Published online Feb 26, 2014. doi: 10.4331/wjbc.v5.i1.12
Revised: December 11, 2013
Accepted: January 6, 2014
Published online: February 26, 2014
Processing time: 160 Days and 13.8 Hours
Interferon production is an important defence against viral replication and its activation is an attractive therapeutic target. However, it has long been known that viruses perpetually evolve a multitude of strategies to evade these host immune responses. In recent years there has been an explosion of information on virus-induced alterations of the host immune response that have resulted from data-rich omics technologies. Unravelling how these systems interact and determining the overall outcome of the host response to viral infection will play an important role in future treatment and vaccine development. In this review we focus primarily on the interferon pathway and its regulation as well as mechanisms by which respiratory RNA viruses interfere with its signalling capacity.
Core tip: Many novel regulators of innate immune signalling pathways, such as the interferon signalling pathway, have been discovered recently. These advances may be in part attributed to high-throughput systems biology techniques including genomic, proteomic, miRNA and siRNA screens, as well as through various confirmatory methods using quantitative polymerase chain reaction, microscopy, and animal models. Collectively, these studies have provided insights into novel drug targets that could boost host innate immunity or could potentially serve as broad-spectrum anti-virals against RNA respiratory viruses.
- Citation: Kroeker AL, Coombs KM. Systems biology unravels interferon responses to respiratory virus infections. World J Biol Chem 2014; 5(1): 12-25
- URL: https://www.wjgnet.com/1949-8454/full/v5/i1/12.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i1.12
Virus-host studies of a wide range of viruses have identified many host changes that occur upon infection, including the induction of a variety of anti-viral pathways. For example, these include autophagy, apoptosis, endoplasmic-reticular stress, nuclear-factor kappa B (NF-κB) and proteasomal degradation pathways as well as the topic of this review, interferon signalling. Some of these studies have utilized global genomic, transcriptomics and proteomic technologies and have led to the characterizations of “infectomes”, “interactomes” and “interferomes”. One of the great advantages to systems biology tools is that they can provide a relatively unbiased “bottom-up” discovery approach such as with global transcriptome and siRNA screens. These have proven useful in the characterization of innate immune responses. Biological tools for detection of specific subsets of the cell are also continually being developed, including probes for specific classes of enzymes, methods to detect different protein post-translational and epigenetic modifications, and subcellular fractionation techniques. As will be discussed below, many studies have begun to characterize gene transcription programs in response to viruses, have identified novel anti-viral proteins and regulators of interferon production and have experimented with novel approaches to treatment of viral infection.
The study of interferons (IFN) is one of the oldest known family of proteins with anti-viral properties. They are produced and released in response to pathogens, such as viruses and bacteria, and function in establishing an anti-viral state in host cells and activating immune cells (for review see[1]). Type I interferons in humans include IFN-α, IFN-β, IFN-ε, IFN-κ and IFN-ω and are classified as such by their ability to bind the IFNAR1-IFNAR2 interferon receptor complex[2]. IFN-γ is a type II interferon and signals through the IFNGR1-IFNGR2 receptor complex. A third class of interferons, type III, has been proposed and would likely contain IFN-λ1, -λ2 and -λ3, which are also known as interleukin-29 (IL-29), IL-28A and IL-28B, respectively, and bind IFNLR1 (also known as IL-28 receptor-α, IL-28Rα) and IL-10Rβ[3]. Effects of interferons are numerous and depend on downstream signaling pathways. The canonical activation of Janus-Kinase-Signal Transduction Activator (JAK-STAT) signalling[4], for example, induces a variety of interferon-stimulated genes (ISGs) of which some have known anti-viral activities. Activation of mitogen-activated protein kinases[5] has also been shown to have anti-viral as well as anti-proliferative effects. In contrast, phosphatidylinositol 3-kinase activation[6] induces cell proliferation and increased protein synthesis (for review see[7]). Autophagy has also been described as an inducer of interferon[8,9] as well as being induced by interferons[10,11]. The interactions and cross-regulation of these pathways are complex and are not well defined but overall, the ability of the host to mount an effective interferon response typically plays a significant protective role against viral pathogenicity.
Regulation of the interferon signalling pathway is influenced by multiple cellular regulatory systems including phosphorylation, ubiquitination and miRNA silencing. In addition, viral components such as viral proteins and viral RNA can also significantly impact interferon production by the infected host cell. Systems biology approaches have substantially contributed to understanding the interactions of these various regulatory networks, the overall outcome of their actions, and their impact on respiratory virus replication. For example, it is becoming increasingly popular to combine various omics technologies such as transcriptome and proteomic screens with functional validation using techniques such as siRNA screens, pPCR and microscopy imaging.
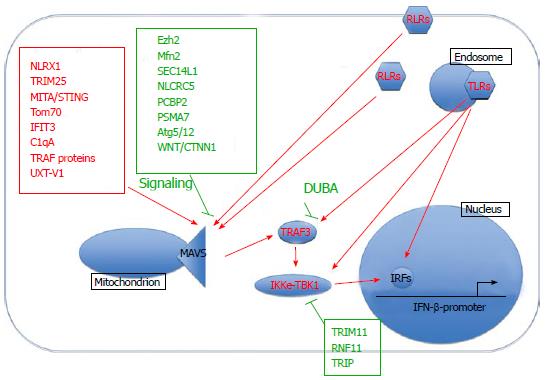
Innate immune responses are initially triggered in response to viral infection through the recognition of highly conserved pathogen association molecular patterns (PAMPs). In terms of RNA viruses this typically involves activation of RIG-like (RLR), Toll-like (TLR) and Nod-like receptors (NLR) in the cytoplasm and at membranous surfaces such as the plasma membrane, endosomes and endoplasmic reticulum. A major outcome of RLR and TLR activation is the production of interferons. This induction, and its regulation, will be the focus of this review (summarized in Figure 1).
An important event following RLR activation consists of the formation of mitochondrion-centric anti-viral signalling complexes that regulate interferon and NF-κB signalling cascades and subsequent immune responses. The mitochondrial anti-viral signaling protein (MAVS)/virus-induced signaling adaptor/interferon-beta promoter stimulator protein 1/Cardif protein is central to this process. Located at the outer mitochondrial membrane, it acts as a scaffolding protein that interacts with a variety of different host proteins that regulate downstream signalling pathways. There are many activators and facilitators of MAVS-mediated signalling and some of the most recently discovered ones include retinoic-acid inducible gene I (RIG-I), nucleotide-binding oligomerization domain, leucine rich repeat containing X1 (NLRX1), MITA/Stimulator of interferon genes protein[12], Tom70[13], interferon-induced protein with tetratricopeptide repeats 3 (IFIT3)[14], C1qA[15], tumor necrosis factor receptor associated factor (TRAF) proteins[16] and UXT-V1[17]. The formation of MAVS-mediated complexes can subsequently lead to the recruitment of tank binding kinase (TBK1) and inhibitor of nuclear factor kappa-B kinase (IKKe). However, this process is also carefully controlled through recruitment of negative regulators such as Ezh2[18], Mfn2[19], SEC14L1[20] and Wnt/beta-catenin (WNT/CTNNB1) signalling[21]. MAVS has also been described to associate with the endoplasmic reticulum[12,22-24], peroxisomes[22], and autophagosomes[25], although the outcome of these events are beyond the scope of this review. For further details we direct readers to a review by Belgnaoui[26]. Overall, MAVS-interacting partners influence the extent of activation or inhibition of downstream interferon and NF-κB anti-viral pathways.
