra KA, Swagemakers SM, Ozgür Z, Corssmit EP, Burggraaf J, van Ijcken WF, van der Spek PJ, Smit JW, Visser TJ. Physiological thyroid hormone levels regulate numerous skeletal muscle transcripts. J Clin Endocrinol Metab. 2009;94:3487-3496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 231. | Zhang D, Wang X, Li Y, Zhao L, Lu M, Yao X, Xia H, Wang YC, Liu MF, Jiang J. Thyroid hormone regulates muscle fiber type conversion via miR-133a1. J Cell Biol. 2014;207:753-766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 232. | Dong H, Paquette M, Williams A, Zoeller RT, Wade M, Yauk C. Thyroid hormone may regulate mRNA abundance in liver by acting on microRNAs. PLoS One. 2010;5:e12136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 233. | Tsai WJ, McCormick KM, Brazeau DA, Brazeau GA. Estrogen effects on skeletal muscle insulin-like growth factor 1 and myostatin in ovariectomized rats. Exp Biol Med (Maywood). 2007;232:1314-1325. [PubMed] |
| 234. | Zhao E, Keller MP, Rabaglia ME, Oler AT, Stapleton DS, Schueler KL, Neto EC, Moon JY, Wang P, Wang IM. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome. 2009;20:476-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 235. | Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009;218:444-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 236. | Dai A, Sun H, Fang T, Zhang Q, Wu S, Jiang Y, Ding L, Yan G, Hu Y. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587:2474-2482. [PubMed] |
| 237. | Huang F, Li ML, Fang ZF, Hu XQ, Liu QM, Liu ZJ, Tang L, Zhao YS, Zhou SH. Overexpression of MicroRNA-1 improves the efficacy of mesenchymal stem cell transplantation after myocardial infarction. Cardiology. 2013;125:18-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 238. | Huang F, Tang L, Fang ZF, Hu XQ, Pan JY, Zhou SH. miR-1-mediated induction of cardiogenesis in mesenchymal stem cells via downregulation of Hes-1. Biomed Res Int. 2013;2013:216286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 239. | Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013;100:7-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 266] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 240. | Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 492] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 241. | Burkly LC, Michaelson JS, Zheng TS. TWEAK/Fn14 pathway: an immunological switch for shaping tissue responses. Immunol Rev. 2011;244:99-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 172] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 242. | Was H, Dulak J, Jozkowicz A. Heme oxygenase-1 in tumor biology and therapy. Curr Drug Targets. 2010;11:1551-1570. [PubMed] |
| 243. | Jia HY, Chen F, Chen JZ, Wu SS, Wang J, Cao QY, Chen Z, Zhu HH. MicroRNA expression profiles related to early stage murine concanavalin A-induced hepatitis. Cell Physiol Biochem. 2014;33:1933-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 244. | Skovgaard K, Cirera S, Vasby D, Podolska A, Breum SØ, Dürrwald R, Schlegel M, Heegaard PM. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2). Innate Immun. 2013;19:531-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 245. | Yue J, Guan J, Wang X, Zhang L, Yang Z, Ao Q, Deng Y, Zhu P, Wang G. MicroRNA-206 is involved in hypoxia-induced pulmonary hypertension through targeting of the HIF-1α/Fhl-1 pathway. Lab Invest. 2013;93:748-759. [PubMed] |
| 246. | Duan X, Zohaib A, Li Y, Zhu B, Ye J, Wan S, Xu Q, Song Y, Chen H, Cao S. miR-206 modulates lipopolysaccharide-mediated inflammatory cytokine production in human astrocytes. Cell Signal. 2015;27:61-68. [PubMed] |
| 247. | Mallappa C, Hu YJ, Shamulailatpam P, Tae S, Sif S, Imbalzano AN. The expression of myogenic microRNAs indirectly requires protein arginine methyltransferase (Prmt)5 but directly requires Prmt4. Nucleic Acids Res. 2011;39:1243-1255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 248. | Torella D, Iaconetti C, Catalucci D, Ellison GM, Leone A, Waring CD, Bochicchio A, Vicinanza C, Aquila I, Curcio A. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ Res. 2011;109:880-893. [PubMed] |
| 249. | Chen M, Herring BP. Regulation of microRNAs by Brahma-related gene 1 (Brg1) in smooth muscle cells. J Biol Chem. 2013;288:6397-6408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 250. | Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 142] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 251. | Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS One. 2009;4:e6880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 252. | Stope MB, Stender C, Schubert T, Peters S, Weiss M, Ziegler P, Zimmermann U, Walther R, Burchardt M. Heat-shock protein HSPB1 attenuates microRNA miR-1 expression thereby restoring oncogenic pathways in prostate cancer cells. Anticancer Res. 2014;34:3475-3480. [PubMed] |
