Published online Aug 26, 2013. doi: 10.4331/wjbc.v4.i3.64
Revised: July 13, 2013
Accepted: July 17, 2013
Published online: August 26, 2013
Processing time: 47 Days and 10.1 Hours
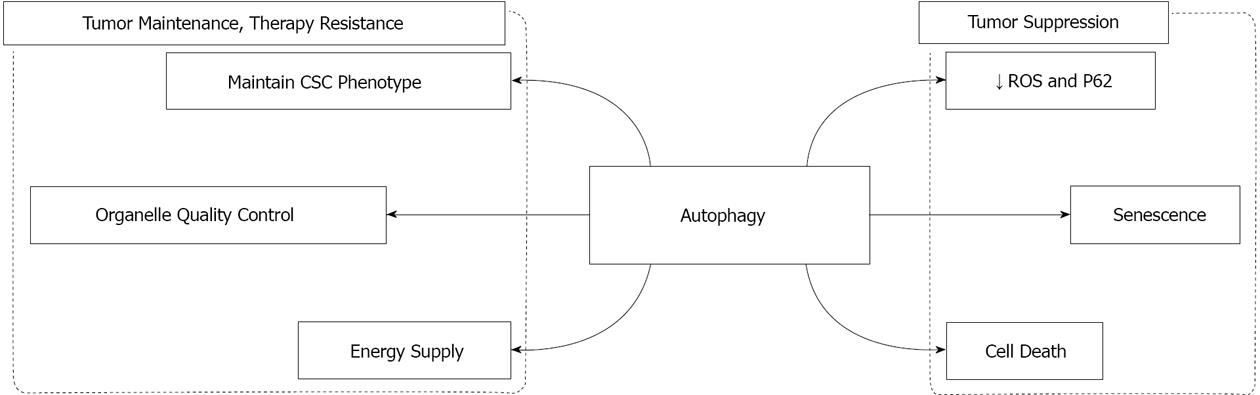
Autophagy is a homeostatic and evolutionarily conserved mechanism of self-digestion by which the cells degrade and recycle long-lived proteins and excess or damaged organelles. Autophagy is activated in response to both physiological and pathological stimuli including growth factor depletion, energy deficiency or the upregulation of Bcl-2 protein expression. A novel role of autophagy in various cancers has been proposed. Interestingly, evidence that supports both a positive and negative role of autophagy in the pathogenesis of cancer has been reported. As a tumor suppression mechanism, autophagy maintains genome stability, induces senescence and possibly autophagic cell death. On the other hand, autophagy participates in tumor growth and maintenance by supplying metabolic substrate, limiting oxidative stress, and maintaining cancer stem cell population. It has been proposed that the differential roles of autophagy in cancer are disease type and stage specific. In addition, substrate selectivity might be involved in carrying out the specific effect of autophagy in cancer, and represents one of the potential directions for future studies.
Core tip: The differential expression of selective autophagic receptors in cancers of different origin and stage might induce the selective removal or preservation of certain cellular components and contribute to either tumor suppression or cancer cell survival.
- Citation: Lu SZ, Harrison-Findik DD. Autophagy and cancer. World J Biol Chem 2013; 4(3): 64-70
- URL: https://www.wjgnet.com/1949-8454/full/v4/i3/64.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v4.i3.64
Autophagy is an evolutionarily conserved catabolic pathway which delivers long-lived proteins and excess or damaged organelles into the lysosome for degradation and recycling[1,2]. Three mechanistically distinguished subtypes including macroautophagy, microautophagy and chaperon-mediated autophagy exist, of which, macroautophagy (hereafter referred to as autophagy) is most studied. Traditionally known as a mechanism to maintain homeostasis and degrade cellular components in response to starvation, further functions have been identified as our understanding of autophagy has progressed. A novel role of autophagy in cancer has also been proposed in recent years. In the current review, we attempt to provide a brief evaluation of the current literature and discuss the potential mechanisms of how autophagy is involved in the pathogenesis of cancer.
The basic machinery and regulation of autophagy has been described in numerous excellent reviews[1,3-6] and will not be discussed in detail here. We will briefly introduce the autophagy process and key players to facilitate our further discussion. Autophagy process is divided into four stages: nucleation, elongation, autophagosome formation and fusion. The nucleation is initiated by the dephosphorylation (i.e., activation) of the unc-51-like kinase (ULK) complex. ULK complex is otherwise kept inactive by the mammalian target of rapamycin (mTOR), a highly conserved serine/threonine protein kinase. mTOR integrates the signal of growth factor and nutrition availability and serves as the pivotal inhibitory regulator of autophagy. In other words, limited growth factor and nutrient inactivates mTOR and release ULK complex from its inhibition. Upon activation, ULK complex induces the re-localization of a phosphatidyl-inositol-3-kinase-class III (PtDIns3K) complex, which is composed of vacuolar protein sorting 34 (Vps34), p150, mAtg14 and Beclin1, to the nucleation site. Beclin1 mediates the cross-talk between autophagy and apoptosis in that it is a binding partner of anti-apoptotic Bcl-2 family proteins (e.g., Bcl-2, Bcl-xl and Mcl-1). Beclin1 can be sequestered by these Bcl-2 proteins, which will prevent the formation of PtDIns3K complex and thereby block the nucleation process. The pro-apoptotic BH-3 only Bcl-2 proteins (e.g., Bnip-3, Bad and Puma) compete with Beclin1 for the binding to anti-apoptotic Bcl-2 proteins and hence promote autophagy. Once formed, PtdIns3K complex catalyzes the production of phosphatidylinositol (3)-phosphate [PtdIns(3)P], which further recruits autophagy related (Atg) proteins. The two interrelated ubiquitin-like conjugation systems, Atg12-Atg5-Atg16 and microtubule-associated protein light chain 3 (LC3)- phosphatidylethanolamine (PE) play a major role in the elongation of the phagophore. The subsequent step, autophagosome formation, is accomplished by the invagination of phagophore membrane and the sequestration of cytosolic contents. In order for its contents to be degraded, the autophagosomes will form autolysosomes by fusing with lysosomes or late inner body.
In recent years, the concept of substrate selectivity in autophagy has gained further recognition. This is quite different from the initial understanding of autophagy, which was regarded as a non-specific self-eating process. However, recent studies have indicated that a specificity for substrate in autophagy is conveyed through different receptor proteins. More importantly, a correlation between the targeted removal of cellular components by autophagy and human diseases has been established[7]. The autophagy receptors, which play a key role in the substrate selectivity[8], tether the substrate of interest to the autophagic machinery (LC3) through a specific sequence called LC3-interacting region (LIT) motif[9-12]. For example, p62/SQSTM1 (p62) participates in aggrephagy (protein aggregate autophagy) and p62 binds ubiquitinated protein aggregates through an ubiquitin-associated (UBA) domain. On the other hand, BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), which is a mitochondrial localized BH-3 only Bcl-2 family protein, is involved in mitophagy (mitochondrial autophagy). Both p62 and BNIP3 induce degradation of their specific target by autophagy via their LIT motifs[10-13].
As a pro-survival pathway, the role of autophagy in cancer has long been speculated. However, significant evidence suggests that autophagy might participate in both tumor suppression and tumor maintenance. Furthermore, the resistance to chemotherapy, which is one of the major obstacles in the treatment of cancers, has been linked to autophagy, as supported by the latest studies. This multiplicity function of autophagy in cancer is discussed in detail below.