RLR and TLR activation culminate in the phosphorylation, activation and nuclear translocation of various IRF transcription factors. Two well-known factors are IRF3 and IRF7, which can be activated by kinases TBK1, IKKi, TAK1, and interleukin-1 receptor-associated kinase. This activation is carefully controlled through ubiquitin-mediated degradation of TBK1, which can be negatively regulated by tripartite motif containing 11 (TRIM11)[27], ring finger protein 11 (RNF11)[28] and thyroid receptor-interacting protein[29]. Interaction with other molecules such as TRAF3, DDX3 [(DEAD (Asp-Glu-Ala-Asp) box polypeptide 3][30] and nef-associated protein 1[31] can also modulate downstream signalling. Interestingly, a recent study using triple IRF3/IRF5/IRF7 knockout mice[32] demonstrated a formerly unappreciated role of IRF5 in interferon induction in myeloid dendritic cells. Genome-wide IRF1 binding sites have also been characterized in primary monocytes[33]. Overall, the IRF family members are essential mediators of interferon signalling in response to RNA viral infection.
Numerous other proteins have been described in regulating interferon production including activators Gab1[34] and suppressors protein tyrosine phosphatase 1[35], forkhead box protein O3[36], and toll/interleukin-1 receptor domain containing adaptor molecule 2 (TRIF) degradation[37]. Several E3 ligases promote interferon signalling such as Pellino1[38], TRIM25[39], TRIM32[40] and Riplet[41]. Other E3 ligases have been characterized with a negative regulatory role in interferon production, such as Smurf1[42], RNF125[43], disintegrin and metalloproteinase domain-containing protein 15[44], TRIM38[37], TRIM11[27] and TRIM21[45]. Finally, several deubiquitinases appear to negatively regulate interferon responses, for example OTUB1[46] and UCHL1[47]. In addition, miRNAs are emerging as important regulators of interferon-mediated anti-viral responses such as miR-155[48], miR-21[49,50], miR-146[51] and miR-466l[52].
Secreted type I interferons bind to interferon receptors at the cell-membrane and induce the janus activated kinase-signal transducer and activator (JAK-STAT) pathway. The bound receptor activates self-catalyzed kinase activity and causes phosphorylation, dimerization and nuclear translocation of STAT proteins. Ubiquitination has also been demonstrated to negatively regulate this pathway, for example, by ubiquitinating JAK1[53] and STAT1[54,55] as well as through binding of suppressor of cytokine signaling and protein inhibitor of activated STAT proteins, which recruit E3 ligases[56]. In addition, mir-19a has been identified as a JAK-STAT regulator[57].
There are many different interferon transcriptional programs that depend on factors such as the receptor and JAK isoforms, as well as the type of STAT dimer[58] that are induced. These in turn are dependent on the stimulus, species, cell type, and co-stimuli. Because of this complexity, the study of interferon-stimulated gene (ISG) transcription patterns has benefited greatly from omics studies and has begun to provide powerful insights into the effects of interferons on host transcription. The response to interferon-gamma, for example, has been a source of recent interest and has been demonstrated to regulate ISGs at both the mRNA[59] and miRNA level[59,60]. A few specific miRNAs that have been identified as interferon regulators include miR-203[61] and miR-9[62]. Genome-wide DNA-binding sites for STAT1 have also been characterized using ChIP-Seq[63]. Many quantitative proteomic studies have also identified altered expression patterns of interferon-induced proteins upon various stimuli, especially after viral infection; some of these genes have also been found to be dependent upon NF-κB signalling[64].
A variety of models have been used to study the induction of innate immune pathways following virus infection, including epithelial cells, productive and abortive infections in macrophages[65], dendritic cells[66] and animal models (see Table 1). Microarrays have been particularly popular for these studies due to its ability to provide a comprehensive analysis of the entire cellular genome with relatively sensitive quantification of gene expression (see[67] for review of microarray technologies). Quantitative proteomic studies have also been important in validating these findings at the protein level and have been useful, for example, in the search for biomarkers. Many respiratory viruses, such as influenza[68-73], reovirus, and rhinovirus[74,75], demonstrate a robust activation of antiviral pathways and pro-inflammatory cytokines. Both genomic and proteomic analyses have demonstrated hubs of gene and protein induction that are induced by key transcriptional factors such as IRFs, STAT proteins, NF-κB and JNK. On the other hand, genomic profiling of respiratory syncytial virus[65] and pathogenic coronaviruses such as severe acute respiratory syndrome (SARS) and EMC strains have been reported to elicit weaker innate immune responses[76-78]. The absence of interferon signalling has also been recapitulated in several proteomic viral-host studies[79-81].
| Cell type | Proteomics | Genomics | |
| Respiratory syncytial virus | Epithelial cells | [95-98] | [99] |
| Macrophages | - | [65,100] | |
| Cord blood | - | [101] | |
| Coronavirus | Epithelial cells | [79,80,102-104] | [76-78] |
| Pro-monocytes | - | ||
| Influenza | Macrophages | [70,73,105,106] | [110] |
| Epithelial cells | [71, 72, 107] | [111] | |
| Mice | [108] | [112,113] | |
| Ferrets | - | [114,115] | |
| Macaques | [109] | [112,116] | |
| Reovirus | Epithelial cells | [117-120] | [64] |
| Myocytes | [119] | - | |
| Mice | - | [121] | |
| Rhinovirus | Epithelial cells | - | [74,122,123] |
| Dendritic cells | [66] | - | |
| Human nasal cells | - | [75] |
Analyses of microRNA expression during influenza have recently begun to emerge in a variety of models including respiratory epithelial cells[82-85], human blood[86], immune cells[87-89] and lung tissue in animal models[90,91]. Collectively these have identified roles for miR-18a[86,92] and miR-223[86,93] in negative regulation of STAT3, mir-29 in IFN-γ1 production[89], and miR-449b as a positive regulator of IFN-β production[85]. miR-23b has also been identified as a novel anti-viral molecule that is induced through RLR signaling during rhinovirus infection[94].
Strain differences: One of the fundamental questions of virology revolves around deciphering factors of pathogenesis. Hence, some studies have attempted to identify pathways that are differentially altered by pathogenic viral strains compared to less pathogenic strains. Influenza has been particularly well studied in this respect and several host factors have been identified that are unique to the replication of strains such as the pathogenic avian H5N1, the p2009 swine flu and the 1918 strain[69,70,108,124]. However, rather than inducing radically different cell responses, many different influenza strains have been found to activate surprisingly similar immune signatures (reviewed in[125]). It was, instead, the degree and timing of activation and resolution[125] of these pathways that was found to significantly impact the severity of disease[126]. Dysregulation of the host inflammatory response in particular is a major determinant of influenza pathogenicity and is influenced by both viral and host factors[127]. Different rhinovirus strains, for example type 14 and 1B[128,129], have also been demonstrated to have different abilities to attenuate interferon production and secretion from epithelial cells. This effect has been attributed to the inhibition of IRF3 dimerization[74,129] but the viral mechanism leading to this is unknown.
Cell type differences: Cell types have also been demonstrated to express different basal levels of interferon and hence, have different innate susceptibilities to viral infection[130,131]. For example, a direct comparison of interferon signaling between primary bronchial lung epithelial cells and the A549 continuous alveolar epithelial cell line suggested differences between either primary and cancer cell lines and/or epithelial cells of different origins in the lung[72]. Additionally, different cell types have been shown to influence the degree of interferon activation after reovirus infection[132].