| 253. | Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162-6170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 550] [Cited by in RCA: 567] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 254. | Leone V, D’Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab. 2011;96:E1388-E1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 255. | Wang F, Song G, Liu M, Li X, Tang H. miRNA-1 targets fibronectin1 and suppresses the migration and invasion of the HEp2 laryngeal squamous carcinoma cell line. FEBS Lett. 2011;585:3263-3269. [PubMed] |
| 256. | Fleming JL, Gable DL, Samadzadeh-Tarighat S, Cheng L, Yu L, Gillespie JL, Toland AE. Differential expression of miR-1, a putative tumor suppressing microRNA, in cancer resistant and cancer susceptible mice. PeerJ. 2013;1:e68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 257. | Jiang C, Chen H, Shao L, Wang Q. MicroRNA-1 functions as a potential tumor suppressor in osteosarcoma by targeting Med1 and Med31. Oncol Rep. 2014;32:1249-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 258. | Furukawa S, Kawasaki Y, Miyamoto M, Hiyoshi M, Kitayama J, Akiyama T. The miR-1-NOTCH3-Asef pathway is important for colorectal tumor cell migration. PLoS One. 2013;8:e80609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 259. | Zhao Q, Zhang B, Shao Y, Chen L, Wang X, Zhang Z, Shu Y, Guo R. Correlation between the expression levels of miR-1 and PIK3CA in non-small-cell lung cancer and their relationship with clinical characteristics and prognosis. Future Oncol. 2014;10:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 260. | Tominaga E, Yuasa K, Shimazaki S, Hijikata T. MicroRNA-1 targets Slug and endows lung cancer A549 cells with epithelial and anti-tumorigenic properties. Exp Cell Res. 2013;319:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 261. | Osaka E, Yang X, Shen JK, Yang P, Feng Y, Mankin HJ, Hornicek FJ, Duan Z. MicroRNA-1 (miR-1) inhibits chordoma cell migration and invasion by targeting slug. J Orthop Res. 2014;32:1075-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 262. | Duan Z, Shen J, Yang X, Yang P, Osaka E, Choy E, Cote G, Harmon D, Zhang Y, Nielsen GP. Prognostic significance of miRNA-1 (miR-1) expression in patients with chordoma. J Orthop Res. 2014;32:695-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 263. | Yoshino H, Enokida H, Chiyomaru T, Tatarano S, Hidaka H, Yamasaki T, Gotannda T, Tachiwada T, Nohata N, Yamane T. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem Biophys Res Commun. 2012;417:588-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 264. | Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari KI, Ogawa D, Balaj L, De Rienzo G, Mineo M, Nakano I. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74:738-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 265. | Tsai KW, Hu LY, Chen TW, Li SC, Ho MR, Yu SY, Tu YT, Chen WS, Lam HC. Emerging role of microRNAs in modulating endothelin-1 expression in gastric cancer. Oncol Rep. 2015;33:485-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 266. | Li D, Yang P, Li H, Cheng P, Zhang L, Wei D, Su X, Peng J, Gao H, Tan Y. MicroRNA-1 inhibits proliferation of hepatocarcinoma cells by targeting endothelin-1. Life Sci. 2012;91:440-447. [PubMed] |
| 267. | Lu JW, Liao CY, Yang WY, Lin YM, Jin SL, Wang HD, Yuh CH. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS One. 2014;9:e85318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 268. | Li D, Liu Y, Li H, Peng JJ, Tan Y, Zou Q, Song XF, Du M, Yang ZH, Tan Y. MicroRNA-1 promotes apoptosis of hepatocarcinoma cells by targeting apoptosis inhibitor-5 (API-5). FEBS Lett. 2015;589:68-76. [PubMed] |
| 269. | Xu L, Zhang Y, Wang H, Zhang G, Ding Y, Zhao L. Tumor suppressor miR-1 restrains epithelial-mesenchymal transition and metastasis of colorectal carcinoma via the MAPK and PI3K/AKT pathway. J Transl Med. 2014;12:244. [PubMed] |
| 270. | Yu QQ, Wu H, Huang X, Shen H, Shu YQ, Zhang B, Xiang CC, Yu SM, Guo RH, Chen L. MiR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells. Biomed Pharmacother. 2014;68:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 271. | Letelier P, García P, Leal P, Álvarez H, Ili C, López J, Castillo J, Brebi P, Roa JC. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin Exp Pathol. 2014;7:1849-1867. [PubMed] |