The first evidence of a tumor suppressive role of autophagy in cancer originated from the observation that heterozygous loss of the Beclin1 encoding gene (Becn1) was detected in breast, ovarian and prostate cancer[14]. Subsequent studies with mouse models further established the role of autophagy in tumor suppression. Becn1 heterozygous knockout mice developed tumors of both benign and malignant nature in various tissues[15,16], suggesting that Becn1 is a haploinsufficient tumor suppressor gene[16]. Similarly, a mouse model with systemic mosaic deletion of Atg5 and the liver-specific homozygous deletion of Atg7 both developed benign liver adenomas[17]. Vice versa, re-introducing beclin-1 into human breast carcinoma cells decreased both the proliferation in vitro and tumorigenesis capacity in vivo[14]. Apart from the experimental evidence, the tumor suppressive role of autophagy is also supported by the observation that other tumor suppressor genes, such as ULK3, UV irradiation resistance-associated gene and Bif-1, frequently participate in autophagy signaling[2,18]. On the other hand, the overexpression of oncogenes usually imposes a negative effect on autophagic activity. For instance, PI3K/AKT pathway, which is activated in various cancers, suppresses autophagy through mTOR phosphorylation (i.e., activation)[19]. The up-regulation of anti-apoptotic Bcl-2 proteins in cancer also suppresses autophagy via Beclin1, as described above.
The knowledge regarding the mechanisms underlying the role of autophagy in tumor suppression is still limited. However, an interesting study by Mathew et al[20] have reported that the allelic loss of Beclin1 results in increased chromosomal instability. They further showed that the altered regulation of nuclear factor κB, which resulted from p62/SQSTM1 (p62) and reactive oxygen species (ROS) accumulation, is responsible for the damage induced by autophagy deficiency[20].
Senescence is also a potential mechanism by which autophagy can exert a tumor suppressive role. Senescence is the status of cell cycle arrest with active metabolism[21]. Autophagy has been shown to activate senescence in cultured human lung fibroblast cells[21]. Similarly, autophagy has been suggested to mediate senescence in primary biliary cirrhosis[22]. By inducing senescence in transformed cells, autophagy can induce cell cycle arrest in transformed cells and prevent tumorigenesis.
Another plausible route of tumor suppression is through autophagy-mediated cell death[23]. Although the definition and mechanisms by which autophagy induces cell death is still under debate, several studies strongly support a role for autophagic cell death in tumor suppression. Gurpinar et al[24] have shown the involvement of autophagy in cell death induced by the treatment of lung adenocarcinoma cells with sulindac sulfide amide. Interestingly, cell death in this system occurred in the absence of caspase activation[24]. In addition, Lamy et al[25] have shown that myeloma cells can avoid cell death by restricting the autophagic activity through the cleavage of autophagic inducer, BCL2-interacting protein BCLAF1, by caspase-10.
Interestingly, a role in promoting and maintaining tumors has also been suggested for autophagy regarding cancer development. The conditions which induce autophagy, such as nutrient deprivation, hypoxia and reactive oxygen species, are also present in the tumor microenvironment, especially in tumors with limited blood supply. Yang et al[26] have shown that the basal level of autophagy is elevated in pancreatic cancers. Blocking autophagy by chemical inhibitors or RNAi methodology inhibits the tumorigenic potential of the cancer cells, as determined by both in vitro and in vivo assays[26]. Autophagy inhibition is also correlated with a decrease in oxidative phosphorylation and ATP production. Similar findings were also reported with Ras-transformed immortal, nontumorigenic mouse kidney epithelial cells isolated from baby mice[27]. In addition, the requirement for a functional autophagy machinery for Ras-induced cellular transformation has also been confirmed in other cell models[28,29].
The understanding of the mechanisms by which autophagy supports oncogenic growth is still in its infancy. One possibility is that autophagy process might be used by cancer cells to meet their energy requirements. As discussed in earlier sections, there is a connection between autophagy inhibition and the depletion of intracellular ATP stores and oxidative phosphorylation[26,27]. However, the requirement of oxidative phosphorylation by cancer cells is unclear because cancer cells have been suggested to be dependent more on glycolysis to fuel their growth (aka Warburg effect) even in the presence of oxygen (i.e., aerobic glycolysis)[30]. Nevertheless, some studies point to the intriguing possibility that cancer cells can stimulate autophagy in the adjacent stromal cells, which in turn provide cancer cells with metabolic substrates[31]. Another potential mechanism may be linked to the organelle quality control function of the autophagy process. Damaged organelles, such as mitochondria, can be targeted for autophagy by the BH-3 only Bcl-2 family members including Pink3, BNIP3 and Nix proteins[12,13,32]. Of note, any damage to mitochondria will induce ROS production and may lead to genomic instability[33,34]. Autophagy has also been shown to be directly involved in the degradation and elimination of oxidized proteins. Despite playing a positive role in the initial stages of tumorigenesis, oxidative stress and genomic instability are detrimental to tumor growth in the later stages[2]. It is therefore feasible that autophagy can mitigate these damages and thereby sustain oncogenic growth[35].
An association between autophagy and the effectiveness of treatment has also been suggested by recent studies. For example, autophagy has been reported to be elevated in pancreatic cancer cells treated with chemotherapeutic drugs[36-38]. It should however be noted that there was no consensus as to whether the increased autophagic activity contributes to cell death[36,37] or facilitates cancer cell survival under stress conditions in pancreatic cancer[38]. On the other hand, in a different model using Myc-induced lymphoma, Amaravadi et al[39] have reported that chemotherapy induces autophagy and that the inhibition of autophagy enhances apoptosis induced by chemotherapy drugs. Furthermore, autophagy inhibitors, such as chloroquine and hydroxychloroquine have been shown to exhibit a synergistic effect with chemotherapy and radiotherapy[39-43]. Autophagy inhibitors are currently being tested in clinical trials as part of the combined therapy approach for various cancers[6].
Tumor maintenance function (see above) of autophagy may also alleviate the stress induced by cancer therapy and thereby induce therapy resistance. Besides, the new role ascribed to autophagy in the regulation of cancer stem cell (CSC) phenotype[44,45] might serve as a potential mechanism for autophagy to promote therapy resistance. The so-called “cancer stem cell theory” has generated lively discussion in recent years. CSCs are a small (< 5%) subpopulation of heterogeneous cancer cells, which are capable of self-renewing and differentiating into the whole spectrum of tumor cell population. CSCs have also been suggested to be resistant to treatment[46-50]. Despite the controversy, which still exists regarding the characteristics of CSC in various solid tumors, a correlation between autophagy and CSC population has been suggested. Autophagy has been shown to be involved in the maintenance of CSCs in breast cancer[51]. Accordingly, inhibiting ATG12 and LC-3 by siRNA methodology or with the pharmacological inhibitors of autophagy altered the phenotype of breast CSCs[51]. Similarly, Rausch et al[52] reported that the autophagic markers co-localize with CSC markers in tumors which were surgically removed from pancreatic cancer patients. The pancreatic cancer cell line, MIA-PaCa2 has been shown to exhibit more prominent stem-like properties (as determined by functional assays) compared to another pancreatic cancer cell line, BxPc-3[53]. In accordance with their stronger stem-like features, MIA-PaCa2 cells also displayed higher autophagic activity[54] and resistance to cell death induced by chemotherapeutic drug gemcitabine[54,55] than that observed with BxPc-3 cells. It is therefore possible that autophagy is associated with the maintenance of the stem cell phenotype of pancreatic CSCs and thereby contributes to the resistance observed with therapy. Nevertheless, the underlying mechanisms by which autophagy modulates CSC phenotype and contributes to drug resistance requires further research.
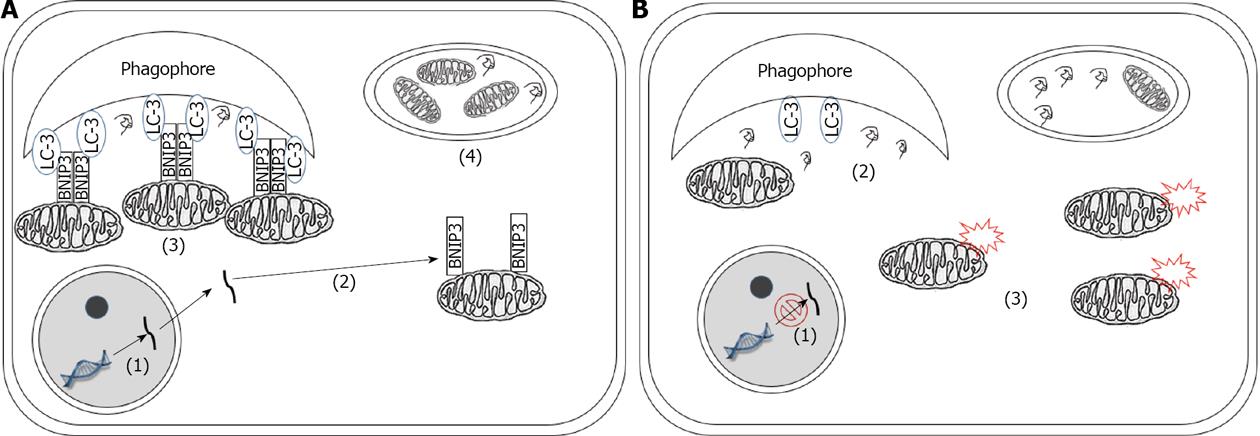
In summary, based on current knowledge, autophagy can act both as a positive and negative regulator of tumor growth in various cancers (Figure 1). Several hypotheses have been proposed to reconcile these seemingly contradictory observations, which can be summarized as follows: (1) The differential effects of autophagy in cancer might be attributed to the tissue specificity. This is supported by the fact that the highest correlation between tumor growth and elevated autophagy is observed in Ras-induced oncogenesis[2]; (2) A dynamic role for autophagy has been proposed in the development of cancer. Namely, autophagy might play a suppressive role in the initiation stages of cancer but support the maintenance of tumor growth in the later stages of tumorigenesis. This hypothesis is supported by the observation that homozygous Atg5 and Atg7 KO mice, which display more significant autophagic inhibition, developed only benign tumors[17]. In contrast, Becn1 KO mice, which exhibit relatively higher level of autophagy, displayed both benign and malignant tumors[2,15,16]; and (3) The substrate selectivity of autophagy has recently emerged as a potential mechanism responsible for the differential roles of autophagy in cancer. Mitophagy has been shown to be activated in Ras transformed cells. Autophagy deficiency results in accumulation of abnormal mitochondria when cells are challenged with starvation[27]. In contrast, in pancreatic cancer cells, initial attempts have failed to detect any significant mitophagic activity[26]. Interestingly, the specific receptor for mitophagy, BNIP3, has been found to be silenced in various pancreatic cancer cell lines[56,57]. Since damaged mitochondria are the major source of ROS which promote tumorigenesis and malignant transformation, it is feasible that mitophagy might serve as a protective mechanism in the initial stage of tumorigenesis (Figure 2A). The loss of this protective role, resulting from the silencing of mitophagic receptor, may promote the tumor to a more advanced stage (Figure 2B). Indeed, immunohistochemical staining of BNIP3 in pancreatic tissues indicated that BNIP3 silencing is a late event in pancreatic cancer pathogenesis[56]. In the early stage pancreatic cancer tissues, BNIP3 exhibits a perinuclear distribution pattern[56]. These findings strongly suggest that mitophagy is activated in the early stages of pancreatic cancer[58-60]. In contrast, this distinct pattern of BNIP3 expression is missing in late stages of pancreatic adenocarcinoma. In addition, Takahashi et al[61] have found that haploinsufficiency of a tumor suppressor gene, Bif-1, attenuates mitophagy and subsequently promotes chromosomal instability in a mouse model of B-cell lymphoma. Similarly, mitochondrial content has been shown to be elevated in breast cancer[62], colorectal cancer[63], and ovarian cancer[64]. Although direct evidence is still lacking for the substrate specificity of autophagy in cancer, further studies are required to understand the importance of this mechanism in various cancers.
Studies so far support both a tumor suppressive and an initiative role for autophagy in cancer. These differential effects of autophagy could be due to several reasons including the tissue specificity of tumors and the different stages of tumorigenesis. The role of substrate specificity of autophagy (e.g., mitophagy) and other potential mechanisms warrant further research. It is of great importance that we improve our understanding of the roles which autophagy plays in cancer. Notably, this will enable the development of individualized treatments for cancer patients according to their cancer type and its progression. In fact, autophagy inhibitors are already being tested in clinical trials and hold promise for combined cancer therapies.
P- Reviewers Iyer G, Saeki K S- Editor Song XX L- Editor A E- Editor Yan JL
| 1. | Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1403] [Cited by in RCA: 1602] [Article Influence: 100.1] [Reference Citation Analysis (0)] |
| 2. | Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 475] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 3. | Zhou S, Zhao L, Kuang M, Zhang B, Liang Z, Yi T, Wei Y, Zhao X. Autophagy in tumorigenesis and cancer therapy: Dr. Jekyll or Mr. Hyde? Cancer Lett. 2012;323:115-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 4. | Kung CP, Budina A, Balaburski G, Bergenstock MK, Murphy M. Autophagy in tumor suppression and cancer therapy. Crit Rev Eukaryot Gene Expr. 2011;21:71-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 5. | Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961-967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1483] [Cited by in RCA: 1456] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 6. | Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 830] [Cited by in RCA: 980] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 7. | Vives-Bauza C, Przedborski S. Mitophagy: the latest problem for Parkinson’s disease. Trends Mol Med. 2011;17:158-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 8. | Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: more than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 9. | Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131-24145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3082] [Cited by in RCA: 3626] [Article Influence: 201.4] [Reference Citation Analysis (0)] |
| 10. | Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847-22857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 639] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 11. | Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 339] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 12. | Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094-19104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 593] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 13. | Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxid Redox Signal. 2012;17:794-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 14. | Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 2699] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 15. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1835] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 16. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077-15082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1534] [Cited by in RCA: 1721] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 17. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 18. | Liang C, Jung JU. Autophagy genes as tumor suppressors. Curr Opin Cell Biol. 2010;22:226-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851-865. [PubMed] |
| 20. | Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1442] [Cited by in RCA: 1418] [Article Influence: 88.6] [Reference Citation Analysis (0)] |
| 21. | Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavaré S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 836] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 22. | Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Autophagy mediates the process of cellular senescence characterizing bile duct damages in primary biliary cirrhosis. Lab Invest. 2010;90:835-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 23. | Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 323] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 24. | Gurpinar E, Grizzle WE, Shacka JJ, Mader BJ, Li N, Piazza NA, Russo S, Keeton AB, Piazza GA. A novel sulindac derivative inhibits lung adenocarcinoma cell growth through suppression of Akt/mTOR signaling and induction of autophagy. Mol Cancer Ther. 2013;12:663-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Lamy L, Ngo VN, Emre NC, Shaffer AL, Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landgren O. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 26. | Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 1188] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 27. | Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1090] [Cited by in RCA: 1052] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 28. | Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 390] [Cited by in RCA: 383] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 29. | Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, Hwang SG, Yoon G, Lee SJ. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924-12932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 182] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 30. | Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3445] [Cited by in RCA: 3710] [Article Influence: 265.0] [Reference Citation Analysis (0)] |
| 31. | Lisanti MP, Martinez-Outschoorn UE, Chiavarina B, Pavlides S, Whitaker-Menezes D, Tsirigos A, Witkiewicz A, Lin Z, Balliet R, Howell A. Understanding the “lethal” drivers of tumor-stroma co-evolution: emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Biol Ther. 2010;10:537-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 32. | Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, Gustafsson ÅB. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721-731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 203] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 33. | Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6484] [Cited by in RCA: 6079] [Article Influence: 379.9] [Reference Citation Analysis (0)] |
| 34. | Kurihara Y, Kanki T, Aoki Y, Hirota Y, Saigusa T, Uchiumi T, Kang D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287:3265-3272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 35. | Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci. 2011;36:30-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 864] [Cited by in RCA: 1004] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 36. | Pardo R, Lo Ré A, Archange C, Ropolo A, Papademetrio DL, Gonzalez CD, Alvarez EM, Iovanna JL, Vaccaro MI. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology. 2010;10:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 37. | Donadelli M, Dando I, Zaniboni T, Costanzo C, Dalla Pozza E, Scupoli MT, Scarpa A, Zappavigna S, Marra M, Abbruzzese A. Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism. Cell Death Dis. 2011;2:e152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 38. | Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, Savaraj N, Lampidis TJ. 2-Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol. 2011;67:899-910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 156] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 39. | Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 832] [Cited by in RCA: 916] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 40. | Degtyarev M, De Mazière A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 358] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 41. | Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 425] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 42. | Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654-666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 727] [Cited by in RCA: 718] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 43. | Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 384] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 44. | Guo W, Lasky JL, Wu H. Cancer stem cells. Pediatr Res. 2006;59:59R-64R. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 45. | Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1172] [Cited by in RCA: 1189] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 46. | Gottschling S, Schnabel PA, Herth FJ, Herpel E. Are we missing the target? Cancer stem cells and drug resistance in non-small cell lung cancer. Cancer Genomics Proteomics. 2012;9:275-286. [PubMed] |
| 47. | Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int J Biochem Cell Biol. 2012;44:2144-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 498] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 48. | Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 415] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 49. | Rebucci M, Michiels C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem Pharmacol. 2013;85:1219-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 295] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 50. | Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839-2845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 579] [Cited by in RCA: 546] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 51. | Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle. 2011;10:3871-3885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 52. | Rausch V, Liu L, Apel A, Rettig T, Gladkich J, Labsch S, Kallifatidis G, Kaczorowski A, Groth A, Gross W. Autophagy mediates survival of pancreatic tumour-initiating cells in a hypoxic microenvironment. J Pathol. 2012;227:325-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 53. | Kallifatidis G, Rausch V, Baumann B, Apel A, Beckermann BM, Groth A, Mattern J, Li Z, Kolb A, Moldenhauer G. Sulforaphane targets pancreatic tumour-initiating cells by NF-kappaB-induced antiapoptotic signalling. Gut. 2009;58:949-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 54. | Pan X, Arumugam T, Yamamoto T, Levin PA, Ramachandran V, Ji B, Lopez-Berestein G, Vivas-Mejia PE, Sood AK, McConkey DJ. Nuclear factor-kappaB p65/relA silencing induces apoptosis and increases gemcitabine effectiveness in a subset of pancreatic cancer cells. Clin Cancer Res. 2008;14:8143-8151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 111] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 55. | Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, Denny WA, Canman CE, Kraker AJ, Lawrence TS. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 56. | Erkan M, Kleeff J, Esposito I, Giese T, Ketterer K, Büchler MW, Giese NA, Friess H. Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene. 2005;24:4421-4432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 169] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 57. | Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338-5346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 176] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 58. | Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837-4848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 697] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 59. | Vives-Bauza C, de Vries RL, Tocilescu M, Przedborski S. PINK1/Parkin direct mitochondria to autophagy. Autophagy. 2010;6:315-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 60. | Marchbank K, Waters S, Roberts RG, Solomon E, Whitehouse CA. MAP1B Interaction with the FW Domain of the Autophagic Receptor Nbr1 Facilitates Its Association to the Microtubule Network. Int J Cell Biol. 2012;2012:208014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 61. | Takahashi Y, Hori T, Cooper TK, Liao J, Desai N, Serfass JM, Young MM, Park S, Izu Y, Wang HG. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood. 2013;121:1622-1632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 62. | Bai RK, Chang J, Yeh KT, Lou MA, Lu JF, Tan DJ, Liu H, Wong LJ. Mitochondrial DNA content varies with pathological characteristics of breast cancer. J Oncol. 2011;2011:496189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 63. | Qu F, Liu X, Zhou F, Yang H, Bao G, He X, Xing J. Association between mitochondrial DNA content in leukocytes and colorectal cancer risk: a case-control analysis. Cancer. 2011;117:3148-3155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 64. | Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95:1087-1091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |