INTRODUCTION
The zinc finger motif was used as a vehicle for the initial discovery of Ikaros in the context of T-cell differentiation[1] and has been central to all subsequent analyses of Ikaros function. The C2H2 zinc finger motif was first described in 1985[2] with the discovery of a total of nine tandem zinc fingers in transcription factor IIIA (TFIIIA) in Xenopus oocytes. A retrospective review of the original discovery, subsequent structural studies, and the application this information in the design of zinc fingers with novel DNA binding functions has recently been provided[3]. Although no complete structures are available for the Ikaros protein or any of its family members, considerable evidence has accumulated about the structure of zinc fingers and the role that this structure plays in the functions of the Ikaros family of proteins.
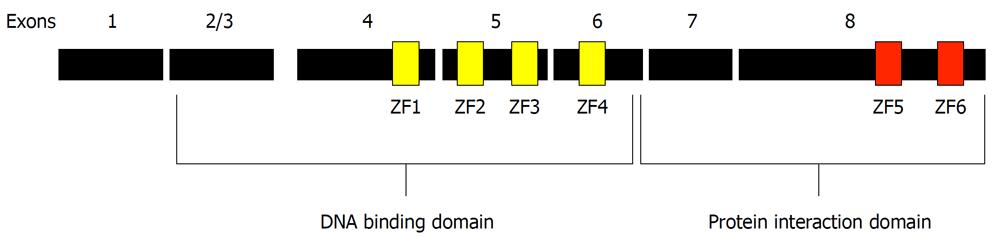
The Ikaros gene is alternately spliced to produce several isoforms that confer diversity of function and consequently have complicated analysis of the function of Ikaros in vivo. The basic architecture of the longest Ikaros isoform [519 amino acids, Gene: IKZF1 (UniProt: gi|3913926), for the full-length human Ikaros] consists of an N-terminal DNA binding domain containing four centrally located C2H2 zinc fingers and a C-terminal protein interaction (dimerization) domain with two additional zinc fingers near the C-terminus of the protein (Figure 1). For purposes of analysis and discussion the zinc fingers have been numbered from N to C: 1-6. The two presumed domains of Ikaros have distinctly different functions and are treated as such here.
Figure 1 Architecture of the Ikaros protein.
The full-length Ikaros is shown in context of the exons present in the longest form of Ikaros. The positions of the six zinc fingers are shown in their approximate locations. Fingers 1-4 are contiguous as well as fingers 5 and 6.
ARCHITECTURE OF THE N-TERMINAL DNA BINDING DOMAIN
To understand the role of the C2H2 zinc finger in Ikaros-DNA interactions, we examine the structure of a C2H2 zinc finger peptide, which was engineered based on C2H2 consensus sequences, during interaction with DNA
(Figures 2 and 3)[4]. This structure has three tandem zinc fingers bound in the major groove of the DNA. Individual fingers have two antiparallel β sheets folded in on an α helix. In the interior of the fingers two cysteines in the β sheets and two histidines in the helix are coordinated with a zinc that confers considerable rigidity to the structure. Deeper in the interior of the fold are three hydrophobic side chains that are also important in maintaining the structure of the finger. This gives a total of seven characteristic amino acids that are essential for the basic C2H2 zinc finger structure. The residues responsible for the sequence-specific DNA interactions generally occur at locations toward the N-side of the helix (positions -1, 2, 3 and 6, which make the start of the helix). The side chains of amino acids at positions -1, 3 and 6 interact with a triplet of nucleotide bases on one strand of the DNA and the position 2 side chain interacts with a base adjacent to the triplet on the opposite strand of the DNA (see[5] and[3] for detailed descriptions of these interactions). The interactions with the nucleotide bases in this binding arrangement are in the major groove. These patterns have allowed construction of a loose code for the sequence-specific protein-DNA interactions to be developed for C2H2 zinc fingers, although considerable variation has been observed in both natural and engineered zinc finger specificity determinants[6]. This makes the zinc finger code a good starting point for analysis but it clearly has limited explanatory power in individual cases (as with ZF1 and ZF4 in Ikaros).
Figure 2 Primary sequence of the six zinc fingers of Ikaros.
The sequences of the six zinc fingers of human Ikaros are shown along with their respective linkers (UniProt: gi|3913926). The letters in red are the consensus Cys and His residues that chelate zinc in the fingers. The blue highlighted letters represent the -1, 2, 3, and 6 positions of the finger helices read from left to right.
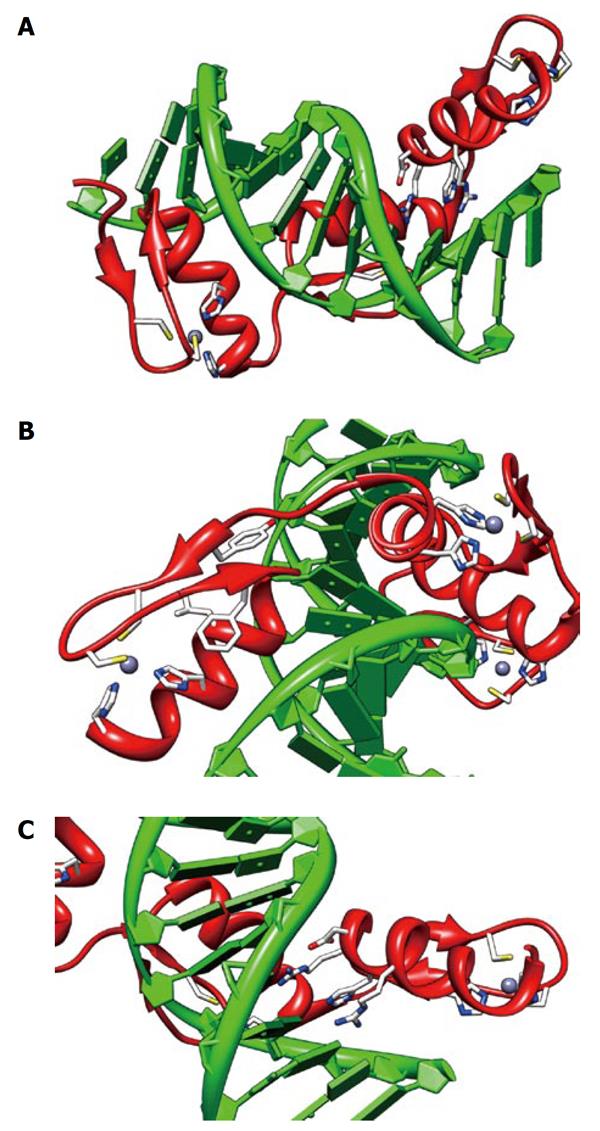
Figure 3 Structure of an engineered peptide with three tandem zinc fingers similar to Ikaros ZF2-3.
The views shown are from 1MEY (pdb), an engineered three tandem zinc finger peptide (shown in red) in complex with cognate DNA[4] shown in green. All three fingers show the zinc (grey sphere) complexed to two sulfurs of cysteine (yellow) and two imidazole nitrogen (blue). A: An overview of the zinc fingers nestled in the major groove of the DNA. The N terminus is on the left. The C-terminal finger has the DNA-interacting side chains shown; B: A view of the N-terminal finger showing the seven essential residues for zinc finger structural integrity; C: A view of the DNA-interacting residues on the C-terminal finger, -1:arg, 2:asp, 3:his, 6:arg. The views were produced using CHIMERA[35].
Numerous studies of both naturally occurring and engineered zinc fingers have shown that modular zinc fingers often appear in tandem separated by short flexible linkers. For a series of two or three tandem fingers, the mode of binding in the major groove described above can easily be maintained. However for four or more tandem fingers in a protein such as Ikaros, the topological constraints imposed by the twist of the B-DNA helix dictates more complex DNA interactions. For short peptides composed of tandem zinc fingers, up to three fingers can follow the major groove of the DNA around the helical axis without introducing undue strain in the DNA other than some underwinding. Several studies have shown that with C2H2 zinc fingers the DNA is slightly under wound to accommodate the zinc finger. This also makes the major groove slightly more open than in B-DNA[6,7]. For four fingers, at least one of the fingers generally binds outside of the canonical binding determinants described above[8,9]. Detailed studies of the zinc-finger domain of the Wilms tumor suppressor protein (WT1, which contains four tandem C2H2 zinc fingers) showed that ZF1 was in the major groove but does not use any of the normal amino acid-base interactions for binding[9]. The conclusion is that ZF1 contributes only to non-specific affinity of WT1 for DNA with the other three zinc fingers that confer sequence-specificity[9]. For larger tandem arrays of six zinc fingers it has been shown that all six can reside in the major groove. However such an arrangement is accompanied by some strain on the DNA and several non-standard DNA-zinc finger interactions[10]. Alternatively, the first and last of a series of zinc fingers can bind to the major groove with intervening fingers placed out of the major groove and interacting primarily with the minor groove. Placing Ikaros in the context of the data from other proteins and synthetic peptides, it is highly likely that at least one of the fingers does not bind in the major groove of the DNA in the classical arrangement. There is strong evidence that the second and third fingers of Ikaros do conform closely to the classical binding mode in the major groove[11] as discussed below. The function and possible DNA binding modes for fingers 1 and 4 are less clear.
The role of the N-terminal tandem zinc fingers (ZF1-ZF4) of Ikaros in targeting pericentromeric heterochromatin has been studied in detail using mutational analysis. Cell lysates from 3T3 cells transfected with mutant Ikaros have been used in gel shift assays with probes derived from multiple Ikaros binding sites to provide a functional assessment[11]. The full-length Ikaros has been shown to tolerate substantial deletions while maintaining DNA targeting behavior. Perhaps due to the natural architecture of Ikaros that supports multiple isoforms, ZF1 and ZF4 are not required for targeting. However, deletions of ZF2 and/or ZF3 and ZF5/ZF6 are not tolerated. To identify the parts of ZF2 and ZF3 that are essential for targeting, several substitution mutants have been generated to replace F2 and/or ZF3 with all or a part of the ZF5 finger (the most divergent in sequence from ZF2 and ZF3). The results showed that the residues most important for DNA binding are located in the region between the second β sheet and the N-terminal half of the α helix in both ZF2 and ZF3. This is the region of the finger that interacts directly with the DNA bases in classic C2H2 fingers. Alanine-scanning mutagenesis has been used to probe further the critical sequences in ZF2 and ZF3. Alanine scanning mutagenesis is coupled with confocal microscopy to assess DNA targeting, and with gel-shift assays to evaluate binding affinity for target DNA sequences. In the substitution and point mutations, gel-shift binding assays have shown high correlation with confocal data, which give confidence to the conclusion that the DNA targeting is dependent on the direct DNA binding of ZF2 and ZF3. To develop a rationale for the role of individual residues of ZF2 and ZF3 in DNA binding, structural models of ZF2 and ZF3 have been produced via homology modeling[11]. For ZF2, the essential residues include those located at positions -1, 2, 3 and 6 of the α helix, along with residues that are a part of the hydrophobic interior of the finger as mentioned earlier for the classical C2H2 zinc finger. ZF3 does not conform as closely to the classic C2H2 zinc finger binding pattern. Only position -1 is essential for binding to all DNA probes tested. Positions 2 and 3 give variable binding results, which depends on which probe is used. Position 6 shows no correlation with DNA binding (presumably because it is a glycine with no side chain). Available data and the models of ZF2 and ZF3 suggest that F2 has higher affinity for DNA and also perhaps greater sequence specificity than ZF3, but no direct quantitative DNA-binding affinity data are available to test this hypothesis at present. The roles of ZF1 and ZF4 are still not well understood, although some data suggest that ZF1 contributes to DNA binding affinity for select DNA probes[11].
FUNCTION OF THE LINKERS BETWEEN ZINC FINGERS
A complete picture of the binding of tandem zinc fingers to DNA (as in the case with Ikaros) must include the role of the linkers[12]. The classic linker consensus sequence is TGEKP as observed in TFIIIA and many other proteins with tandem zinc fingers. In fact, the above linker consensus is highly conserved, occurring in several thousand proteins. In the absence of DNA, the linkers are fairly flexible but become much more ordered when flanking zinc fingers are bound to DNA[6]. The linker is important in increasing DNA-binding affinity of tandem zinc fingers but the exact sequence of the linker can vary widely and still serve that function.
The linkers between Ikaros ZF1-ZF2, ZF2-ZF3 and ZF3-ZF4, have the sequences TGERP, SGEKP and SVGKP respectively. A striking feature of these linkers is that all three have been shown to be phosphorylated in cells arrested at the G2/M boundary of the cell cycle as they enter mitosis[13]. Studies of several mutants, including phosphomimetic mutants with charged residues that mimic phosphorylation at the threonine and serine in position 1 of the linker, indicate that phosphorylation causes Ikaros to dissociate from the pericentromeric heterochromatin[13]. Gel-shift assays of the phosphomimetic mutants have shown that this behavior is correlated with decreased affinity for DNA. Furthermore, the low affinity of Ikaros for DNA that is observed in cells arrested in the G2 stage of the cell cycle is increased dramatically upon treatment with phosphatase[13]. This phosphorylation phenomena has also been observed with sp1, a transcription factor that is also inactivated in G2 arrested cells[14]. This has led Dovat et al[13] to propose that phosphorylation of linkers is a fundamental mechanism by which the affinity of tandem zinc fingers for DNA is modulated during mitosis and a potentially important mechanism for the regulation of transcription in general.
The above conclusion is supported by studies of YY1, a widely distributed transcription factor with a C-terminal DNA binding domain containing four tandem zinc fingers[15]. The DNA binding domain of YY1 is phosphorylated in the first position of the linkers between fingers 2 and 3 (both threonine) in addition to a third site at a serine outside of the DNA binding domain. Phosphorylation (or phosphomimetic mutation) at either threonine in the linker dramatically decreases DNA binding and prevents nuclear localization of YY1. The phosphorylated serine outside the DNA binding domain lacks the above effects. Thus YY1 function has also been postulated to be regulated by linker phosphorylation[15].
Insight into the mechanism by which linker phosphorylation causes dissociation from DNA has been provided by a study of the effects of linker phosphorylation on DNA binding affinity using direct measurement of fluorescence anisotropy of fluorescent probes attached to synthetic DNA[16,17]. This method has been used to measure the affinity of all possible phosphorylation states of a synthetic three-zinc-finger protein for a DNA probe[16]. Direct measurements of dissociation constants have confirmed that phosphorylation at the first position of either of the two linkers decreases DNA binding affinity by 30-49-fold while phosphorylation of both linkers produces a 130-fold decrease in affinity, thus supporting the hypothesis that linker phosphorylation can modulate DNA binding affinity in vivo. Investigations of the crystal structures of tandem zinc fingers complexed with cognate DNA have revealed that the addition of the phosphate should not actually pose any serious steric problems[16]. Other studies have indicated that the hydroxyl of the threonine or serine at position 1 of the linker forms an H-bond with the last residue of the α-helix of the zinc finger providing a “cap” to the helix[18]. This may also be important in establishing the orientation of the two flanking zinc fingers with respect to each other. Indeed, simply substituting an alanine for the threonine or serine at position 1 of the linker is sufficient to disrupt DNA binding of Ikaros[13], which supports the idea of an important structural role for the hydroxyl of threonine or serine at position 1 of the linker.
Several other phosphorylation sites on Ikaros have been demonstrated to exist outside of the zinc finger regions. These appear to have indirect effects on the DNA-binding affinity of Ikaros[19-21]. A detailed analysis of the interplay between phosphorylation and Ikaros function awaits a complete 3D structure of Ikaros or one of its family members.
FUNCTION OF THE C-TERMINAL ZINC FINGERS
The C-terminal protein interaction (dimerization) domain of Ikaros supports both homodimerization and heterodimerization with close family members[22-24], including Aiolos[25], Helios[24,26-28], Eos[29,30], and Pegasus[29]. Although a single molecule of Ikaros binds DNA as a module, the full biological function of Ikaros requires the C terminus of Ikaros that encompasses zinc fingers 5 and 6. This segment of the protein is both necessary and sufficient for Ikaros dimerization[22]. McCarty et al[22] have produced a construct that contains a short N-terminal sequence fused to a 64-amino-acid peptide that contains only fingers 5 and 6 which they have termed the dimerization zinc finger (DZF) domain. This DZF domain can duplicate all the normal dimerization behavior of Ikaros as measured by gel filtration, co-immunoprecipitation and cross-linking. Several other constructs have been made by exchanging zinc fingers 2 and/or 3 for fingers 5 and/or 6. Only constructs with intact zinc fingers 5 and 6 support dimerization. Experiments have been performed with homologs, Hunchback (from Drosophila)[31] and TRPS-1 (from human)[32], both of which homodimerize but do not dimerize with Ikaros[22]. A series of mutants have been produced that identified amino acids critical for binding. Important positions reside largely in the N-terminal portion of the α helices of both F5 and F6 with the selectivity region of Ikaros extending from the C portion of the helix in F5 to the middle of the helix in F6. McCarty et al[22] have also been able to produce a chimeric DZF that selectively homodimerizes, which suggests that the DZF comes in direct contact with the corresponding DZF in the binding partner. Homology modeling of the DZF has shown that the critical amino acids cluster on one face of the model. Precisely how the two DZF domains on separate subunits interact remains unknown.
INTERPLAY BETWEEN DNA BINDING AND PROTEIN INTERACTION DOMAINS
With a protein such as Ikaros that has multiple binding partners and phosphorylation sites there is certain to be linkage between the DNA binding domain and the protein interaction domains with each modulating the respective affinities of the other. A picture of this interplay can be developed from examining a synthetic construct between a zinc finger DNA-binding domain and a leucine zipper protein interaction domain (designated Zif268-GCN4)[33]. This protein selected by phage display techniques is capable of dimerizing with high affinity and binding a bipartite DNA probe that is cognate to the zinc finger domains[33]. This protein has two tandem zinc fingers that bind to DNA as a dimer, which recognizes overlapping DNA sequences on opposite strands of the DNA. The second zinc finger serves as a transition from the DNA binding domain to the protein interaction domain by interacting with both DNA and the other subunit of Zif268-GCN4. A high-affinity probe for Ikaros (BS-4)[34] has wider spacing between the cognate DNA sequences (three base pairs) than Zif268-GCN4. However, binding ZF2 and ZF3 of Ikaros to the BS-4 DNA is likely to place ZF4 in close proximity with the corresponding ZF4 from the interacting Ikaros partner with probably the same orientation as Zif268-GCN4. Thus ZF4 of Ikaros could serve a dual function of promoting DNA binding and the formation of the terniary Ikaros2-DNA complex, providing a crucial linkage between the Ikaros domains.
CONCLUSION
Ikaros and its family members are an interesting study in the application of both the reductionist and the holistic approaches to studying important cellular functions. The ultimate answers to the biological questions are clearly rooted in the fine-tuned molecular interactions such as those discussed here. The zinc finger domains form a functional core around which multiple cellular effects are manifest through the interactions/activities mediated by the remainder of the protein.
ACKNOWLEDGMENTS
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Peer reviewer: Hoyun Lee, Senior Scientist & Professor, Tumor Biology Group, Regional Cancer Program, Sudbury Regional Hospital, 41 Ramsey Lake Road, Sudbury, Ontario P3E 5J1, Canada
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM