INTRODUCTION
The primary structure of eukaryotic chromatin is composed of nucleosomes connected by linker DNA sequences. This beads-on-a-string structure is organized into higher order structures, which is facilitated by linker histone H1 that associates with linker DNA and stabilizes the nucleosomes. Formation of chromatin compacts DNA and preserves genome stability, but also presents an obstacle to DNA transactions such as transcription, replication, and repair. To address this issue, cells have evolved various mechanisms to modify/remodel chromatin structure resulting in chromatin states that are suitable for genome functions. The high mobility group box (HMGB) family proteins are non-histone chromatin architectural factors characterized by one or more HMGB motifs that bind DNA in a sequence nonspecific fashion. They play a major role in chromatin dynamics that impact various cellular functions. The yeast Hmo1 protein is classified as an HMGB protein as it contains two HMGB motifs. However, unlike a canonical HMGB protein that has a stretch of acidic amino acids at the C-terminus, Hmo1 ends with a lysine rich, basic, C-terminus, resembling linker histone H1. It is therefore not surprising that Hmo1 seems to exhibit characteristics of both HMGB proteins and linker histones in its multiple functions including genome maintenance, chromatin dynamics, gene transcription, and DNA damage response. On one hand, it has been well established that, like canonical HMGB proteins, Hmo1 plays a key role in promoting transcription by RNA polymerases I and II. On the other hand, there is evidence suggesting that Hmo1, like linker histones, makes yeast chromatin more compact/stable. Recent studies demonstrated that Hmo1 destabilizes/disrupts nucleosome similarly as other HMGB proteins in vitro and revealed an intriguing role of Hmo1 in maintaining a common topological architecture of genes in yeast genome.
ROLE OF HMO1 IN REGULATING TRANSCRIPTION BY RNA POLYMERASE I
HMGB proteins such as human upstream binding factor 1 (UBF1) play a critical role in ribosome biogenesis necessary for cell growth and proliferation[1]. This function is shared by yeast Hmo1 protein, as Hmo1 facilitates the transcription of genes coding for ribosomal RNAs (rRNAs) and ribosomal proteins (RPs)[1-3]. Yeast genome contains about 150 tandemly arranged ribosomal DNA (rDNA) repeats each encoding the 35S rRNA (precursor of 25S, 18S and 5.8S rRNAs) transcribed by RNA polymerase I (RNAPI) and 5S rRNA transcribed by RNA polymerase III (RNAPIII). In yeast, as in human cells, each rDNA repeat exists in either a closed or open chromatin state. The closed state is characterized by a dormant 35S rRNA gene with stable nucleosome occupancy, whereas the open state is associated with an actively transcribed 35S rRNA gene that largely lacks nucleosomes[4,5]. The balance between the closed and open rDNA chromatin states is controlled by an equilibrium of DNA replication-mediated nucleosome assembly and RNAPI transcription-mediated nucleosome removal[5].
Hmo1 binds throughout the entire 35S rDNA sequence[6-8], suggesting that it may play a role in the formation and/or maintenance of rDNA chromatin state. Importantly, Hmo1 specifically associates with the open, nucleosome-free, rDNA repeats[4,5]. Hmo1 association with rDNA is dependent on RNAPI transcription and Hmo1 stays on rDNA even after RNAPI has departed from the template[5,8]. Acetylation of lysine 56 of histone H3 (H3-K56-Ac) has been shown to aid in Hmo1 binding to rDNA[9]. Given that H3-K56-Ac destabilizes nucleosome/chromatin structure[10,11], it may facilitate the eviction of nucleosomes from rDNA, thereby promoting Hmo1 binding. Outside of S phase of the cell cycle, Hmo1 bound to a rDNA repeat counteracts replication-independent nucleosome assembly to maintain the open state of rDNA[5]. In vitro Hmo1-DNA binding studies led to a model in which Hmo1 maintains the stability of nucleosome-free chromatin regions by forming complex DNA structures via DNA bridging and looping mediated by Hmo1-Hmo1 interaction[12].
There is ample evidence supporting the notion that Hmo1 is a functional homolog of human UBF1 regarding regulation of rRNA synthesis[1,3]. Like Hmo1, human UBF1 associates with the entire rRNA genes on rDNA including the promoter and the coding region, and UBF1-bound rDNA repeats assume an open, nucleosome-free, chromatin state[13,14]. The conserved function of maintaining a nucleosome-free state of rDNA by Hmo1 or UBF1 is believed to allow a swift restart of rDNA transcription without the need to overcome the nucleosomal barrier after rDNA transcription is repressed by environmental signals[15-18]. The functional conservation between Hmo1 and UBF1 is further supported by the finding that ectopic expression of UBF1 in yeast partially substitutes for Hmo1’s function in regulating rDNA transcription, whereas Hmo1 expressed in human cells is directly targeted to rDNA chromatin[19].
UBF1 has long been known to facilitate both the initiation and elongation of RNAPI transcription with well-defined mechanisms[20-23]. However, how Hmo1 promotes RNAPI transcription has just begun to be elucidated. Huffines and Schneider recently found that Hmo1 promotes efficient transcriptional elongation by RNAPI, possibly by helping RNAPI counter pausing in G-rich regions of rDNA[24]. Whether Hmo1 also acts to facilitate RNAPI initiation has not been resolved.
The evolutionarily conserved target of rapamycin complex 1 (TORC1) is a central regulator of gene expression in response to environmental cues such as nutrient abundance[25]. Both rDNA and ribosome protein genes (RPGs) are subject to TORC1 regulation: Their transcription is induced by TORC1 when nutrients are available and repressed when TORC1 is inactivated upon nutrient depletion. TORC1 directly associates with the promoter of 35 rRNA gene and activates its transcription[26]. TORC1 inhibition resulted from nutrient starvation leads to its own exit from nucleus and the departure of RNAPI from the nucleolus[26,27]. Notably, TORC1 inhibition causes a release of Hmo1 from rDNA, suggesting a role of Hmo1 in TORC1-mediated regulation of rDNA transcription[7].
ROLE OF HMO1 IN REGULATING TRANSCRIPTION BY RNA POLYMERASE II
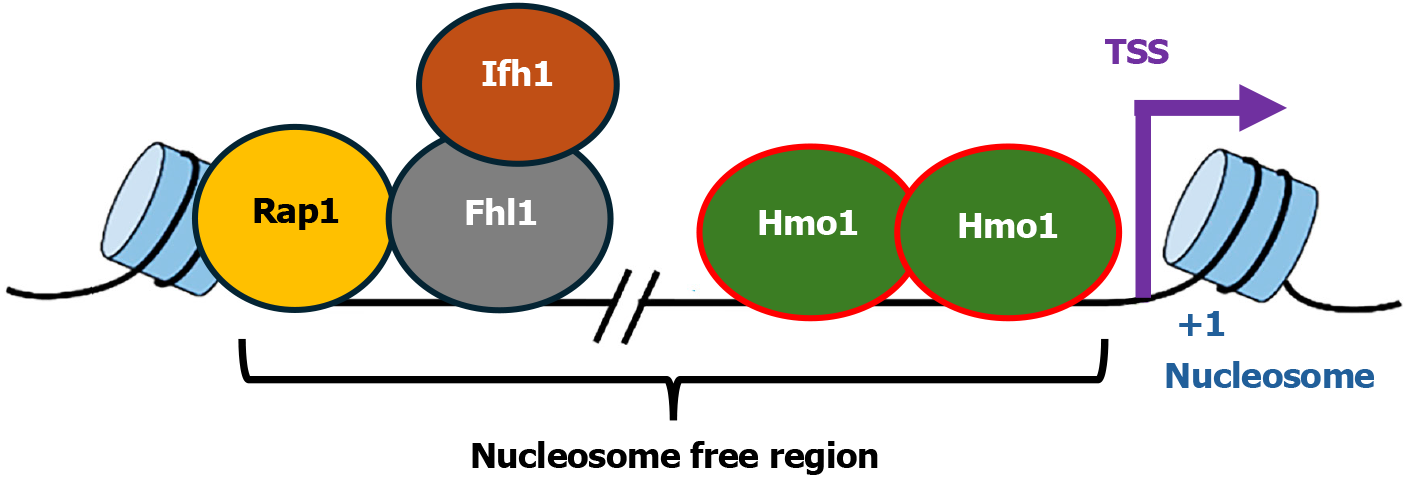
Hmo1 participates in the regulation of genes encoding proteins involved in ribosome biogenesis especially RPs. The 79 yeast RPs are encoded by 138 RPGs, many of which are duplicated genes. The promoters of RPGs are heterogeneous regarding their association with transcription factors (TFs). TFs that bind promoters of RPGs include Rap1, Fhl1 and Ifh1[6,8,28] (Figure 1). Hmo1 binds about half of RPGs promoters[6,8,29-31]. At an RPG promoter, Rap1 serves as a pioneer TF whose binding is required for the recruitment of other TFs[29,30]. Rap1 interacts with Fhl1 and recruits it to RPG promoters, and both Rap1 and Fhl1 associate with the promoters constitutively regardless of the transcriptional states of the genes[28]. Binding of Ifh1 is dependent on Fhl1[32]. Fhl1 and Ifh1 interact with each other, which is critical for active transcription of RPGs[32]. Given that Rap1-binding induces nucleosome removal[33], it is possible that Hmo1 binds the nucleosome free region in an RPG promoter induced by Rap1-binding[29,30] (Figure 1).
Figure 1 Diagram of the organization of Hmo1 associated Category I ribosome protein gene promoters.
The transcription factors, Rap1, Fhl1, Ifh1, and Hmo1, associated with the promoter are shown. The transcription start site, +1 nucleosome of the gene, and the nucleosome free region, are indicated. TSS: Transcription start site.
Hmo1-bound RPG promoters have been collectively designated Category I (Cat I)[29,31]. There is evidence that Hmo1 promotes Fhl1 recruitment to Cat I promoters[6,8] and Hmo1 physically interacts with transcription factor IID (TFIID) and facilitates TFIID occupancy at a group of RPG promoters[34]. Hmo1 deletion causes an upstream shift in transcriptional start sites of at least nine RPGs including RPS5, indicating that Hmo1 can participate in start site selection of RPGs[34]. A detailed analysis of the impact of Hmo1 on RPS5 promoter revealed evidence indicating that Hmo1 directs the assembly of preinitiation complex (PIC) by controlling the range of the nucleosome free region in which PIC can form at the appropriate position[35] (Figure 1). There is evidence that this also occurs at other Cat I promoters[29]. Lack of Hmo1 allows PIC to form ectopically in the now unprotected Hmo1-bidning region of a promoter, which is believed to cause a shift in transcriptional start site[35]. Interestingly, Hmo1, together with Fhl1 and Ifh1, also regulates the expression of HMO1 gene and all three factors associate with HMO1 promoter[6,7,36,37]. Notably, Hmo1 promotes Fhl1 recruitment to HMO1 promoter as it does at Cat I promoters[26]. Therefore, Hmo1 can regulate transcription initiation of RPGs and HMO1 gene via facilitating TF recruitment and/or modulating promoter architecture.
Like rDNA transcription by RNAPI, transcription of RPGs by RNA polymerase II (RNAPII) is also regulated by TORC1 in response to changes in nutrient availability[25]. TORC1 modulates the occupancy of TFs on RPG promoters[7,32]. Whereas active TORC1 promotes targeting of Ifh1 to promoters to facilitate RPG transcription, inactivation of TORC1 leads to Ifh1 removal from promoters and transcription repression[32]. The dissociation of Ifh1 from RPG promoters is caused by the disruption of Ifh1-Fhl1 interaction upon TORC1 inactivation[32]. TORC1 inhibition also leads to the departure of Hmo1 from RPGs, and Hmo1 deletion results in the alleviation of TORC1 dependent repression of RPGs[28]. Similar to the expression of RPGs, HMO1 transcription is also subject to TORC1 regulation[36]. TORC1 inactivation triggers the dissociation of Hmo1 and Ifh1 from HMO1 promoter[36,37]. Therefore, Hmo1 plays a key role in TORC1 mediated regulation of RPGs and HMO1 gene. This, together with the involvement of Hmo1 in the regulation of rDNA transcription by TROC1 mentioned earlier, suggests that Hmo1 may coordinate TORC1-dependent regulation of expression of rRNAs and RPGs[3,38].
FUNCTIONAL DOMAINS OF HMO1
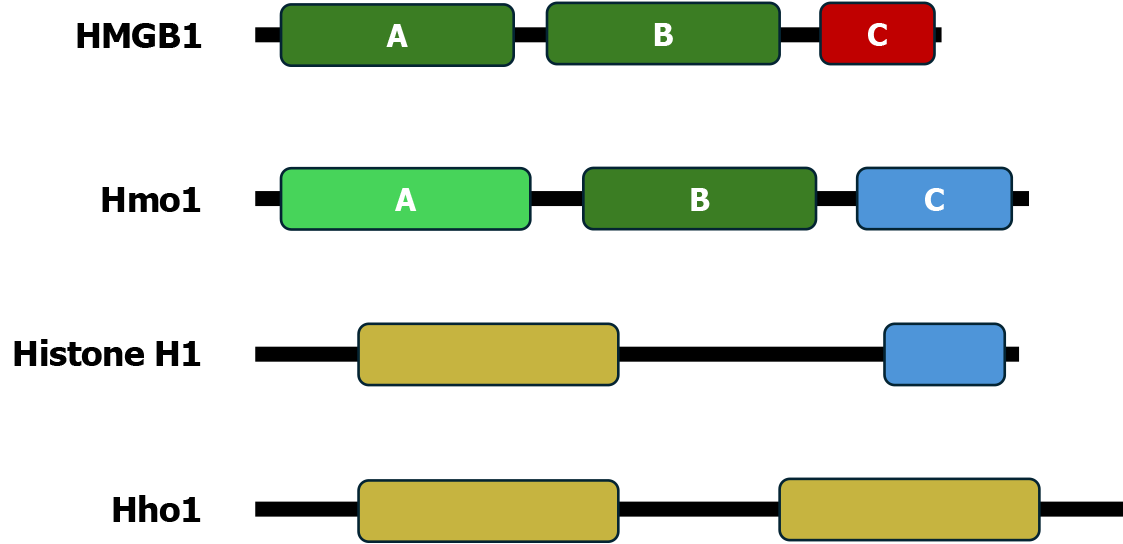
The HMGB family of chromosomal architecture proteins such as human HMGB1 protein are characterized by one or more HMGB motifs and an acidic carboxyl (C)-terminal domain (CTD)[39] (Figure 2). A canonical HMGB motif is a DNA binding domain that is sequence nonspecific, but has preference for distorted DNA structures, such as hemicatenates and four-way junctions[39]. The 246 amino acid long Hmo1 protein is organized into three functional domains designated A, B and C[3,40] (Figure 2). Domain A (residues 12-90) is considered an HMGB-like domain as it has only limited homology to canonical HMGB motif[40,41]. Domain B (residues 106-179) is a canonical HMGB motif. Domain C, or CTD (219-246), is a lysine-rich, basic region at the C-terminus. Note that the boundaries of the C domain have been defined slightly differently by different investigators.
Figure 2 Domain structures of Hmo1 and related chromosomal architecture factors.
Illustrated are the high mobility group boxes (HMGBs) A and B and acidic C-terminal domain (CTD) of human HMGB1; the HMGB-like domain A, the HMGB homolog domain B, and the basic CTD of Hmo1; the globular domain and basic CTD of human linker histone H1.4; and the two globular domains of Hho1, yeast ortholog of linker histone H1. HMGB: High mobility group boxes.
Domain A of Hmo1 binds DNA weakly with a modest specificity for altered DNA conformations including DNA with nicks, gaps, overhangs, or loops, as well as for 4-way junction structures and supercoiled DNA[19,41]. Therefore, domain A may participate in the recognition of structural features of target binding sites[42]. Domain A also mediates Hmo1 dimerization and is sometimes referred to as a dimerization domain[19,41-43]. Domain A has also been shown to mediate the formation of Hmo1 oligomers of more than two proteins on DNA in vivo[44]. Domain A-mediated self-assembly of Hmo1 contributes to its binding to DNA in vitro and in vivo[44]. Hmo1-Hmo1 interaction stabilizes DNA bridges and loops in vitro[12]. There is evidence suggesting that the principal role of domain A is to assemble multiple domain Bs in the appropriate orientation for stable DNA binding[44]. Domain B of Hmo1 binds DNA with high affinity and a limited preference for structural features of DNA[19,41].
The CTD of Hmo1 is highly basic as it is lysine-rich, a feature opposite to the acidic CTD of a canonical HMGB protein (Figure 2)[39,40]. Removal of CTD has little effect on DNA-binding affinity of the truncated Hmo1 protein (designated hmo1-AB)[41,43,45]. As such, CTD is not required for DNA binding by Hmo1. Hmo1 can also bend DNA, which is dependent on CTD in vitro[43,45]. There is evidence that DNA bending by Hmo1 requires the interaction between domains A and C[43]. Hmo1 has been shown to facilitate end joining of linear DNA, which is dependent on its CTD[45]. This can be explained by assuming that Hmo1 compacts DNA in a CTD-dependent fashion[45].
How the properties of the three domains of Hmo1 summarized above are related to its multiple functions in vivo has only been partially elucidated. Both domains A and B are required for the function of Hmo1 in stimulating RNAPI transcription in vivo[19]. Hmo1-CTD is dispensable for autoregulation of HMO1 gene by Hmo1[36]. Whether CTD is also not required for Hmo1-mediated regulation of RPGs has yet to be examined. There is evidence suggesting that a combined nuclear and nucleolar localization signal resides in Hmo1-CTD[19,46]. However, CTD deletion does not seem to compromise localization of hmo1-AB to the nucleus[19,36]. It was proposed that the small size of hmo1-AB allows its passive import/diffusion into the nuclear/nucleolar compartment[19]. Notably, whereas Hmo1 deletion results in a severe growth defect in yeast, cells expressing hmo1-AB exhibit no growth defect[36,40]. As such, the function of Hmo1 that when deleted leads to a slow growth phenotype does not reside within the CTD[36,40].
ROLE OF HMO1 IN REGULATING DNA DAMAGE RESPONSE
DNA damage response recognizes DNA lesions, triggers the activation of checkpoints to arrest cell cycle progression, stabilizes DNA replication forks, and promotes DNA damage repair[47,48]. As DNA damage response in eukaryotes operates in the context of chromatin, it is intimately linked to chromatin structure and dynamics[49,50]. Therefore, it is not surprising that Hmo1, like other chromatin architecture proteins, has been shown to play a role in DNA damage response[3].
Hmo1 is involved in yeast response to double-strand DNA break (DSB). Cellular response to DSB involves a series of early events that modify/remodel chromatin and process DNA ends including histone H2A phosphorylation (aka gH2A, equivalent to human gH2AX), recruitment of chromatin remodelers, and removal of histones, followed by DNA end resection[49,51]. These events are made more efficient/faster by deletion of Hmo1[52], which is consistent with the notion that Hmo1 compacts/stabilizes chromatin, making it more resistant to remodeling/disruption. Notably, Hmo1 mediated inhibition of DSB response events is dependent on its CTD[52]. Since the function of linker histone H1 in chromatin compaction also requires its CTD, it may be Hmo1-CTD induced DNA bending and compaction that promotes chromatin stability. Hmo1 is evicted along with core histones during cellular response to DSBs[52]. These findings are consistent with a model in which Hmo1 compacts/stabilizes chromatin via DNA bending and compaction by its CTD and must be removed before efficient DSB repair could occur[3,52].
In the absence of external DNA-damaging agents, there exist endogenous DNA double-strand breaks (EDSBs), or replication-independent (RIND)-EDSBs in non-dividing eukaryotic cells. Hmo1 deletion downregulates the level of RIND-EDSB in yeast[53]. Likewise, RIND-EDSB level is decreased in human cells lacking HMGB1[53]. Therefore, there seems to be an evolutionarily conserved presence of RIND-EDSBs in eukaryotes whose level is modulated by HMGB proteins. RIND-EDSBs are repaired by DSB repair pathways[53]. The reduction of RIND-EDSB abundance in cells lacking Hmo1 may be due to more efficient DSB repair as a result of the elimination of Hmo1-mediated inhibition of DSB repair processes[3].
