Published online May 26, 2011. doi: 10.4331/wjbc.v2.i5.90
Revised: April 27, 2011
Accepted: May 4, 2011
Published online: May 26, 2011
AIM: To determine if and how a loop region in the peptide deformylase (PDF) of Chlamydia trachomatis regulates enzyme function.
METHODS: Molecular dynamics simulation was used to study a structural model of the chlamydial PDF (cPDF) and predict the temperature factor per residue for the protein backbone atoms. Site-directed mutagenesis was performed to construct cPDF variants. Catalytic properties of the resulting variants were determined by an enzyme assay using formyl-Met-Ala-Ser as a substrate.
RESULTS: In silico analysis predicted a significant increase in atomic motion in the DGELV sequence (residues 68-72) of a loop region in a cPDF mutant, which is resistant to PDF inhibitors due to two amino acid substitutions near the active site, as compared to wild-type cPDF. The D68R and D68R/E70R cPDF variants demonstrated significantly increased catalytic efficiency. The E70R mutant showed only slightly decreased efficiency. Although deletion of residues 68-72 resulted in a nearly threefold loss in substrate binding, this deficiency was compensated for by increased catalytic efficiency.
CONCLUSION: Movement of the DGELV loop region is involved in a rate-limiting conformational change of the enzyme during catalysis. However, there is no stringent sequence requirement for this region for cPDF enzyme activity.
- Citation: Oey CB, Bao X, Lewis C, Kerrigan JE, Fan H. High tolerance to mutations in a Chlamydia trachomatis peptide deformylase loop. World J Biol Chem 2011; 2(5): 90-97
- URL: https://www.wjgnet.com/1949-8454/full/v2/i5/90.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v2.i5.90
Bacterial infection is a leading cause of death worldwide. Bacteria are increasingly resistant to antibiotics, but few new antibiotics have been developed in the past 3-4 decades. As a result, the number of effective antibiotic pipelines for several infectious diseases has now been reduced to one or two. This underscores the importance of identification and characterization of potential new antibacterial targets.
The protein sequence of newly synthesized bacterial proteins starts with an N-formyl methionine. Sequential removal of the N-formyl group and methionine residue by peptide deformylase (PDF) and N-methionine peptidase is essential for either maturation into functional proteins or the degradation of functional formylated proteins as required for bacterial survival[1]. Studies have shown that PDF is a promising therapeutic target of a variety of bacterial pathogens. Accordingly, there have been tremendous efforts to develop new antibiotics that target PDF[2,3].
Conceptually, two types of PDF inhibitors can be therapeutically useful. The first type is broad spectrum inhibitors that are effective against a large variety of pathogens. Most (if not all) PDF inhibitors developed to date fall into this category since they are pseudosubstrate-based compounds[3,4]. Although the PDF proteins from different bacteria share relatively low homology, they have a conserved tertiary structure[5-8]. As a result, pseudosubstrate-based inhibitors, either naturally occurring or developed for the PDF from a specific species, can also inhibit the enzyme from multiple species. However, for the prevention and treatment of certain infections, narrow-spectral PDF inhibitors that target specific pathogens may be more desirable. For example, successful treatment of tuberculosis requires months of medication, therefore, highly specific anti-mycobacterials would minimize the risk of normal microflora disruption and the selection of resistant strains in other species. Furthermore, specific mycobacterial PDF (mPDF) inhibitors may also reduce host cell toxicity because mammals, including humans, also encode a PDF (although the function of this enzyme is not well understood). Interestingly, mPDF contains three consecutive Arg residues in a loop region, which is unique among all PDFs examined. It has been shown that mutation of the Arg residues significantly reduces the catalytic properties of mPDF[9]. Therefore, this mPDF loop is a potential specific anti-tuberculosis target.
Sexually transmitted infection is another highly challenging health problem. Chlamydia trachomatis (C. trachomatis) is a leading sexually transmitted pathogen. Most patients with C. trachomatis infection exhibit only mild or even no symptoms, therefore, they do not seek medical treatment. However, a significant proportion of the untreated patients develop severe complications such as pelvic inflammatory disease and infertility[10]. In the absence of an effective vaccine, chemopreventive measures are being sought after. Since re-infection is very common, as a result of the existence of multiple C. trachomatis serovars and the inability of the human body to mount lasting protective immunity against the pathogens, chemoprevention needs to be implemented among the sexually active population as long as they practice unprotected sex. For this reason, only antichlamydials with little or no effects on other microbial species and the human host should be used for the prevention of sexually transmitted chlamydial infection.
PDF is also essential for C. trachomatis. PDF inhibitors effectively inhibit chlamydial replication not only in cell culture, but also in a genital infection mouse model, suggesting that chlamydial PDF (cPDF) is a potential prophylactic and therapeutic target[11,12]. In characterizing a cPDF variant, we found that mutation of two amino acids resulted in a significant increase in atomic motion in the loop region corresponding to the mPDF loop region that contained the three critical Arg residues. However, our analysis showed that radical substitutions in the cPDF loop region either had little effect on enzyme kinetics or resulted in improved catalytic efficiency, indicating high sequence flexibility in the enzyme.
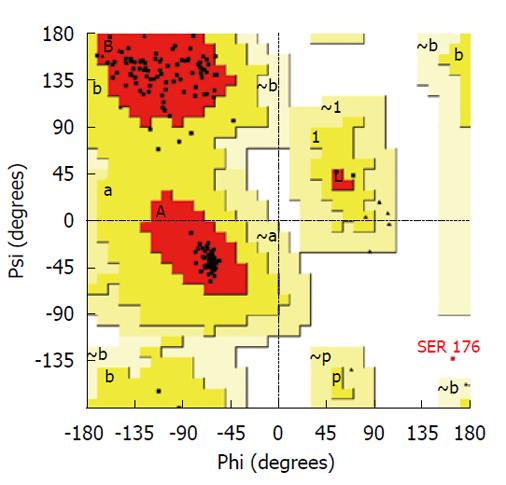
All graphics/workstation-level computations were performed on a Sun Ultra 24 Linux workstation. Graphical illustrations were prepared using the VMD program[13]. A multiple template approach was used to build a homology model of C. trachomatis PDF using the Modeller (9v5) program[14]. The following PDF structures were used as templates: 1RL4A of Plasmodium falciparum[15]; 1XEOA of Escherichia coli (E. coli)[16]; 1N5NA of Thermus thermophilus[6]; and 1V3YA of Pseudomonas aeruginosa[17]. Results of alignment of amino acid sequences of these proteins with cPDF are shown in Figure 1. All of these templates have a sequence identity to cPDF of 42% or higher. The high-energy loop region (residues 61-76) was refined using the loop refinement algorithm in Modeller. The Procheck program[18] was used for stereochemical analysis of the model. The Ramachandran plot analysis of the final lowest energy model revealed that 99.4% of residues were in core or allowable regions (Figure 2). The G-factor analysis revealed no unusual dihedrals or other properties.
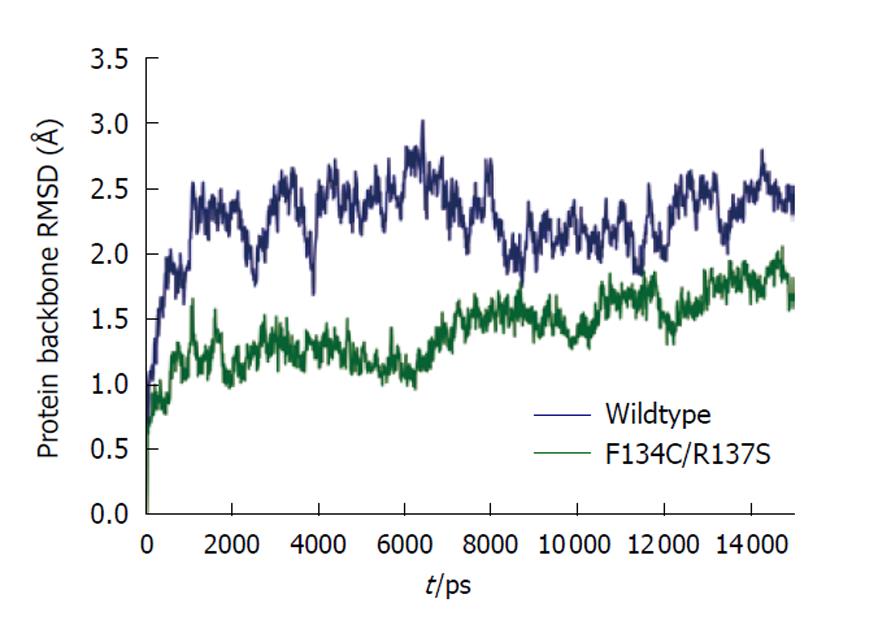
The Amber 10 suite of biomolecular simulation programs was used for all molecular dynamics calculations and results analysis[19]. All molecular dynamics computations were performed on the Ranger supercomputer at the Texas Advanced Computer Center, University of Texas at Austin. The force field of Duan et al[20] was used to model the proteins for all molecular mechanics calculations. The cationic dummy atom approach was used to model the tetrahedral metal ion in the active site of the enzyme[21,22]. The initial models were energy minimized in vacuo to remove bad steric contacts. The model was solvated in a truncated octahedral periodic box of SPC/E water[23], with the overall charge of the system neutralized by insertion of the appropriate number of sodium ions. A 1-nm short-range cutoff was used for all computations. Long-range electrostatics in the model were treated using the particle mesh Ewald method[24,25]. Each system was subjected to energy minimization of the solvent only, followed by minimization of the entire system before dynamics. A 2-fs time step was used for all molecular dynamics work. All bonds to hydrogen were restrained using the “shake” method[26]. A preliminary dynamics step restraining motion of the protein while gradually increasing temperature from 0 K to 300 K was run for 100 ps. This step was followed by a constant temperature (300 K) and constant pressure (1 bar) production run for a time period of 15 ns. As shown in Figure 3, at the end of this 15-ns period, the protein backbones of both wild-type cPDF and the F134C/R137S mutant had fully stabilized. Structural averages of the proteins were calculated from the last 3 ns of simulation and were refined using in vacuo energy minimization. The average per residue temperature factors were calculated from atomic root mean square fluctuation data obtained from the last 2 ns of simulation trajectory.
A pET21-based expression vector for wild-type cPDF carrying a carboxyl terminal (His)6-tag has been previously described[11]. This vector was used as the template for the construction of the D68R, E70R, D68R/E70R or ΔD68-V72 cPDF variants using a QuickChange site-directed mutagenesis kit (Stratagene). Sequence authenticity of the cPDF-coding sequence in the vectors was verified by automated DNA sequencing. Production and purification of the recombinant proteins were carried out following published procedures with modifications[11]. Plasmid-transformed ArcticExpress E. coli was cultured on a shaker at 30°C. When the A600 of the culture reached about 0.8, the culture temperature was lowered to 13°C, isopropyl β-D-1-thiogalactopyranoside (final concentration: 1 mmol/L) and CoCl2 (final concentration: 100 μmol/L) were added to the culture to induce cPDF gene transcription and subsequent synthesis of cobalt-containing cPDF enzyme. After overnight culture at 13°C, the bacteria were collected by centrifugation, and lysed by a French Press. Cell debris was removed by centrifugation at 25 000 g for 30 min. cPDFs were purified with the Talon affinity metal Agarose (Clontech) and stored at -80°C after the addition of glycerol to a final concentration of 10%.
An assay previously developed for the E. coli PDF[27] was modified to measure the activity of the chlamydial enzyme. The assay mix, in a total of 50 μL reaction volume, contained 50 mmol/L HEPES (pH 7.2), 10 mmol/L NaCl, 125 ng PDF, 0-50 mmol/L formyl-Met-Ala-Ser (fMAS) and 50-500 nmol/L GM6001. The deformylation reaction was allowed to proceed at 37°C for 10 min and then terminated by heating at 95°C for 2 min. The amount of MAS produced was reported by 2,4,6 trinitrobenzene sulfonic acid (TNBSA) that reacts with the free amine group to form a chromogenic peptide conjugate. Briefly, 550 μL of 0.0036% TNBSA, freshly prepared in 0.1 mol/L NaHCO3, was added to the reaction and incubated at 37°C for 1 h. Following the addition of 200 μL SDS and 100 μL 1.0 mol/L HCl, the peptide-TNBSA conjugate was quantified by measuring A335. All A335 values were within the range of free methionine standards. Kinetic constants were calculated from the Lineweaver-Burk plots. Data presented are mean ± SD of three experiments.
We have previously demonstrated that chlamydiae are susceptible to hydroxamate-based metalloprotease inhibitors. By selecting and genome sequencing of the resistant mutants, we have identified cPDF as the target of the inhibitors[11]. One of the mutants has two nucleotide changes in the coding region, resulting in two amino acid substitutions (F134C and R137S) in the cPDF protein (observations from lab of Fan H to be published elsewhere). The mutant protein carrying a carboxyl terminal (His)6-tag (i.e. F134C/R137S cPDF) was expressed in E. coli and purified to apparent homogeneity. Compared to wild-type cPDF, the F134C/R137S variant had a nearly 10-fold higher Km when fMAS was used as a substrate, a sevenfold higher kcat, a nearly 30% lower kcat/Km, and > 6-fold higher Ki for the inhibitor GM6001 (observations from lab of Fan H to be published elsewhere).
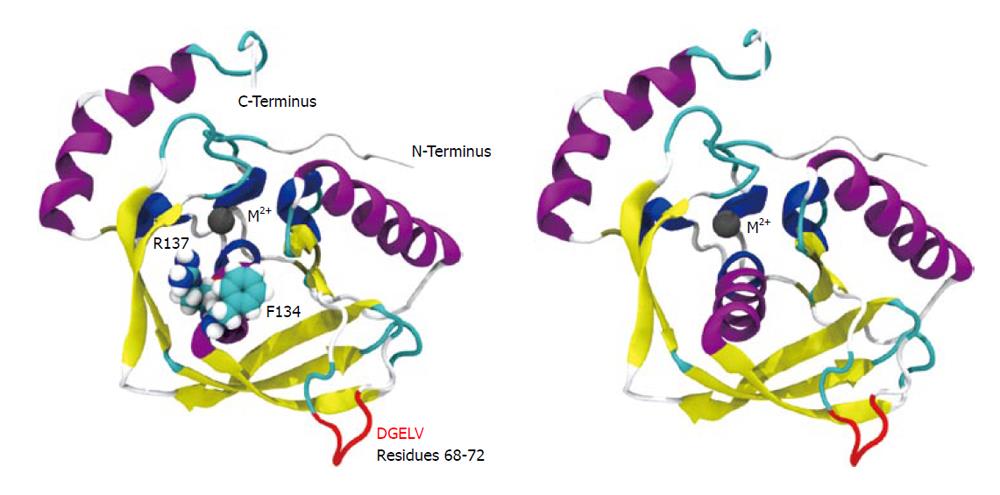
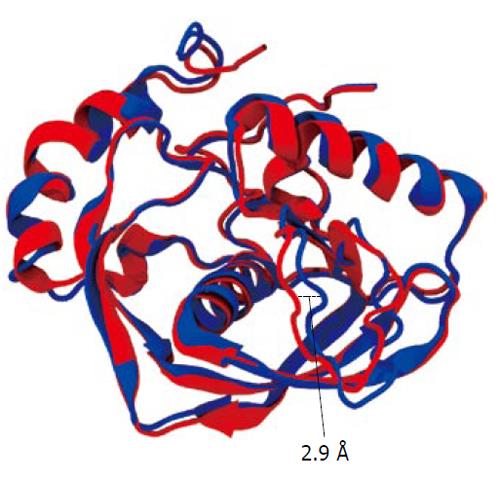
We conducted molecular simulation to understand how the amino acid sequence change affects enzyme catalysis. We first built a homology model of cPDF, because its experimental structure does not exist. The structures of PDFs from several other species are well known and were used to build the model of cPDF. A Ramachandran plot based on an analysis of 118 structures of resolution of at least 0.2 nm and R-factors no greater than 20% is shown in Figure 2. In the plot, 148 of the total 181 residues (93.1%) in cPDF were found in the most favored region, and 10 residues (6.3%) in an additional allowed region, and only one residue (0.6%), Ser176 at the C terminus, in a disallowed region. These data suggest that our model was of excellent quality because a good model requires only 90% of residues distributed in the most favored regions. According to our model, cPDF shares a conserved overall tertiary structure with the enzyme from other species studied (Figure 4), which is consistent with findings with other PDFs previously studied[5-8].
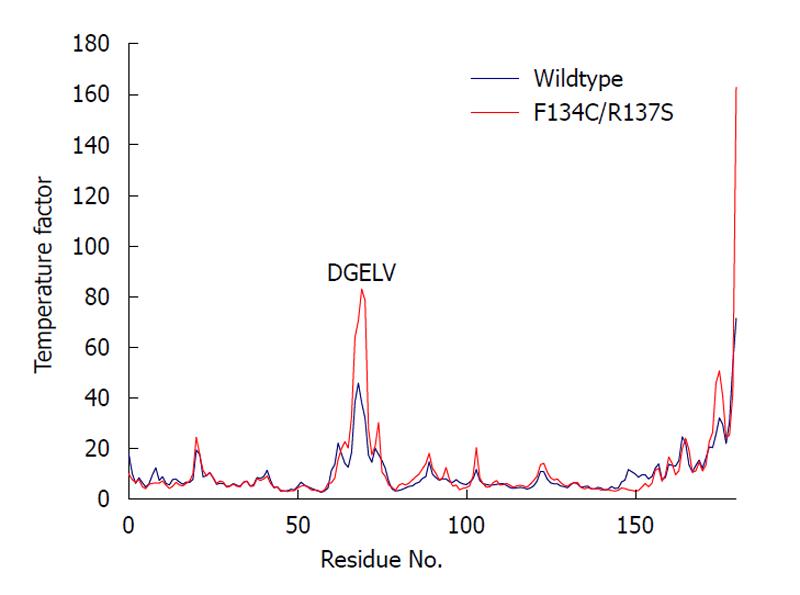
We computed the average temperature factor per residue for the protein backbone atoms to examine protein residue backbone mobility in the model. Temperature factor is an indicator of dynamic mobility, with higher factors indicating high mobility and low factors indicating low mobility. As shown in Figure 5, in addition to the anticipated high mobility at the C-terminal end, we also observed a significant amount of motion in a loop region spanning residues 61-76. Interestingly, the F134C/R137S cPDF showed a further sharp increase in the backbone atom motion in DGELV residues (68-72) within the loop (Figure 5), as compared to wild-type protein. The mutated residues 134 and 137 were located on a stable α helix forming the enzyme active site and their backbone atom motion remained stable and unaffected by mutations (Figure 4). These data suggest that amino acid substitutions at or near the active site of cPDF could alter the stability of the loop region.
It has previously been demonstrated that mutation of any of three consecutive Asp residues to Lys in a loop region in mPDF, which corresponds to the loop region containing the DGELV sequence in cPDF, causes moderate decreases to severe loses in the enzyme activity[9,28]. Molecular simulation indicates structural changes in the substrate-binding pocket in the loop-mutated mPDF enzymes[9]. The mPDF study and our prediction of increased motion in the catalytically inefficient F134C/R137S cPDF prompted us to hypothesize that there may also be interdependence between the active site and the loop region in cPDF.
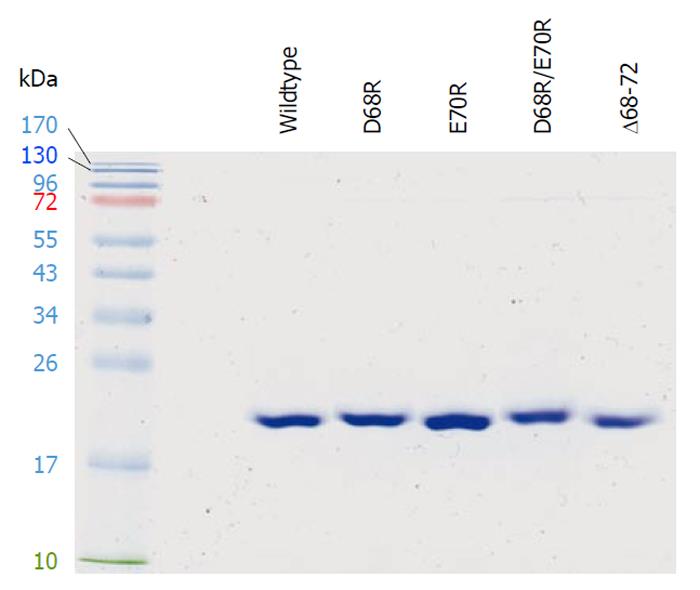
To test this hypothesis, we performed site-directed mutagenesis in the DGELV region of cPDF on the basis of a pET21 expression vector[11]. Thus, the negatively charged Asp68 and Glu70 residues were mutated, individually or in combination, to the positively charged Arg. Furthermore, a cPDF variant with the deletion of the DGELV sequence was also constructed. The cPDF variants as well as wild-type cPDF were expressed in E. coli and purified to apparent near homogeneity for kinetic analyses. A Coomassie-blue-stained gel with purified recombinant loop variants as well as the wild-type cPDF protein is shown in Figure 6. The Km values of D68R, E70R and D68R/E70R cPDF were very similar to that of the wild-type enzyme (Table 1). Both the D68R and D68R/E70R variants exhibited significantly higher levels of kcat and kcat/Km, whereas the D70E cPDF displayed a relatively small but statistically significant 26% decrease in kcat, and an even smaller and statistically insignificant 17% drop in kcat/Km (Table 1). Compared to the wild-type enzyme, Δ68-72 cPDF had a fivefold higher kcat and a twofold higher kcat/Km, although these were accompanied by a nearly threefold increase in Km (Table 1). These data suggest that the cPDF enzyme is highly tolerant to radical mutations in the loop region.
| Wild-type | D68R | E70R | D68R/E70R | Δ68-72 | |
| Km | |||||
| mM (mean ± SD) | 6.8 ± 0.7 | 7.4 ± 1.2 | 6.1 ± 1.7 | 6.7 ± 0.4 | 18.6 ± 2.9 |
| % control | 100 | 109 | 91 | 99 | 275 |
| kcat | |||||
| s-1 (mean ± SD) | 37.9 ± 2.6 | 93.1 ± 8.4b | 27.9 ± 4.3a | 113.1 ± 10.9b | 197.0 ± 29.0b |
| % control | 100 | 246 | 74 | 299 | 520 |
| kcat/Km | |||||
| M-1s-1 (mean ± SD) | 5605 ± 357 | 12764 ± 970b | 4652 ± 677 | 16781 ± 678b | 10605 ± 301b |
| % control | 100 | 228 | 83 | 299 | 189 |
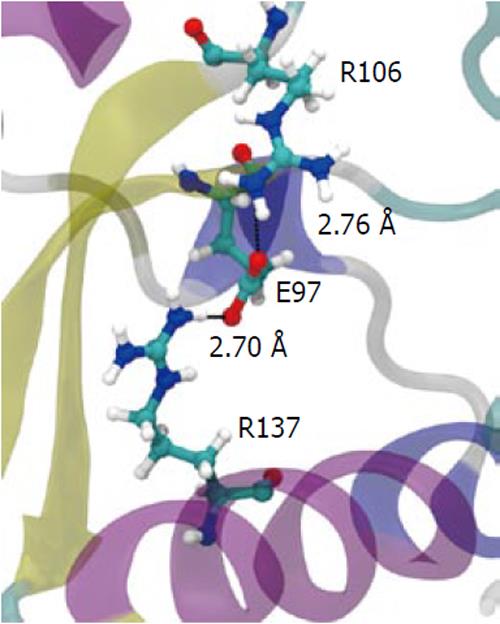
To understand how the atomic motion in the DGELV loop region is increased in the F134C/R137S cPDF variant, we examined the effect of the two amino acid substitutions on the structure of the enzyme. Our structural model showed that the combination of the two residue mutations led to an increase in space for the loop region to occupy. The R137S mutation broke the salt-bridge “sandwich” of Arg137-Glu97-Arg106 in the wild-type (Figure 7). The mutations caused Glu97 to shift over to the vacated space and brought Arg106 more tightly into the active site, which freed up space already made available by the F134C mutation in proximity to the loop (Figure 4). This resulted in the Glu63 to Glu67 region of the loop shifting towards the active site by 0.29 nm. This shift allowed for a higher degree of motion in the DGELV region in the F134C/R137S cPDF (Figure 8). Accordingly, there was an increase in atomic temperature factor in the dynamics simulation of the mutant (Figure 5).
A loop region in the mPDF has been shown to be critical for enzyme activity[9,28]. Accordingly, that enzyme has an extremely low level of tolerance to mutation of three consecutive Arg residues in this region. For one of the Arg residues, even a conserved Arg→Lys mutation results in severe loss of enzyme activity[9]. However, how important the corresponding loop is in other PDFs has not been examined. The increase in atomic motion in the DGELV region of the F134C/R137S mutant prompted us to assess the role of the equivalent cPDF loop region in enzyme function. Kinetic analyses showed that, in contrast to the mPDF, the cPDF tolerated radical mutations individually or in combination at Asp68 and Glu70. In fact, the single D68R and double D68R/E70R mutants appeared to be more efficient enzymes than wild-type cPDF, as indicated by significantly increased kcat and kcat/Km values in the mutants. Clearly, cPDF has a high degree of sequence flexibility in the DGELV loop region. However, the observed kinetic changes in the loop mutants, together with the increased mobility of the DGELV region of the loop in the active site F134C/R137S mutant, do suggest that movement of this loop is involved in a rate-limiting conformational change of the enzyme during catalysis. Furthermore, our data do not exclude the possibility that the cPDF has more stringent sequence requirements in other regions of the loop.
It is particularly interesting why nature has not selected better enzymes such as the D68E mutant during evolution. Perhaps this is because the PDF enzyme activity needs to be controlled at the organismal level, as is the case with many other enzymes and proteins. Although peptide deformylation is required, it is not known what adverse effects too much PDF activity may have. Interestingly, the chlamydial vacuole of GR10, a cPDF-overexpressing mutant[11], has different morphology, compared to that of the parental strain. The mutant appears to generate a large aberrant replicating form (unpublished observations). cPDF is the only gene with a mutation in that mutant (unpublished observations from lab of Fan H), therefore, the abnormal morphology must have resulted from cPDF overexpression. Characterization of such aberrant structures will be useful to the understanding of a possible need for maintaining PDF activity at an appropriate level in normal chlamydial physiology, and perhaps, in general bacterial physiology as well.
Massive bacteria-based overexpression systems may result in incompletely deformylated proteins. This problem can be alleviated by PDF co-expression[29]. Owing to its higher catalytic efficiency, the D68R/E70R cPDF variant would be particularly useful if such a situation of incomplete deformylation of target protein arises, especially if the target protein is a chlamydial protein.
By removing the formyl group from the leading Met of newly synthesized proteins, peptide deformylase (PDF) is an essential enzyme for protein maturation in bacteria. PDF is a promising therapeutic target for infectious diseases including sexually transmitted infection caused by Chlamydia trachomatis. It has recently been discovered that substitution of two amino acids near the active site of the chlamydial PDF (cPDF) contributes to resistance.
Characterization of individual PDFs is of significance for the development of narrow-spectrum PDF inhibitors that target specific pathogens. Elucidation of resistance mechanisms may provide invaluable information for the design of new inhibitors with improved targeting efficacy.
By combining simulation, site-directed mutagenesis and kinetic analysis, this study revealed a role of the DGELV loop region in the catalysis and inhibition efficiency in cPDF, and a remarkably high level of tolerance to radical mutations in the loop region. It is believed that this is the only study that has examined the role of the loop in catalysis by the PDF enzyme, besides the mycobacterial PDF (mPDF). The sequence flexibility in cPDF is in striking contrast to that of mPDF.
The findings presented in this report have implications for the development of PDF-inhibitor-based antibacterials, including those for chlamydial infection and tuberculosis. The cPDF variants with increased catalytic efficiency may be useful for heterologous protein expression requiring improved N-deformylation efficiency.
Temperature factor is an indicator of dynamic mobility. Higher factors indicate high mobility and low factors indicate low mobility.
In this study, Oey et al studied the function of a loop region in cPDF using molecular modeling and biochemical analysis. This work is related to the development of new anti-infective agents, a topic that is both timely and important. The introduction is well presented. The methods are adequate and vigorous. Data are robust and properly discussed. Additional references for the nature of current PDF inhibitors should be provided; as should data documenting the purity of recombinant cPDFs.
Peer reviewers: Gerardo Corzo, PhD, Department of Molecular Medicine and Bioprocess, Instituto de Biotecnología /UNAM, Av. Universidad #2001, Col. Chamilpa C.P. 62210, Cuernavaca, Morelos, Mexico; Andrea Trabocchi, PhD, Department of Chemistry “Ugo Schiff”, University of Florence, Via della Lastruccia 13, I-50019, Sesto Fiorentino, Florence, Italy
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM
| 1. | Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374-1377. |
| 2. | Sharma A, Khuller GK, Sharma S. Peptide deformylase--a promising therapeutic target for tuberculosis and antibacterial drug discovery. Expert Opin Ther Targets. 2009;13:753-765. |
| 3. | Chen D, Yuan Z. Therapeutic potential of peptide deformylase inhibitors. Expert Opin Investig Drugs. 2005;14:1107-1116. |
| 4. | Leeds JA, Dean CR. Peptide deformylase as an antibacterial target: a critical assessment. Curr Opin Pharmacol. 2006;6:445-452. |
| 5. | Chan MK, Gong W, Rajagopalan PT, Hao B, Tsai CM, Pei D. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry. 1997;36:13904-13909. |
| 6. | Kreusch A, Spraggon G, Lee CC, Klock H, McMullan D, Ng K, Shin T, Vincent J, Warner I, Ericson C. Structure analysis of peptide deformylases from Streptococcus pneumoniae, Staphylococcus aureus, Thermotoga maritima and Pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase. J Mol Biol. 2003;330:309-321. |
| 7. | Dirk LM, Schmidt JJ, Cai Y, Barnes JC, Hanger KM, Nayak NR, Williams MA, Grossman RB, Houtz RL, Rodgers DW. Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture. Biochem J. 2008;413:417-427. |
| 8. | Cai J, Han C, Hu T, Zhang J, Wu D, Wang F, Liu Y, Ding J, Chen K, Yue J. Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation. Protein Sci. 2006;15:2071-2081. |
| 9. | Saxena R, Kanudia P, Datt M, Dar HH, Karthikeyan S, Singh B, Chakraborti PK. Three consecutive arginines are important for the mycobacterial peptide deformylase enzyme activity. J Biol Chem. 2008;283:23754-23764. |
| 10. | Schachter J. Infection and disease epidemiology. Chlamydia: intracellular biology, pathogenesis, and immunity. Washington DC: ASM Press 1999; 139-169. |
| 11. | Balakrishnan A, Patel B, Sieber SA, Chen D, Pachikara N, Zhong G, Cravatt BF, Fan H. Metalloprotease inhibitors GM6001 and TAPI-0 inhibit the obligate intracellular human pathogen Chlamydia trachomatis by targeting peptide deformylase of the bacterium. J Biol Chem. 2006;281:16691-16699. |
| 12. | Balakrishnan A, Wang L, Li X, Ohman-Strickland P, Malatesta P, Fan H. Inhibition of chlamydial infection in the genital tract of female mice by topical application of a peptide deformylase inhibitor. Microbiol Res. 2009;164:338-346. |
| 13. | Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33-38, 27-28. |
| 14. | Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5:Unit 5.6. |
| 15. | Robien MA, Nguyen KT, Kumar A, Hirsh I, Turley S, Pei D, Hol WG. An improved crystal form of Plasmodium falciparum peptide deformylase. Protein Sci. 2004;13:1155-1163. |
| 16. | Jain R, Hao B, Liu RP, Chan MK. Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal. J Am Chem Soc. 2005;127:4558-4559. |
| 17. | Kamo M, Kudo N, Lee WC, Motoshima H, Tanokura M. Crystallization and preliminary X-ray crystallographic analysis of peptide deformylase from Thermus thermophilus HB8. Acta Crystallogr D Biol Crystallogr. 2004;60:1299-1300. |
| 18. | Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283-291. |
| 19. | Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668-1688. |
| 20. | Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999-2012. |
| 21. | Pang YP, Xu K, Yazal JE, Prendergas FG. Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci. 2000;9:1857-1865. |
| 22. | Pérez L, Kerrigan JE, Li X, Fan H. Substitution of methionine 435 with leucine, isoleucine, and serine in tumor necrosis factor alpha converting enzyme inactivates ectodomain shedding activity. Biochem Cell Biol. 2007;85:141-149. |
| 23. | Berendsen HJ, Grigera J, Straatsma T. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269-6271. |
| 24. | Darden T, York D, Pedersen L. Particle Mesh Ewald: An N-log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089-10092. |
| 25. | Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L. A smooth particle mesh ewald potential. J Chem Phys. 1995;103:8577-8592. |
| 26. | Miyamoto S, Kollman P. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithms for Rigid Water Models. J Comp Chem. 1992;13:952-962. |
| 27. | Groche D, Becker A, Schlichting I, Kabsch W, Schultz S, Wagner AF. Isolation and crystallization of functionally competent Escherichia coli peptide deformylase forms containing either iron or nickel in the active site. Biochem Biophys Res Commun. 1998;246:342-346. |
| 28. | Saxena R, Chakraborti PK. Identification of regions involved in enzymatic stability of peptide deformylase of Mycobacterium tuberculosis. J Bacteriol. 2005;187:8216-8220. |