Correlation of interferon signaling with pathogenesis: Generally interferon production is considered protective against viral infections. It has been shown numerous times that cells that produce less interferon, such as Vero cells, are more susceptible to viral infection and produce high titers of the virus[133]. The extent of interferon inhibition by the influenza non-structural (NS)-1 protein[134] and RSV NS1 and NS2 proteins[135,136] has also been extensively studied and correlates negatively with pathogenicity[137,138]. Similarly, models in which interferon signaling has been disrupted, such as by deleting IFNR, can produce high viral titers[139] and display increased lung tissue pathology[140]. Conversely, type I interferon signaling has also been shown to contribute to secondary bacterial infections[141,142]. In some studies the degree of interferon induction correlated positively with the degree of pathogenicity. For example, the reovirus T3D strain is considered more pathogenic the T1L strain, but the T3D strain was found to induce higher levels of innate immunity proteins[64,117,118]. The role of interferons in these situations is not currently understood.
Altered innate immune responses in chronic lung diseases: Many studies with rhinovirus have investigated differences in the immune response between healthy and non-healthy donor cells. In one study, infection of chronic obstructive pulmonary disorder (COPD) epithelial cells induced higher transcription levels of cytokines, chemokines, RNA helicases, interferons and increased apoptosis compared to infection of healthy control cells. In addition, basal levels of several antiviral signalling pathways were altered in COPD patients[128]. Similarly, asthma-derived epithelial cells also showed altered expression of several immunity genes both at basal levels and during rhinovirus infection[122,143]. Modulation of rhinovirus-induced host responses has also been investigated in the presence of Echinacea extracts and cigarette smoke[123].
Core innate immune responses shared by multiple respiratory viruses: While many studies that have been discussed in this review have focused on identifying global host responses towards a single virus, a few studies have directly compared viruses from multiple families. For example, Smith el al[144]. identified common gene networks that were activated in response to seven respiratory viruses: influenza A virus, respiratory syncytial virus, rhinovirus, SARS-coronavirus, metapneumonia virus, coxsackievirus and cytomegalovirus[144]. Among those responses were pathways associated with a general immune response including interferon signalling[144]. A second study also identified core immune responses to four respiratory viruses including apoptosis induction, endoplasmic reticulum stress and interferon signalling[98]. In addition several host interferon-induced proteins have been tested against multiple families and strains of viruses. For example, IFIT1[145], Interferon-inducible transmembrane (IFITM) proteins[146], ISG15[147,148] and Viperin[149-152] protect against multiple virus families.
Overall, microarrays and quantitative proteomics have allowed sensitive and comprehensive analyses of the host genome, and have contributed substantially to understanding the types and kinetics of signaling pathways that are activated upon viral infections.
Interactomes, viral-mediated antagonism of interferon signaling: As many viruses encode interferon-antagonizing proteins, there has been significant interest in defining their interacting partners in the host cell. Several studies have also been undertaken to identify host proteins that recognize dsRNA and 5’pppRNA. This has, for example, led to the discovery and characterization of the IFIT family[145] and their role anti-viral innate immunity.
Influenza: The influenza NS1 protein is a well-known antagonist of interferon signalling and is able to interfere with multiple anti-viral pathways. Viral-host studies have identified additional host proteins that interact with the influenza NS1 protein, using either plasmid-based expression of NS1[153-155] or during whole virus infection[153,156]. Collectively, the integration of multiple interactome studies has allowed networks such as Flu-Pol to be established which provide the basis for comparing differences and commonalities between influenza strains and cell types and are useful for targeted drug design.
RSV: RSV proteins NS1 and NS2 strongly inhibit IFN α/β by preventing the phosphorylation of the IFN regulatory factor-3[157,158] as well as activation of NLRX1 and RIG-I[35]. Additionally, the RSV NS1 protein interferes with interferon signaling through interaction with an elonginC-cullin2 E3 ligase complex that ubiquitinates and degrades STAT2[97,159]. RSV NS1 and NS2 have also been shown to alter miRNA expression, which can contribute to antagonism of interferon and NF-κB responses[160].
Coronavirus: In studies with coronaviruses, it has been previously proposed that the viral deubiquitinase, PLpro, plays a major role in suppressing interferon-alpha induction. In support of this idea, Li et al[161] recently demonstrated that PLpro overexpression mediated the down-regulation of mitogen-activated protein kinase and up-regulation of the ubiquitinase Ubiquitin ligase (UBC E2-25k). The open reading frame 6 protein has also been shown to attenuate antiviral responses by sequestering host nuclear impact factors including STAT1[162], vitamin D receptor, cyclic AMP-responsive element-binding protein 1, mothers against decapentaplegic homolog 4, p53, Epas I and Oct3/4[163].
Rhinovirus: Despite induction of interferon gene transcription, rhinovirus (type 14) infection can strongly attenuate interferon secretion from epithelial cells. This effect has been attributed to the inhibition of IRF3 dimerization[74,129] but the viral mechanism leading to this is unknown. In contrast, rhinovirus 1B readily stimulated interferon production in bronchial smooth muscle cells[164], suggesting different interferon regulation between strains and/or cell types.
Reovirus: The degree of IFN-α/β induction after reovirus infection has been attributed to both host and viral factors but is not well understood. However, repression of interferon signaling has been mapped to the M1, L2 and S2[132,165] genes.
Knockdown/Knockout studies: siRNA technology has been important in testing functional effects of interferon-induced proteins. Both whole genome siRNA screens, and individual knockdown experiments have discovered and validated anti-viral effects of many including interferon-induced proteins such as the IFITM1-3 proteins[166], IRF3 and IRF2 (Shapira), ISG15[147] and Viperin[167]. In contrast, several interferon pathway members have been assigned pro-viral functions such as MxB[168] and IFIT2[156,168].
Knock-out animals have also underscored the protective effects of interferon signaling during respiratory virus infections, for example, ISG15-/-[147,169], IFNAR-/-[170], and MxA-/-[171]. In addition to its role in innate immunity, interferons have also been demonstrated to have profound effects on the adaptive immune system, for example, by priming CD+ T-cells during influenza infection[172] and inhibiting neurotropism of reovirus infection[173,174]. Although discussion of the effects of interferon on whole host immunity is beyond the scope of this review, further discussion can be found in several comprehensive reviews[175,176].
Collectively, these studies have provided fundamental insights into how cells respond to RNA virus infection and have highlighted the importance of interferon induction in restricting virus replication and activating an appropriate host immune response. Many new and unexpected regulators of interferon signalling have been discovered and have demonstrated how multiple anti-viral networks interact such as ubiquitin-mediated regulation of interferon signalling molecules. As large omics studies move forward, it will become possible to compare and draw connections between anti-viral networks that are induced by different viruses.
Using interferons therapeutically has been most extensively studied in models of hepatitis. However, it has also shown some promise in protecting against a variety of other virus families, including the respiratory viruses discussed in this review. For example, exogenous IFN-alpha treatment has proven effective against influenza[177-179], rhinovirus[128,180] and coronavirus[181-183]. Interferons are also important in protecting against reovirus infections[184]. The role of type III interferons is generally not as well understood as type I but may also afford protection against respiratory viruses[185].
Interferons can also be endogenously elicited through a variety of RLR and TLR agonists. 5’pppRNA, for example, is a well-known and potent RIG-I agonist and has been demonstrated to protect against both RNA and DNA viruses, including Dengue virus, influenza, hepatitis C and human immunodeficiency virus-1[186]. Similarly, TLR agonists such as dsRNA[187,188] or inosine-containing ssRNA[189] have been shown to protect against coronavirus, influenza, and respiratory syncytial virus infections in mice. A commercial compound, Arbidol, has also had some success in neutralizing various respiratory viruses such as influenza, rhinovirus, adenovirus, coxsackie virus and RSV[190]. Additional small molecules that induce type I interferons have recently been identified using high-throughput screens[191,192]. Alternatively, inhibiting antagonists of interferon signaling can also boost the production of interferon. As discussed above, these antagonists can either be host molecules or viral proteins, and inhibitors to each have been described[193]. Interestingly, ribavirin treatment of RSV-infected epithelial cells was shown to enhance interferon-stimulated gene expression[194] and treating RSV-infected macrophages with lovastatin was shown to blunt pro-inflammatory cytokine gene expression[100]. These therapies may have potential for broad-spectrum anti-viral properties.
Despite successfully treating some viral infection with interferon, it has also been noted that interferon stimulation can increase lung inflammation. Many gene array studies have also positively correlated pathogenicity or cytopathology with the induction of interferon and/or inflammatory genes. For example, the severe pathology of the 1918 influenza pandemic and of H5N1 (bird flu) viruses has been attributed to a “cytokine storm” (reviewed by[125]). It is therefore important to identify the mechanisms behind interferon-dependent protection against viruses. Numerous studies, for example, have suggested that MxA is a major effector of INF-α pre-treatment against influenza[195-197]; other newly identified interferon-induced anti-viral proteins include IFITM proteins[146,198], ISG15[147] and Viperin[149-152]. It may also be useful to combine interferon treatment with anti-inflammatory compounds such as curcumin [199-201], resveratrol[202], S1P agonists[203,204], COX-2 inhibitors[205,206] and statins[100,207].
The study of immune responses to viral infection has benefited greatly from viral proteomic studies. However, knowledge of proteomic subsets is still limited and future studies could provide more detailed insight into the dynamics of protein localization, activity and regulation through post-translational modifications during virus infection. Based on current technologies and identified networks, it may be beneficial to also investigate alterations of the phosphoproteome, ubiquitome, and the activity of proteasomes after viral infection. The development of broad-spectrum anti-virals has also shown some potential and could benefit from comparative analyses of multiple viruses.
P- Reviewers: Carter WG, Faik A S- Editor: Wen LL L- Editor: A E- Editor: Wu HL
| 1. | Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255:25-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 2. | de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: biochemistry and biological functions. J Biol Chem. 2007;282:20053-20057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 290] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 3. | Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1416] [Cited by in RCA: 1484] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 4. | Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 5. | Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1029] [Cited by in RCA: 1369] [Article Influence: 114.1] [Reference Citation Analysis (0)] |
| 6. | Uddin S, Yenush L, Sun XJ, Sweet ME, White MF, Platanias LC. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3’-kinase. J Biol Chem. 1995;270:15938-15941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 158] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2205] [Cited by in RCA: 2621] [Article Influence: 131.1] [Reference Citation Analysis (0)] |
| 8. | Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 285] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 9. | Law AH, Lee DC, Yuen KY, Peiris M, Lau AS. Cellular response to influenza virus infection: a potential role for autophagy in CXCL10 and interferon-alpha induction. Cell Mol Immunol. 2010;7:263-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Schmeisser H, Fey SB, Horowitz J, Fischer ER, Balinsky CA, Miyake K, Bekisz J, Snow AL, Zoon KC. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy. 2013;9:683-696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 11. | Kalvakolanu DV, Gade P. IFNG and autophagy: a critical role for the ER-stress mediator ATF6 in controlling bacterial infections. Autophagy. 2012;8:1673-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1214] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 13. | Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20:994-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 14. | Liu XY, Chen W, Wei B, Shan YF, Wang C. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J Immunol. 2011;187:2559-2568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 15. | Wang Y, Tong X, Zhang J, Ye X. The complement C1qA enhances retinoic acid-inducible gene-I-mediated immune signalling. Immunology. 2012;136:78-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 292] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 17. | Huang Y, Liu H, Ge R, Zhou Y, Lou X, Wang C. UXT-V1 facilitates the formation of MAVS antiviral signalosome on mitochondria. J Immunol. 2012;188:358-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Chen CH, Zhang XQ, Lo CW, Liu PF, Liu YT, Gallo RL, Hsieh MF, Schooley RT, Huang CM. The essentiality of alpha-2-macroglobulin in human salivary innate immunity against new H1N1 swine origin influenza A virus. Proteomics. 2010;10:2396-2401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 19. | Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 197] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 20. | Li MT, Di W, Xu H, Yang YK, Chen HW, Zhang FX, Zhai ZH, Chen DY. Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. J Virol. 2013;87:10037-10046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Baril M, Es-Saad S, Chatel-Chaix L, Fink K, Pham T, Raymond VA, Audette K, Guenier AS, Duchaine J, Servant M. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog. 2013;9:e1003416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 22. | Horner SM, Liu HM, Park HS, Briley J, Gale M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:14590-14595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 401] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 23. | Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653-8658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 699] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 24. | Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2812] [Cited by in RCA: 2682] [Article Influence: 157.8] [Reference Citation Analysis (0)] |
| 25. | Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050-14055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 476] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 26. | Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 27. | Lee Y, Song B, Park C, Kwon KS. TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1. PLoS One. 2013;8:e63255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Charoenthongtrakul S, Gao L, Parvatiyar K, Lee D, Harhaj EW. RING finger protein 11 targets TBK1/IKKi kinases to inhibit antiviral signaling. PLoS One. 2013;8:e53717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Zhang M, Wang L, Zhao X, Zhao K, Meng H, Zhao W, Gao C. TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med. 2012;209:1703-1711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 116] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 30. | Gu L, Fullam A, Brennan R, Schröder M. Human DEAD box helicase 3 couples I Kappa B kinase Epsilon to interferon regulatory factor 3 activation. Mol Cell Biol. 2013;33:2004-2015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Zhao W, Wang L, Zhang M, Wang P, Yuan C, Qi J, Meng H, Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-β production and antiviral response by targeting NAP1. J Immunol. 2012;188:5311-5318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 32. | Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 33. | Shi L, Perin JC, Leipzig J, Zhang Z, Sullivan KE. Genome-wide analysis of interferon regulatory factor I binding in primary human monocytes. Gene. 2011;487:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Zheng Y, An H, Yao M, Hou J, Yu Y, Feng G, Cao X. Scaffolding adaptor protein Gab1 is required for TLR3/4- and RIG-I-mediated production of proinflammatory cytokines and type I IFN in macrophages. J Immunol. 2010;184:6447-6456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Xu H, An H, Hou J, Han C, Wang P, Yu Y, Cao X. Phosphatase PTP1B negatively regulates MyD88- and TRIF-dependent proinflammatory cytokine and type I interferon production in TLR-triggered macrophages. Mol Immunol. 2008;45:3545-3552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 36. | Luron L, Saliba D, Blazek K, Lanfrancotti A, Udalova IA. FOXO3 as a new IKK-Epsilon-controlled check-point of regulation of IFN-β expression. Eur J Immunol. 2012;42:1030-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Xue Q, Zhou Z, Lei X, Liu X, He B, Wang J, Hung T. TRIM38 negatively regulates TLR3-mediated IFN-β signaling by targeting TRIF for degradation. PLoS One. 2012;7:e46825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Enesa K, Ordureau A, Smith H, Barford D, Cheung PC, Patterson-Kane J, Arthur JS, Cohen P. Pellino1 is required for interferon production by viral double-stranded RNA. J Biol Chem. 2012;287:34825-34835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Castanier C, Zemirli N, Portier A, Garcin D, Bidère N, Vazquez A, Arnoult D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012;10:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 40. | Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646-28655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 325] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 41. | Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 298] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 42. | Wang Y, Tong X, Ye X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol. 2012;189:5304-5313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 43. | Luo H, Zhang Z, Zheng Z, Ke X, Zhang X, Li Q, Liu Y, Bai B, Mao P, Hu Q. Human bocavirus VP2 upregulates IFN-β pathway by inhibiting ring finger protein 125-mediated ubiquitination of retinoic acid-inducible gene-I. J Immunol. 