There has been intriguing evidence implicating Hmo1 in DNA damage tolerance (DDT). DDT promotes the bypass of single strand DNA (ssDNA) lesions on DNA template during DNA replication, avoiding the stalling of DNA replication[54]. DDT can be achieved via two mechanistically distinct pathways: Translesion synthesis (TLS) and template switching (TS). TLS is mechanistically simple but error-prone and is known to be a major source of cellular mutagenesis. TS, on the other hand, is more complex but error-free, and is therefore believed to be the preferable process. TS involves the switching of nascent DNA strand from damaged template to undamaged sister strand for extension past the lesion. This leads to the formation of a key intermediate of TS, a joint DNA molecule called sister chromatid junction (SCJ). Cells have evolved intricate mechanisms for choosing DTT pathways. There is genetic evidence suggesting a role of Hmo1 in channeling DNA lesions to TS pathway, and this function is dependent on its CTD[55,56]. Moreover, Hmo1 acts in TS by facilitating SCJ formation possibly by binding and stabilizing sister chromatid bridges and hemicatenanes formed at stalled replication forks bearing ssDNA gaps[55]. Therefore, Hmo1 seems to direct DNA lesions toward error-free TS pathway of DTT and participates in TS by helping form SCJ[55,56]. This provides an explanation for the finding that cells lacking Hmo1 exhibit increased mutagenesis frequency[57].
ROLE OF HMO1 IN MAINTAINING THE TOPOLOGICAL ARCHITECTURE OF GENES
A recent comprehensive survey of DNA supercoiling across RNAPII transcribed genes in yeast revealed that DNA in the coding region (gene body) is generally positively supercoiled (overwound), whereas DNA at gene boundaries is generally negatively supercoiled (underwound)[58]. Notably, this arrangement does not depend on transcription by RNAPII or the cell cycle. The shared topological architecture of genes is mediated by DNA topoisomerases I and II (Top1 and Top2) and Hmo1. Specifically, Top1 maintains positive supercoiling within the gene body, whereas Top2 and Hmo1 preserves negative supercoiling at gene boundaries that flank the gene body. Disruption of the topological architecture results in a reduction in histone abundance and, likely, nucleosome redistribution across the gene. As such, it was posited that that negative supercoiling at gene boundaries prevents supercoil diffusion and nucleosome repositioning in gene body[58]. Consistent with the roles of Top1, Top2 and Hmo1 in the topological architecture of genes, Top1 is enriched within gene body, whereas Top2 and Hmo1 are enriched in gene boundaries in S phase cells[58]. The topological architecture of genes is also maintained in G1[58], but Top2 is only recruited after G1 phase[59]. As such, Top2 is not responsible for DNA negative supercoiling at gene boundaries. Hmo1 is proposed to lock/stabilize gene boundaries independently of Top2, and modulate gene architecture with Top2, which helps to retain the memory of the topological profile of genes regardless of their transcriptional state[58].
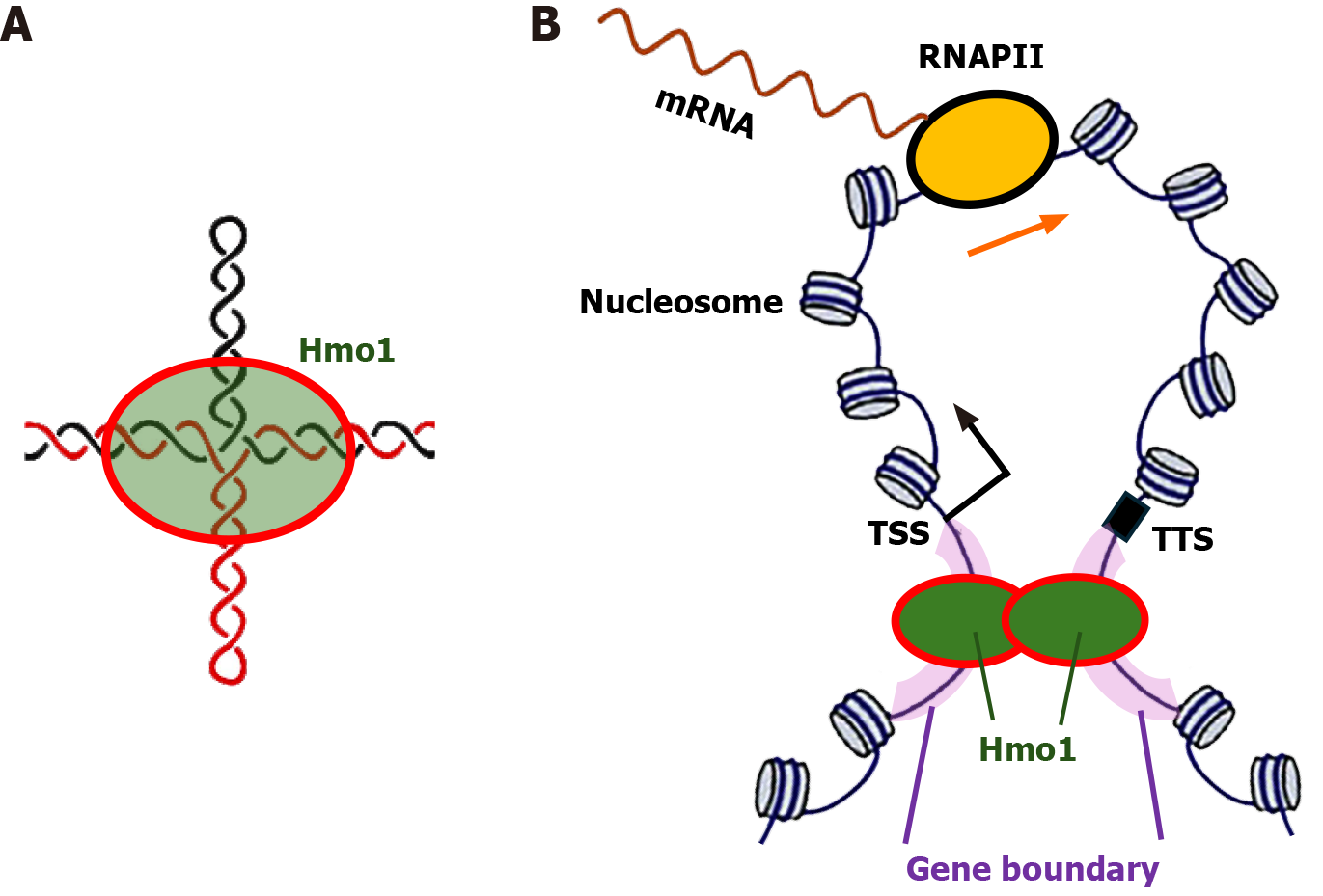
The mechanism underlying the gene boundary function of Hmo1 has not been resolved, but it is likely related to the fact that gene boundaries are negatively supercoiled and Hmo1 prefers binding DNA with altered conformations including supercoiled DNA[19,41]. Negatively supercoiled DNA can adopt pseudo-cruciform structures[60], and Hmo1, like other HMGB proteins, has high affinity for four-way junctions[41,61]. Achar et al[58] hypothesized that Hmo1 binds and stabilizes/locks cruciform structures formed at gene boundaries (Figure 3A). There has been evidence that RNAPII transcribed genes are organized in gene loops[62] and that the integrity of these loops in S phase depends on the concerted action of Top2 and Hmo1[59]. Gene boundaries may help to insulate the topological architecture of gene loops (Figure 3), thereby facilitating efficient recycling of RNAPII from transcription termination site to transcription start site. Consistent with this notion, Top2 is known to promote DNA loop formation and dimerization/oligomerization of Hmo1 also promotes DNA looping[12,63] (Figure 3).
Figure 3 Model for the gene boundary function of Hmo1.
A: Hmo1 binds cruciform structure that may form in negatively supercoiled gene boundaries; B: Diagram of a gene loop structure. The transcriptional start site, transcriptional termination site and gene boundaries are indicated. An RNA polymerase II (RNAPII) transcribing the gene is shown. Note, more than one RNAPII may transcribe the gene at the same time. Hmo1 binds gene boundaries and help maintain the negative supercoiling of DNA in them. Dimerization/oligomerization of Hmo1 promotes the formation and maintenance of gene loop. RNAPII: RNA polymerase II; TSS: Transcriptional start site; TTS: Transcriptional termination site.
ROLE OF HMO1 IN MODULATING NUCLEOSOME/CHROMATIN STRUCTURE
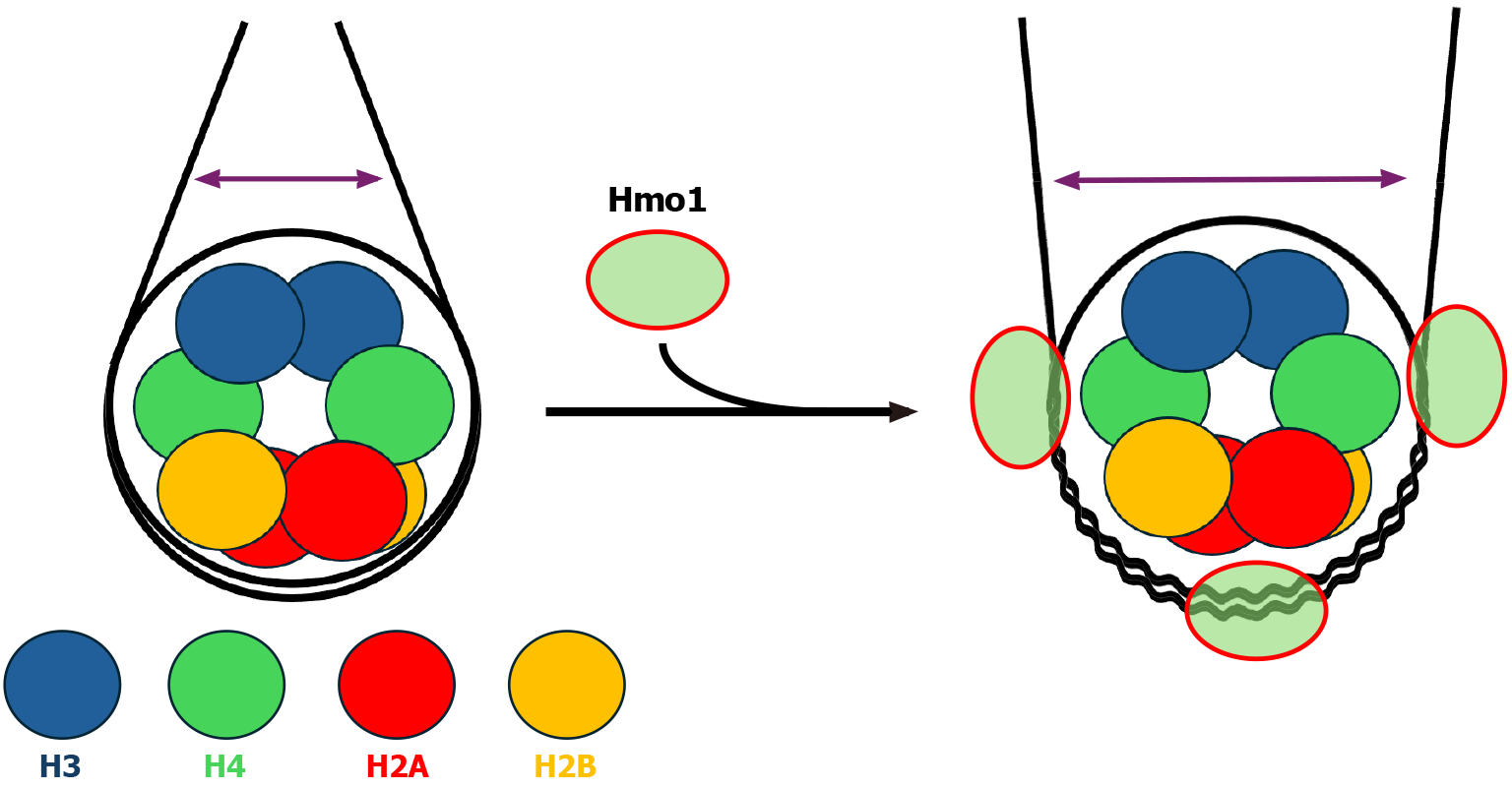
HMGB binding is generally linked to more dynamic chromatin structure that facilitates transcription. For example, association of mammalian HMGB1 to nucleosomes weakens histone-DNA interaction, rendering nucleosomal DNA more accessible[39]. The acidic C-terminal tail of HMGB1 can interact with the basic N-terminal tail of histone H3 that normally associates with linker DNA near the DNA entry/exit point of the nucleosome[64]. This is believed to help position HMGB1 on the linker DNA and enhance its binding to the nucleosome. The fact that Hmo1, unlike canonical HMGB proteins, has a basic CTD[43,45] (Figure 2) that interacts with domain A to promote DNA bending led to the hypothesis that when Hmo1 associates with a nucleosome, its CTD binds DNA instead of histone H3 N-terminal tail[3]. In this model, domain A of Hmo1 binds DNA near the nucleosome dyad axis as it has greater preference for four-way junctions, which frees CTD to interact with linker DNA, thereby preventing DNA bending and stabilizing nucleosomes[3]. This hypothesis is consistent with the finding that deletion of Hmo1, or its CTD alone, makes yeast chromatin more sensitive to nuclease digestion[3,40,65]. However, this model is not supported by results from two recent studies directly examining the impact of Hmo1 binding on nucleosome structure[66,67]. McCauley et al[66] utilized atomic force microscopy and optical tweezers to demonstrate that Hmo1 binds nucleosomes on a reconstituted nucleosomal array with a strong preference for nucleosomal core over linker DNA (Figure 4). Notably, Hmo1 binding destabilizes nucleosomes as it separates DNA from the H2A-H2B dimers and releases half of the DNA associated with the histone H3/H4 tetramer[66] (Figure 4). As such, Histone-DNA contacts throughout the nucleosome is destabilized by Hmo1[66]. Malinina et al[67] employed gel mobility shift assay and single particle Förster resonance energy transfer microscopy to show that Hmo1 binding to nucleosome induces a significant lengthening of both the distance between the gyres of core nucleosomal DNA and the distance between linker DNA segments (Figure 4). The latter is opposite to the enhanced proximity of linkers induced by linker histone H1[67]. Therefore, despite having a basic CTD similarly as linker histone H1 (Figure 2), Hmo1 destabilizes nucleosomes like a canonical HMGB protein (Figure 4).
Figure 4 Impact of Hmo1 on nucleosome structure.
Left: Diagram of a nucleosome. Core histones H2A, H2B, H3 and H4 are shown as color-coded circles. The distance between linker DNA segments is represented by a double headed arrow. Right: Diagram of a Hmo1-bound nucleosome based on results from references[66,67]. Wiggly lines represent DNA strands that are separated from, or interact weakly with, core histones.
Consistent with their ability to destabilize nucleosomes, HMGB proteins generally facilitate the recruitment of gene regulators and aid in nucleosome/chromatin remodeling by chromatin remodelers[39]. For instance, HMGB1 facilitates the binding of chromatin remodeling factors ATP-utilizing chromatin assembly and remodeling factor and chromatin accessibility complex to chromatin and their nucleosome sliding activities[68]. Nucleosome sliding promoted by an HMGB protein would make regulatory sequences available for TFs to bind[69]. Hmo1 helps chromatin remodeler SWItch/Sucrose Non-Fermentable (SWI/SNF) bind the nucleosome and stimulate the transient exposure of nucleosomal DNA and octamer transfer catalyzed by SWI/SNF[70]. It also stimulates the nucleosome sliding activity of SWI/SNF, a function shared by another yeast HMGB protein, Nhp6. Interestingly, the stimulatory effect of Hmo1 on the chromatin remodeling activity of SWI/SNF requires its CTD and the presence of linker DNA[70]. Stimulation of the nucleosome sliding activity of a chromatin remodeler appears to be a shared feature of HMGB proteins. Moreover, SWI/SNF often colocalizes with Hmo1 to gene promoters in yeast, and Hmo1 deletion reduces the level of SWI/SNF bound to the promoters[71]. As such, Hmo1 can both stimulate SWI/SNF activity and assist its recruitment to gene regulatory regions. Consistently, Hmo1 has been shown to physically interact with SWI/SNF[72]. Hmo1 also interacts with chromatin remodeling complexes remodeling the structure of chromatin and imitation Switch 1 a and stimulates their activities[72]. Hmo1 also supports large-scale reversible nucleosome unfolding by the histone chaperone facilitates chromatin transcription (FACT) complex in vitro, and functionally interacts with FACT in vivo[67].
LINKER HISTONE H1-LIKE FEATURES OF HMO1
Hmo1 is not a typical HMGB protein based on its domain structure as it ends with a lysine rich basic CTD instead of an acidic one (Figure 2). Since a basic CTD is a hallmark of canonical linker histone H1 (Figure 2), it has been postulated that Hmo1, as a chromatin architectural protein, may be a functional homolog of histone H1[3]. This is an attractive notion especially because the linker histone ortholog in yeast, Hho1, has a unique structure with an extra globular DNA/nucleosome-binding domain instead of an unstructured, basic, C-terminal tail (or CTD) (Figure 2), and is found not to function as a linker histone in many prior studies[3]. On the other hand, Hmo1 has been found to exhibit linker histone like characteristics[3,65]. Lack of Hmo1 makes yeast chromatin hypersensitive to micrococcal nuclease digestion[40,65]. This indicates that Hmo1, like linker histone H1, promotes chromatin compaction, thereby protecting linker DNA from nuclease digestion[40,65]. Moreover, ectopic expression of human linker histone H1 reverses the nuclease sensitivity of chromatin in cells lacking Hmo1[65]. Like CTD of histone H1, the CTD of Hmo1 is required for its chromatin compaction function[40,65]. Moreover, chromatin remodeling during DSB repair occurs faster in Hmo1 null cells or cells with Hmo1 truncated for its CTD, which is suppressed by ectopic expression of human histone H1[65]. It is believed that association with Hmo1 creates a less dynamic chromatin environment that inhibits DSB repair.
In a recent single-molecule study, Hmo1, but not Hho1, was shown to promote chromatin assembly in yeast nucleoplasmic extract, suggesting that it shares this function with linker histone H1[73]. In addition, Hmo1, but not Hho1, forms condensates with double-stranded DNA (dsDNA) via reversible phase separation, similarly as linker histone H1[73]. Hmo1-CTD is essential for the ability of Hmo1 to facilitate chromatin assembly and form condensate with dsDNA[73]. In short, Hmo1 possesses some functionality similar to that of a linker histone H1, which depends on its basic CTD. However, this functionality is not required for normal cell growth as deletion of CTD of Hmo1 does not lead to the slow growth phenotype of yeast lacking Hmo1[40].
CONCLUSION
Since its first identification as a HMGB protein nearly three decades ago, the yeast Hmo1 protein has been found to exhibit many characteristics of well-characterized HMGB proteins including destabilizing nucleosome structure, facilitating chromatin remodeling, and promoting transcription by RNAPI and RNAPII. Consistently, Hmo1 associated yeast genomic sequences are mostly devoid of nucleosomes. These sequences include active rDNA repeats and the promoters of a group of RNAPII genes. On the other hand, there is also evidence suggesting that, like linker histone H1, Hmo1 facilitates the formation of compact chromatin structure that resists nuclease attack and hinders DSB repair processes. This raises the question of how, unlike linker histone H1, Hmo1 compacts chromatin without binding nucleosomes in the genome. The answer to this question may lie in the recently identified role of Hmo1 in maintaining the boundaries of RNAPII genes and safeguarding the topological architecture of these genes[58]. Disrupting gene boundaries by Hmo1 deletion was found to cause a decrease in histone abundance (and possible redistribution of nucleosomes) in the gene body[58]. It is possible that this makes the gene body more sensitive to nuclease digestion. As RNAPII genes collectively represent the majority (> 70%) of the yeast genome sequence[74], their increased sensitivity to nuclease digestion would make the genome as a whole appear to be more susceptible to nuclease attack. The hypothesis that Hmo1 protects coding sequences of RNAPII genes from nuclease attack via preserving gene boundaries can be tested by examining the effect of Hmo1 deletion on genome wide chromatin accessibility. The model that Hmo1 mediates gene loop (or chromatin loop containing more than one gene) formation[58] (Figure 3) is in line with the recent finding that HMBG1 and HMBG2 associate with the boundaries of a subset of topologically associating domains in human cells[75,76]. This model can be tested by investigating the impact of Hmo1 deletion on chromatin interaction map in yeast.
ACKNOWLEDGEMENTS
I thank the Department of Biology at University of Rochester for support and Ray Sze, Meeta Dahake and Jiwoo Han for reading the manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country of origin: United States
Peer-review report’s classification
Scientific Quality: Grade B
Novelty: Grade B
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Sun XG S-Editor: Fan M L-Editor: A P-Editor: Zheng XM