| 272. | Kinoshita T, Nohata N, Watanabe-Takano H, Yoshino H, Hidaka H, Fujimura L, Fuse M, Yamasaki T, Enokida H, Nakagawa M. Actin-related protein 2/3 complex subunit 5 (ARPC5) contributes to cell migration and invasion and is directly regulated by tumor-suppressive microRNA-133a in head and neck squamous cell carcinoma. Int J Oncol. 2012;40:1770-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 273. | Cheng Z, Liu F, Wang G, Li Y, Zhang H, Li F. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal. 2014;26:2667-2673. [PubMed] |
| 274. | Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883-891. [PubMed] |
| 275. | Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962-3974. [PubMed] |
| 276. | Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 277. | Dong Y, Zhao J, Wu CW, Zhang L, Liu X, Kang W, Leung WW, Zhang N, Chan FK, Sung JJ. Tumor suppressor functions of miR-133a in colorectal cancer. Mol Cancer Res. 2013;11:1051-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 278. | Wang H, An H, Wang B, Liao Q, Li W, Jin X, Cui S, Zhang Y, Ding Y, Zhao L. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer. 2013;49:3924-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 279. | Chekhun VF, Lukyanova NY, Burlaka CA, Bezdenezhnykh NA, Shpyleva SI, Tryndyak VP, Beland FA, Pogribny IP. Iron metabolism disturbances in the MCF-7 human breast cancer cells with acquired resistance to doxorubicin and cisplatin. Int J Oncol. 2013;43:1481-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 280. | Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, Yang XQ, Zhang GH, Xu XC, Zhu T, Wu Q. Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC Cancer. 2012;12:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 281. | Yamamoto H, Kohashi K, Fujita A, Oda Y. Fascin-1 overexpression and miR-133b downregulation in the progression of gastrointestinal stromal tumor. Mod Pathol. 2013;26:563-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 282. | Patron JP, Fendler A, Bild M, Jung U, Müller H, Arntzen MØ, Piso C, Stephan C, Thiede B, Mollenkopf HJ. MiR-133b targets antiapoptotic genes and enhances death receptor-induced apoptosis. PLoS One. 2012;7:e35345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 283. | Zhao Y, Huang J, Zhang L, Qu Y, Li J, Yu B, Yan M, Yu Y, Liu B, Zhu Z. MiR-133b is frequently decreased in gastric cancer and its overexpression reduces the metastatic potential of gastric cancer cells. BMC Cancer. 2014;14:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 284. | Chen XN, Wang KF, Xu ZQ, Li SJ, Liu Q, Fu DH, Wang X, Wu B. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70. [PubMed] |
| 285. | Chen Z, Saad R, Jia P, Peng D, Zhu S, Washington MK, Zhao Z, Xu Z, El-Rifai W. Gastric adenocarcinoma has a unique microRNA signature not present in esophageal adenocarcinoma. Cancer. 2013;119:1985-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 286. | Kowalewska M, Bakula-Zalewska E, Chechlinska M, Goryca K, Nasierowska-Guttmejer A, Danska-Bidzinska A, Bidzinski M. microRNAs in uterine sarcomas and mixed epithelial-mesenchymal uterine tumors: a preliminary report. Tumour Biol. 2013;34:2153-2160. [PubMed] |
| 287. | Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284:31921-31927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 179] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 288. | Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 148] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 289. | Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 259] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 290. | Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143-4164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 291. | Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5:e15224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 470] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 292. | Liu H, Cao YD, Ye WX, Sun YY. Effect of microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in MDA-MB-231 cells. Tumori. 2010;96:751-755. [PubMed] |
| 293. | Leivonen SK, Mäkelä R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellström K, Sahlberg N. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 294. | Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132-1147. [PubMed] |
| 295. | Lin F, Yao L, Xiao J, Liu D, Ni Z. MiR-206 functions as a tumor suppressor and directly targets K-Ras in human oral squamous cell carcinoma. Onco Targets Ther. 2014;7:1583-1591. [PubMed] |
| 296. | Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol. 2009;23:1215-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 297. | Yang Q, Zhang C, Huang B, Li H, Zhang R, Huang Y, Wang J. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol. 2013;25:953-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 298. | Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 299. | Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, Bai M, Liu J, Shang X, Wu K. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332:94-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 300. | Liu X, He M, Hou Y, Liang B, Zhao L, Ma S, Yu Y, Liu X. Expression profiles of microRNAs and their target genes in papillary thyroid carcinoma. Oncol Rep. 2013;29:1415-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 301. | Vickers MM, Bar J, Gorn-Hondermann I, Yarom N, Daneshmand M, Hanson JE, Addison CL, Asmis TR, Jonker DJ, Maroun J. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis. 2012;29:123-132. [PubMed] |
| 302. | Liu W, Xu C, Wan H, Liu C, Wen C, Lu H, Wan F. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med. 2014;34:420-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 303. | Ren J, Huang HJ, Gong Y, Yue S, Tang LM, Cheng SY. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014;4:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 304. | Georgantas RW, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW, Richman L. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res. 2014;27:275-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 305. | Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, Williams C. miR-206 inhibits cell migration through direct targeting of the actin-binding protein coronin 1C in triple-negative breast cancer. Mol Oncol. 2014;8:1690-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 306. | Ali S, Karki N, Bhattacharya C, Zhu R, MacDuff DA, Stenglein MD, Schumacher AJ, Demorest ZL, Harris RS, Matin A. APOBEC3 inhibits DEAD-END function to regulate microRNA activity. BMC Mol Biol. 2013;14:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 307. | Bao YP, Yi Y, Peng LL, Fang J, Liu KB, Li WZ, Luo HS. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev. 2013;14:3751-3755. [PubMed] |
| 308. | Munagala R, Aqil F, Vadhanam MV, Gupta RC. MicroRNA ‘signature’ during estrogen-mediated mammary carcinogenesis and its reversal by ellagic acid intervention. Cancer Lett. 2013;339:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 309. | Taulli R, Foglizzo V, Morena D, Coda DM, Ala U, Bersani F, Maestro N, Ponzetto C. Failure to downregulate the BAF53a subunit of the SWI/SNF chromatin remodeling complex contributes to the differentiation block in rhabdomyosarcoma. Oncogene. 2014;33:2354-2362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 310. | Lee MJ, Yoon KS, Cho KW, Kim KS, Jung HS. Expression of miR-206 during the initiation of mammary gland development. Cell Tissue Res. 2013;353:425-433. [PubMed] |
| 311. | Keklikoglou I, Hosaka K, Bender C, Bott A, Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2014;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 312. | Yamasaki T, Yoshino H, Enokida H, Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Fuse M, Seki N, Nakagawa M. Novel molecular targets regulated by tumor suppressors microRNA-1 and microRNA-133a in bladder cancer. Int J Oncol. 2012;40:1821-1830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 313. | Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol. 2012;30:434-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 314. | Chen WS, Leung CM, Pan HW, Hu LY, Li SC, Ho MR, Tsai KW. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol Rep. 2012;28:1069-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 315. | Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 316] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 316. | Hidaka H, Seki N, Yoshino H, Yamasaki T, Yamada Y, Nohata N, Fuse M, Nakagawa M, Enokida H. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;3:44-57. [PubMed] |
| 317. | Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2014;7:4194-4203. [PubMed] |
| 318. | de Cubas AA, Leandro-García LJ, Schiavi F, Mancikova V, Comino-Méndez I, Inglada-Pérez L, Perez-Martinez M, Ibarz N, Ximénez-Embún P, López-Jiménez E. Integrative analysis of miRNA and mRNA expression profiles in pheochromocytoma and paraganglioma identifies genotype-specific markers and potentially regulated pathways. Endocr Relat Cancer. 2013;20:477-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 319. | Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477-4487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 215] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 320. | Rosell R, Wei J, Taron M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin Lung Cancer. 2009;10:8-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 321. | Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 253] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 322. | Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Fujimura L, Yoshino H, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M. Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol. 2011;38:209-217. [PubMed] |
| 323. | Mutallip M, Nohata N, Hanazawa T, Kikkawa N, Horiguchi S, Fujimura L, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M. Glutathione S-transferase P1 (GSTP1) suppresses cell apoptosis and its regulation by miR-133α in head and neck squamous cell carcinoma (HNSCC). Int J Mol Med. 2011;27:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 324. | Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967-1975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 325. | Hrdličková R, Nehyba J, Bargmann W, Bose HR. Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One. 2014;9:e86990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 326. | Akanuma N, Hoshino I, Akutsu Y, Murakami K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110:189-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 327. | Luo J, Zhou J, Cheng Q, Zhou C, Ding Z. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol Lett. 2014;7:1043-1048. [PubMed] |
| 328. | Novello C, Pazzaglia L, Cingolani C, Conti A, Quattrini I, Manara MC, Tognon M, Picci P, Benassi MS. miRNA expression profile in human osteosarcoma: role of miR-1 and miR-133b in proliferation and cell cycle control. Int J Oncol. 2013;42:667-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 329. | Wen D, Li S, Ji F, Cao H, Jiang W, Zhu J, Fang X. miR-133b acts as a tumor suppressor and negatively regulates FGFR1 in gastric cancer. Tumour Biol. 2013;34:793-803. [PubMed] |
| 330. | Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou JY, Du J, Lv L, Gao K, Zhang Y. TAp63 suppress metastasis via miR-133b in colon cancer cells. Br J Cancer. 2014;110:2310-2320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 331. | Duan FT, Qian F, Fang K, Lin KY, Wang WT, Chen YQ. miR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol Cancer. 2013;12:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 332. | Xiang KM, Li XR. MiR-133b acts as a tumor suppressor and negatively regulates TBPL1 in colorectal cancer cells. Asian Pac J Cancer Prev. 2014;15:3767-3772. [PubMed] |
| 333. | Cristobal I, Madoz-Gurpide J, Martin-Aparicio E, Carames C, Aguilera O, Rojo F, Garcia-Foncillas J. Comment on ‘TAp63 suppress metastasis via miR-133b in colon cancer cells’. Br J Cancer. 2014;111:2369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 334. | Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168-1177. [PubMed] |
| 335. | Wu D, Pan H, Zhou Y, Zhou J, Fan Y, Qu P. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol Med Rep. 2014;9:2491-2498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 336. | Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H. Distinct expressions of microRNAs that directly target estrogen receptor α in human breast cancer. Breast Cancer Res Treat. 2011;130:331-339. [PubMed] |
| 337. | Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 338. | Min W, Wang B, Li J, Han J, Zhao Y, Su W, Dai Z, Wang X, Ma Q. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 339. | Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29-42. [PubMed] |
| 340. | Wei W, Hu Z, Fu H, Tie Y, Zhang H, Wu Y, Zheng X. MicroRNA-1 and microRNA-499 downregulate the expression of the ets1 proto-oncogene in HepG2 cells. Oncol Rep. 2012;28:701-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 341. | Wang XW, Xi XQ, Wu J, Wan YY, Hui HX, Cao XF. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol Rep. 2015;33:1402-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 342. | Guo L, Bai H, Zou D, Hong T, Liu J, Huang J, He P, Zhou Q, He J. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res. 2014;33:99. [PubMed] |
| 343. | Zheng K, Liu W, Liu Y, Jiang C, Qian Q. MicroRNA-133a suppresses colorectal cancer cell invasion by targeting Fascin1. Oncol Lett. 2015;9:869-874. [PubMed] |
| 344. | Murakami Y, Tanahashi T, Okada R, Toyoda H, Kumada T, Enomoto M, Tamori A, Kawada N, Taguchi YH, Azuma T. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One. 2014;9:e106314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (1)] |