2013;191:660-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 44. | Ahmed S, Maratha A, Butt AQ, Shevlin E, Miggin SM. TRIF-mediated TLR3 and TLR4 signaling is negatively regulated by ADAM15. J Immunol. 2013;190:2217-2228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 45. | Young JA, Sermwittayawong D, Kim HJ, Nandu S, An N, Erdjument-Bromage H, Tempst P, Coscoy L, Winoto A. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J Biol Chem. 2011;286:6521-6531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 46. | Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, Ran Y, Tien P, Shu HB. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291-4297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 156] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 47. | Karim R, Tummers B, Meyers C, Biryukov JL, Alam S, Backendorf C, Jha V, Offringa R, van Ommen GJ, Melief CJ. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog. 2013;9:e1003384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 48. | Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226-6233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 358] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 49. | Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108-8116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 50. | Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362-3373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 296] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 51. | Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150-2158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 608] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 52. | Li Y, Fan X, He X, Sun H, Zou Z, Yuan H, Xu H, Wang C, Shi X. MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell Mol Immunol. 2012;9:497-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Lee Y, Hyung SW, Jung HJ, Kim HJ, Staerk J, Constantinescu SN, Chang EJ, Lee ZH, Lee SW, Kim HH. The ubiquitin-mediated degradation of Jak1 modulates osteoclastogenesis by limiting interferon-beta-induced inhibitory signaling. Blood. 2008;111:885-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717-1719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 327] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 55. | Yeh HM, Yu CY, Yang HC, Ko SH, Liao CL, Lin YL. Ubiquitin-specific protease 13 regulates IFN signaling by stabilizing STAT1. J Immunol. 2013;191:3328-3336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 56. | Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci. 2002;27:235-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 333] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 57. | Collins AS, McCoy CE, Lloyd AT, O’Farrelly C, Stevenson NJ. miR-19a: an effective regulator of SOCS3 and enhancer of JAK-STAT signalling. PLoS One. 2013;8:e69090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 58. | Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623-2629. [PubMed] |
| 59. | Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41:2817-2831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 60. | Reinsbach S, Nazarov PV, Philippidou D, Schmitt M, Wienecke-Baldacchino A, Muller A, Vallar L, Behrmann I, Kreis S. Dynamic regulation of microRNA expression following interferon-γ-induced gene transcription. RNA Biol. 2012;9:978-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Buggele WA, Horvath CM. MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56. J Virol. 2013;87:9260-9270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, Zhang YQ, Shi JW, Lin XL, Yang S. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;431:610-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 63. | Satoh J, Tabunoki H. A Comprehensive Profile of ChIP-Seq-Based STAT1 Target Genes Suggests the Complexity of STAT1-Mediated Gene Regulatory Mechanisms. Gene Regul Syst Bio. 2013;7:41-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 64. | O’Donnell SM, Holm GH, Pierce JM, Tian B, Watson MJ, Chari RS, Ballard DW, Brasier AR, Dermody TS. Identification of an NF-kappaB-dependent gene network in cells infected by mammalian reovirus. J Virol. 2006;80:1077-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 65. | Ravi LI, Li L, Sutejo R, Chen H, Wong PS, Tan BH, Sugrue RJ. A systems-based approach to analyse the host response in murine lung macrophages challenged with respiratory syncytial virus. BMC Genomics. 2013;14:190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 66. | Gundacker NC, Haudek VJ, Wimmer H, Slany A, Griss J, Bochkov V, Zielinski C, Wagner O, Stöckl J, Gerner C. Cytoplasmic proteome and secretome profiles of differently stimulated human dendritic cells. J Proteome Res. 2009;8:2799-2811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 67. | Coombs A. Transcriptomics and quantitative proteomics in virology. Future Virol. 2012;7:1193-1204. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 68. | Baskin C. The role and contributions of systems biology to the non-human primate model of influenza pathogenesis and vaccinology. Curr Top Microbiol Immunol. 2013;363:69-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 69. | Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martínez-Sobrido L, Schwemmle M, García-Sastre A. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. MBio. 2011;2:pii: e00151-11. [PubMed] |
| 70. | Cheung CY, Chan EY, Krasnoselsky A, Purdy D, Navare AT, Bryan JT, Leung CK, Hui KP, Peiris JS, Katze MG. H5N1 virus causes significant perturbations in host proteome very early in influenza virus-infected primary human monocyte-derived macrophages. J Infect Dis. 2012;206:640-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 71. | Coombs KM, Berard A, Xu W, Krokhin O, Meng X, Cortens JP, Kobasa D, Wilkins J, Brown EG. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J Virol. 2010;84:10888-10906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 124] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 72. | Kroeker AL, Ezzati P, Halayko AJ, Coombs KM. Response of primary human airway epithelial cells to influenza infection: a quantitative proteomic study. J Proteome Res. 2012;11:4132-4146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 73. | Lietzén N, Ohman T, Rintahaka J, Julkunen I, Aittokallio T, Matikainen S, Nyman TA. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 2011;7:e1001340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 74. | Peng T, Kotla S, Bumgarner RE, Gustin KE. Human rhinovirus attenuates the type I interferon response by disrupting activation of interferon regulatory factor 3. J Virol. 2006;80:5021-5031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 75. | Proud D, Turner RB, Winther B, Wiehler S, Tiesman JP, Reichling TD, Juhlin KD, Fulmer AW, Ho BY, Walanski AA. Gene expression profiles during in vivo human rhinovirus infection: insights into the host response. Am J Respir Crit Care Med. 2008;178:962-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 76. | Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4:e00165-e00113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 77. | Lau SK, Lau CC, Chan KH, Li CP, Chen H, Jin DY, Chan JF, Woo PC, Yuen KY. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679-2690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 305] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 78. | Tang BS, Chan KH, Cheng VC, Woo PC, Lau SK, Lam CC, Chan TL, Wu AK, Hung IF, Leung SY. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180-6193. [PubMed] |
| 79. | Jiang XS, Tang LY, Dai J, Zhou H, Li SJ, Xia QC, Wu JR, Zeng R. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol Cell Proteomics. 2005;4:902-913. [PubMed] |
| 80. | Lai CC, Jou MJ, Huang SY, Li SW, Wan L, Tsai FJ, Lin CW. Proteomic analysis of up-regulated proteins in human promonocyte cells expressing severe acute respiratory syndrome coronavirus 3C-like protease. Proteomics. 2007;7:1446-1460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 81. | Zhang L, Zhang X, Ma Q, Ma F, Zhou H. Transcriptomics and proteomics in the study of H1N1 2009. Genomics Proteomics Bioinformatics. 2010;8:139-144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 82. | Lam WY, Yeung AC, Ngai KL, Li MS, To KF, Tsui SK, Chan PK. Effect of avian influenza A H5N1 infection on the expression of microRNA-141 in human respiratory epithelial cells. BMC Microbiol. 2013;13:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Terrier O, Textoris J, Carron C, Marcel V, Bourdon JC, Rosa-Calatrava M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J Gen Virol. 2013;94:985-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Buggele WA, Johnson KE, Horvath CM. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J Biol Chem. 2012;287:31027-31040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 85. | Buggele WA, Krause KE, Horvath CM. Small RNA profiling of influenza A virus-infected cells identifies miR-449b as a regulator of histone deacetylase 1 and interferon beta. PLoS One. 2013;8:e76560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 86. | Tambyah PA, Sepramaniam S, Mohamed Ali J, Chai SC, Swaminathan P, Armugam A, Jeyaseelan K. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS One. 2013;8:e76811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 87. | Rosenberger CM, Podyminogin RL, Navarro G, Zhao GW, Askovich PS, Weiss MJ, Aderem A. miR-451 regulates dendritic cell cytokine responses to influenza infection. J Immunol. 2012;189:5965-5975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 88. | Song H, Wang Q, Guo Y, Liu S, Song R, Gao X, Dai L, Li B, Zhang D, Cheng J. Microarray analysis of microRNA expression in peripheral blood mononuclear cells of critically ill patients with influenza A (H1N1). BMC Infect Dis. 2013;13:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 89. | Fang J, Hao Q, Liu L, Li Y, Wu J, Huo X, Zhu Y. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J Virol. 2012;86:1010-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 90. | Langlois RA, Varble A, Chua MA, García-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci USA. 2012;109:12117-12122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 91. | Wu Z, Hao R, Li P, Zhang X, Liu N, Qiu S, Wang L, Wang Y, Xue W, Liu K. MicroRNA expression profile of mouse lung infected with 2009 pandemic H1N1 influenza virus. PLoS One. 2013;8:e74190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 92. | Brock M, Trenkmann M, Gay RE, Gay S, Speich R, Huber LC. MicroRNA-18a enhances the interleukin-6-mediated production of the acute-phase proteins fibrinogen and haptoglobin in human hepatocytes. J Biol Chem. 2011;286:40142-40150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 93. | Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, Lithgow GJ, Campisi J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY). 2009;1:402-411. [PubMed] |
| 94. | Ouda R, Onomoto K, Takahasi K, Edwards MR, Kato H, Yoneyama M, Fujita T. Retinoic acid-inducible gene I-inducible miR-23b inhibits infections by minor group rhinoviruses through down-regulation of the very low density lipoprotein receptor. J Biol Chem. 2011;286:26210-26219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 95. | Munday DC, Emmott E, Surtees R, Lardeau CH, Wu W, Duprex WP, Dove BK, Barr JN, Hiscox JA. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol Cell Proteomics. 2010;9:2438-2459. [PubMed] |
| 96. | Munday DC, Hiscox JA, Barr JN. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus subgroup B using SILAC coupled to LC-MS/MS. Proteomics. 2010;10:4320-4334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 97. | Ternette N, Wright C, Kramer HB, Altun M, Kessler BM. Label-free quantitative proteomics reveals regulation of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) and 5’-3’-exoribonuclease 2 (XRN2) during respiratory syncytial virus infection. Virol J. 2011;8:442. [PubMed] |
| 98. | van Diepen A, Brand HK, Sama I, Lambooy LH, van den Heuvel LP, van der Well L, Huynen M, Osterhaus AD, Andeweg AC, Hermans PW. Quantitative proteome profiling of respiratory virus-infected lung epithelial cells. J Proteomics. 2010;73:1680-1693. [PubMed] |
| 99. | Huang YC, Li Z, Hyseni X, Schmitt M, Devlin RB, Karoly ED, Soukup JM. Identification of gene biomarkers for respiratory syncytial virus infection in a bronchial epithelial cell line. Genomic Med. 2008;2:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 100. | Ravi LI, Li L, Wong PS, Sutejo R, Tan BH, Sugrue RJ. Lovastatin treatment mitigates the pro-inflammatory cytokine response in respiratory syncytial virus infected macrophage cells. Antiviral Res. 2013;98:332-343. [PubMed] |
| 101. | Fjaerli HO, Bukholm G, Skjaeret C, Holden M, Nakstad B. Cord blood gene expression in infants hospitalized with respiratory syncytial virus bronchiolitis. J Infect Dis. 2007;196:394-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 102. | Emmott E, Smith C, Emmett SR, Dove BK, Hiscox JA. Elucidation of the avian nucleolar proteome by quantitative proteomics using SILAC and changes in cells infected with the coronavirus infectious bronchitis virus. Proteomics. 2010;10:3558-3562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 103. | Vogels MW, van Balkom BW, Kaloyanova DV, Batenburg JJ, Heck AJ, Helms JB, Rottier PJ, de Haan CA. Identification of host factors involved in coronavirus replication by quantitative proteomics analysis. Proteomics. 2011;11:64-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 104. | Zhang L, Zhang ZP, Zhang XE, Lin FS, Ge F. Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication. J Virol. 2010;84:6050-6059. [PubMed] |
| 105. | Liu L, Zhou J, Wang Y, Mason RJ, Funk CJ, Du Y. Proteome alterations in primary human alveolar macrophages in response to influenza A virus infection. J Proteome Res. 2012;11:4091-4101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 106. | Ohman T, Rintahaka J, Kalkkinen N, Matikainen S, Nyman TA. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J Immunol. 2009;182:5682-5692. [PubMed] |
| 107. | Kroeker AL, Ezzati P, Coombs KM, Halayko AJ. Influenza A infection of primary human airway epithelial cells up-regulates proteins related to purine metabolism and ubiquitin-related signaling. J Proteome Res. 2013;12:3139-3151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 108. | Zhao D, Liang L, Li Y, Liu L, Guan Y, Jiang Y, Chen H. Proteomic analysis of the lungs of mice infected with different pathotypes of H5N1 avian influenza viruses. Proteomics. 2012;12:1970-1982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 109. | Baas T, Baskin CR, Diamond DL, García-Sastre A, Bielefeldt-Ohmann H, Tumpey TM, Thomas MJ, Carter VS, Teal TH, Van Hoeven N. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80:10813-10828. [PubMed] |
| 110. | Wang J, Nikrad MP, Travanty EA, Zhou B, Phang T, Gao B, Alford T, Ito Y, Nahreini P, Hartshorn K. Innate immune response of human alveolar macrophages during influenza A infection. PLoS One. 2012;7:e29879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 111. | Yang XX, Du N, Zhou JF, Li Z, Wang M, Guo JF, Wang DY, Shu YL. Gene expression profiles comparison between 2009 pandemic and seasonal H1N1 influenza viruses in A549 cells. Biomed Environ Sci. 2010;23:259-266. [PubMed] |
| 112. | Kash JC, Basler CF, García-Sastre A, Carter V, Billharz R, Swayne DE, Przygodzki RM, Taubenberger JK, Katze MG, Tumpey TM. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol. 2004;78:9499-9511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 132] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 113. | Belisle SE, Tisoncik JR, Korth MJ, Carter VS, Proll SC, Swayne DE, Pantin-Jackwood M, Tumpey TM, Katze MG. Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J Virol. 2010;84:12576-12588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 114. | Rowe T, León AJ, Crevar CJ, Carter DM, Xu L, Ran L, Fang Y, Cameron CM, Cameron MJ, Banner D. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology. 2010;401:257-265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 115. | Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008;82:11308-11317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 116. | Baskin CR, García-Sastre A, Tumpey TM, Bielefeldt-Ohmann H, Carter VS, Nistal-Villán E, Katze MG. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina). J Virol. 2004;78:10420-10432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 117. | Berard AR, Cortens JP, Krokhin O, Wilkins JA, Severini A, Coombs KM. Quantification of the host response proteome after mammalian reovirus T1L infection. PLoS One. 2012;7:e51939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 118. | Coombs KM. HeLa cell response proteome alterations induced by mammalian reovirus T3D infection. Virol J. 2013;10:202. [PubMed] |
| 119. | Li L, Sevinsky JR, Rowland MD, Bundy JL, Stephenson JL, Sherry B. Proteomic analysis reveals virus-specific Hsp25 modulation in cardiac myocytes. J Proteome Res. 2010;9:2460-2471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 120. | Jiang J, Opanubi KJ, Coombs KM. Non-Biased Enrichment Does Not Improve Quantitative Proteomic Delineation of Reovirus T3D-Infected HeLa Cell Protein Alterations. Front Microbiol. 2012;3:310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 121. | Tyler KL, Leser JS, Phang TL, Clarke P. Gene expression in the brain during reovirus encephalitis. J Neurovirol. 2010;16:56-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 122. | Bochkov YA, Hanson KM, Keles S, Brockman-Schneider RA, Jarjour NN, Gern JE. Rhinovirus-induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal Immunol. 2010;3:69-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 123. | Proud D, Hudy MH, Wiehler S, Zaheer RS, Amin MA, Pelikan JB, Tacon CE, Tonsaker TO, Walker BL, Kooi C. Cigarette smoke modulates expression of human rhinovirus-induced airway epithelial host defense genes. PLoS One. 2012;7:e40762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 124. | Go JT, Belisle SE, Tchitchek N, Tumpey TM, Ma W, Richt JA, Safronetz D, Feldmann H, Katze MG. 2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models. BMC Genomics. 2012;13:627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 125. | Korth MJ, Tchitchek N, Benecke AG, Katze MG. Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses. Semin Immunol. 2013;25:228-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 126. | Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, Øien NC, McClain MT, Varkey JB, Nicholson B. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza a infection. PLoS Genet. 2011;7:e1002234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 163] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 127. | Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011;23:481-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 201] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 128. | Baines KJ, Hsu AC, Tooze M, Gunawardhana LP, Gibson PG, Wark PA. Novel immune genes associated with excessive inflammatory and antiviral responses to rhinovirus in COPD. Respir Res. 2013;14:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 129. | Kotla S, Peng T, Bumgarner RE, Gustin KE. Attenuation of the type I interferon response in cells infected with human rhinovirus. Virology. 2008;374:399-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 130. | Stewart MJ, Smoak K, Blum MA, Sherry B. Basal and reovirus-induced beta interferon (IFN-beta) and IFN-beta-stimulated gene expression are cell type specific in the cardiac protective response. J Virol. 2005;79:2979-2987. [PubMed] |
| 131. | van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361-372. [PubMed] |
| 132. | Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol. 2009;83:2178-2187. [PubMed] |
| 133. | Montomoli E, Khadang B, Piccirella S, Trombetta C, Mennitto E, Manini I, Stanzani V, Lapini G. Cell culture-derived influenza vaccines from Vero cells: a new horizon for vaccine production. Expert Rev Vaccines. 2012;11:587-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 134. | Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 265] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 135. | Boyapalle S, Wong T, Garay J, Teng M, San Juan-Vergara H, Mohapatra S, Mohapatra S. Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS One. 2012;7:e29386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 136. | Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734-3742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 175] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 137. | Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, Graham BS. The role of IFN in respiratory syncytial virus pathogenesis. J Immunol. 2002;168:2944-2952. [PubMed] |
| 138. | Wolff T, Ludwig S. Influenza viruses control the vertebrate type I interferon system: factors, mechanisms, and consequences. J Interferon Cytokine Res. 2009;29:549-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 139. | Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81:2025-2030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 153] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 140. | Arimori Y, Nakamura R, Yamada H, Shibata K, Maeda N, Kase T, Yoshikai Y. Type I interferon limits influenza virus-induced acute lung injury by regulation of excessive inflammation in mice. Antiviral Res. 2013;99:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 141. | Li W, Moltedo B, Moran TM. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of γδ T cells. J Virol. 2012;86:12304-12312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 142. | Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910-1920. [PubMed] |
| 143. | Sykes A, Macintyre J, Edwards MR, Del Rosario A, Haas J, Gielen V, Kon OM, McHale M, Johnston SL. Rhinovirus-induced interferon production is not deficient in well controlled asthma. Thorax. 2014;69:240-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 144. | Smith SB, Dampier W, Tozeren A, Brown JR, Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One. 2012;7:e33174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 145. | Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat Immunol. 2011;12:624-630. [PubMed] |
| 146. | Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 436] [Cited by in RCA: 504] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 147. | Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, García-Sastre A. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104:1371-1376. [PubMed] |
| 148. | Yángüez E, García-Culebras A, Frau A, Llompart C, Knobeloch KP, Gutierrez-Erlandsson S, García-Sastre A, Esteban M, Nieto A, Guerra S. ISG15 regulates peritoneal macrophages functionality against viral infection. PLoS Pathog. 2013;9:e1003632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 149. | Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 375] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 150. | Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NK, Koh EG, Chow A, Leo YS. Viperin restricts chikungunya virus replication and pathology. J Clin Invest. 2012;122:4447-4460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 151. | Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, Wilkinson J, Bye CR, Wright TK, Rambukwelle D. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120:778-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 152. | Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, McCartney EM, Jangra RK, Lemon SM, Beard MR. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 153. | Tafforeau L, Chantier T, Pradezynski F, Pellet J, Mangeot PE, Vidalain PO, Andre P, Rabourdin-Combe C, Lotteau V. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J Virol. 2011;85:13010-13018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 154. | de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot PE, Ciancia C. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 2013;9:e1003440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 155. | Mok BW, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau SY, Wu WL, Matsumoto K. The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection. J Virol. 2012;86:12695-12707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 156. | Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 520] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 157. | Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol. 2007;81:1401-1411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 256] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 158. | Rudd BD, Burstein E, Duckett CS, Li X, Lukacs NW. Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J Virol. 2005;79:3350-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 213] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 159. | Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol. 2007;81:3428-3436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 150] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 160. | Thornburg NJ, Hayward SL, Crowe JE. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. MBio. 2012;3. [PubMed] |
| 161. | Li SW, Lai CC, Ping JF, Tsai FJ, Wan L, Lin YJ, Kung SH, Lin CW. Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J Gen Virol. 2011;92:1127-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 162. | Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81:9812-9824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 423] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 163. | Sims AC, Tilton SC, Menachery VD, Gralinski LE, Schäfer A, Matzke MM, Webb-Robertson BJ, Chang J, Luna ML, Long CE. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol. 2013;87:3885-3902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 164. | Calvén J, Yudina Y, Uller L. Rhinovirus and dsRNA induce RIG-I-like receptors and expression of interferon β and λ1 in human bronchial smooth muscle cells. PLoS One. 2013;8:e62718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 165. | Irvin SC, Zurney J, Ooms LS, Chappell JD, Dermody TS, Sherry B. A single-amino-acid polymorphism in reovirus protein μ2 determines repression of interferon signaling and modulates myocarditis. J Virol. 2012;86:2302-2311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 166. | Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 1055] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 167. | Tan KS, Olfat F, Phoon MC, Hsu JP, Howe JL, Seet JE, Chin KC, Chow VT. In vivo and in vitro studies on the antiviral activities of viperin against influenza H1N1 virus infection. J Gen Virol. 2012;93:1269-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 168. | Tran AT, Rahim MN, Ranadheera C, Kroeker A, Cortens JP, Opanubi KJ, Wilkins JA, Coombs KM. Knockdown of specific host factors protects against influenza virus-induced cell death. Cell Death Dis. 2013;4:e769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 169. | Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, García-Sastre A, Zhang DE, Lenschow DJ. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83:1147-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 170. | Bartlett NW, Slater L, Glanville N, Haas JJ, Caramori G, Casolari P, Clarke DL, Message SD, Aniscenko J, Kebadze T. Defining critical roles for NF-κB p65 and type I interferon in innate immunity to rhinovirus. EMBO Mol Med. 2012;4:1244-1260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 171. | Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 397] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 172. | Wei J, Waithman J, Lata R, Mifsud NA, Cebon J, Kay T, Smyth MJ, Sadler AJ, Chen W. Influenza A infection enhances cross-priming of CD8+ T cells to cell-associated antigens in a TLR7- and type I IFN-dependent fashion. J Immunol. 2010;185:6013-6022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 173. | Shmulevitz M, Pan LZ, Garant K, Pan D, Lee PW. Oncogenic Ras promotes reovirus spread by suppressing IFN-beta production through negative regulation of RIG-I signaling. Cancer Res. 2010;70:4912-4921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 174. | Li S, Sun F, Zhang YB, Gui JF, Zhang QY. Identification of DreI as an antiviral factor regulated by RLR signaling pathway. PLoS One. 2012;7:e32427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 175. | Yoo JK, Kim TS, Hufford MM, Braciale TJ. Viral infection of the lung: host response and sequelae. J Allergy Clin Immunol. 2013;132:1263-176; quiz 1277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 176. | See H, Wark P. Innate immune response to viral infection of the lungs. Paediatr Respir Rev. 2008;9:243-250. [PubMed] |
| 177. | Haasbach E, Droebner K, Vogel AB, Planz O. Low-dose interferon Type I treatment is effective against H5N1 and swine-origin H1N1 influenza A viruses in vitro and in vivo. J Interferon Cytokine Res. 2011;31:515-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 178. | Matzinger SR, Carroll TD, Fritts L, McChesney MB, Miller CJ. Exogenous IFN-alpha administration reduces influenza A virus replication in the lower respiratory tract of rhesus macaques. PLoS One. 2011;6:e29255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 179. | Jiang H, Yang H, Kapczynski DR. Chicken interferon alpha pretreatment reduces virus replication of pandemic H1N1 and H5N9 avian influenza viruses in lung cell cultures from different avian species. Virol J. 2011;8:447. [PubMed] |
| 180. | Gaajetaan GR, Geelen TH, Vernooy JH, Dentener MA, Reynaert NL, Rohde GG, Beuken EV, Grauls GE, Bruggeman CA, Stassen FR. Interferon-β induces a long-lasting antiviral state in human respiratory epithelial cells. J Infect. 2013;66:163-169. [PubMed] |
| 181. | Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 377] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 182. | de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RW, Posthuma CC, van der Meer Y, Bárcena M, Haagmans BL. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J Gen Virol. 2013;94:1749-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 280] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 183. | Kuri T, Weber F. Interferon interplay helps tissue cells to cope with SARS-coronavirus infection. Virulence. 2010;1:273-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 184. | Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, Kelsall BL. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J Exp Med. 2007;204:1349-1358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 185. | Empey KM, Orend JG, Peebles RS, Egaña L, Norris KA, Oury TD, Kolls JK. Stimulation of immature lung macrophages with intranasal interferon gamma in a novel neonatal mouse model of respiratory syncytial virus infection. PLoS One. 2012;7:e40499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 186. | Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9:e1003298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 187. | Zhao J, Wohlford-Lenane C, Zhao J, Fleming E, Lane TE, McCray PB, Perlman S. Intranasal treatment with poly(I•C) protects aged mice from lethal respiratory virus infections. J Virol. 2012;86:11416-11424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 188. | Boukhvalova MS, Sotomayor TB, Point RC, Pletneva LM, Prince GA, Blanco JC. Activation of interferon response through toll-like receptor 3 impacts viral pathogenesis and pulmonary toll-like receptor expression during respiratory syncytial virus and influenza infections in the cotton rat Sigmodon hispidus model. J Interferon Cytokine Res. 2010;30:229-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 189. | Liao JY, Thakur SA, Zalinger ZB, Gerrish KE, Imani F. Inosine-containing RNA is a novel innate immune recognition element and reduces RSV infection. PLoS One. 2011;6:e26463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 190. | Brooks MJ, Burtseva EI, Ellery PJ, Marsh GA, Lew AM, Slepushkin AN, Crowe SM, Tannock GA. Antiviral activity of arbidol, a broad-spectrum drug for use against respiratory viruses, varies according to test conditions. J Med Virol. 2012;84:170-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 191. | Shi L, Xiong H, He J, Deng H, Li Q, Zhong Q, Hou W, Cheng L, Xiao H, Yang Z. Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch Virol. 2007;152:1447-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 192. | Martínez-Gil L, Ayllon J, Ortigoza MB, García-Sastre A, Shaw ML, Palese P. Identification of small molecules with type I interferon inducing properties by high-throughput screening. PLoS One. 2012;7:e49049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 193. | Teng MN. The non-structural proteins of RSV: targeting interferon antagonists for vaccine development. Infect Disord Drug Targets. 2012;12:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 194. | Zhang Y, Jamaluddin M, Wang S, Tian B, Garofalo RP, Casola A, Brasier AR. Ribavirin treatment up-regulates antiviral gene expression via the interferon-stimulated response element in respiratory syncytial virus-infected epithelial cells. J Virol. 2003;77:5933-5947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 195. | Kugel D, Kochs G, Obojes K, Roth J, Kobinger GP, Kobasa D, Haller O, Staeheli P, von Messling V. Intranasal administration of alpha interferon reduces seasonal influenza A virus morbidity in ferrets. J Virol. 2009;83:3843-3851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 196. | Matzinger SR, Carroll TD, Dutra JC, Ma ZM, Miller CJ. Myxovirus resistance gene A (MxA) expression suppresses influenza A virus replication in alpha interferon-treated primate cells. J Virol. 2013;87:1150-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 197. | Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol. 2013;87:13053-13058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 198. | Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J Biol Chem. 2013;288:17261-17271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 199. | Narayanan A, Kehn-Hall K, Senina S, Lundberg L, Van Duyne R, Guendel I, Das R, Baer A, Bethel L, Turell M. Curcumin inhibits Rift Valley fever virus replication in human cells. J Biol Chem. 2012;287:33198-33214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 200. | Si X, Wang Y, Wong J, Zhang J, McManus BM, Luo H. Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J Virol. 2007;81:3142-3150. [PubMed] |
| 201. | Obata K, Kojima T, Masaki T, Okabayashi T, Yokota S, Hirakawa S, Nomura K, Takasawa A, Murata M, Tanaka S. Curcumin prevents replication of respiratory syncytial virus and the epithelial responses to it in human nasal epithelial cells. PLoS One. 2013;8:e70225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 202. | Xie XH, Zang N, Li SM, Wang LJ, Deng Y, He Y, Yang XQ, Liu EM. Resveratrol Inhibits respiratory syncytial virus-induced IL-6 production, decreases viral replication, and downregulates TRIF expression in airway epithelial cells. Inflammation. 2012;35:1392-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 203. | Carr JM, Mahalingam S, Bonder CS, Pitson SM. Sphingosine kinase 1 in viral infections. Rev Med Virol. 2013;23:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 204. | Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50-60. [PubMed] |
| 205. | Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JC, Vogel SN. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 2010;3:291-300. [PubMed] |
| 206. | Verma S, Kumar M, Nerurkar VR. Cyclooxygenase-2 inhibitor blocks the production of West Nile virus-induced neuroinflammatory markers in astrocytes. J Gen Virol. 2011;92:507-515. [PubMed] |
| 207. | Nabatov AA, Pollakis G, Linnemann T, Paxton WA, de Baar MP. Statins disrupt CCR5 and RANTES expression levels in CD4(+) T lymphocytes in vitro and preferentially decrease infection of R5 versus X4 HIV-1. PLoS One. 2007;2:e470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |