Published online Mar 27, 2023. doi: 10.4331/wjbc.v14.i2.13
Peer-review started: November 21, 2022
First decision: December 13, 2022
Revised: December 20, 2022
Accepted: February 13, 2023
Article in press: February 13, 2023
Published online: March 27, 2023
Processing time: 122 Days and 17.2 Hours
Early-onset colorectal cancer (EOCRC) has been rising in global prevalence and incidence over the past several decades. Environmental influences, including generational lifestyle changes and rising obesity, contribute to these increased rates. While the rise in EOCRC is best documented in western countries, it is seen throughout the world, although EOCRC may have distinct genetic mutations in patients of different ethnic backgrounds. Pathological and molecular characterizations show that EOCRC has a distinct presentation compared with later-onset colorectal cancer (LOCRC). Recent studies have identified DNA, RNA, and protein-level alterations unique to EOCRC, revealing much-needed biomarkers and potential novel therapeutic targets. Many molecular EOCRC studies have been performed with Caucasian and Asian EOCRC cohorts, however, studies of other ethnic backgrounds are limited. In addition, certain molecular characterizations that have been conducted for LOCRC have not yet been repeated in EOCRC, including high-throughput analyses of histone modifications, mRNA splicing, and proteomics on large cohorts. We propose that the complex rela
Core Tip: Early-onset colorectal cancer (EOCRC) has a considerably different clinical presentation and genetic profile compared with later-onset colorectal cancer. Furthermore, molecular alterations in EOCRC tumors differ in patients from separate geographical locations and distinct ethnic groups. Small human cohorts and the lack of a suitable mouse model system limit EOCRC studies, however, several actionable clinical targets and biomarkers specific to EOCRC have been identified. In this review, we discuss molecular alterations in EOCRC tumors at the DNA, RNA, and protein levels, and suggest future work to examine how these changes contribute to EOCRC pathogenesis.
- Citation: Marx O, Mankarious M, Yochum G. Molecular genetics of early-onset colorectal cancer. World J Biol Chem 2023; 14(2): 13-27
- URL: https://www.wjgnet.com/1949-8454/full/v14/i2/13.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v14.i2.13
Cancers of the colon and rectum are the third most commonly found in both men and women globally[1]. Colon cancer screenings have increased early detection in patients over the age of 50-year-old and have contributed to the overall decline in global rates of colorectal cancer (CRC)[1]. However, the population of early-onset CRC (EOCRC) patients, under 50-year-old, has been steadily rising over the past several decades[2,3], and by 2030, the rates of early-onset colon and rectal cancers are expected to increase by 27.7%, and 46.0%, respectively[4]. Unfortunately, 10%-15% of CRC patients are diagnosed before the age of average-risk screening recommendation (before 2018, 50-year-old)[5]. Due to a lack of screenings and a delay in the diagnosis of younger patients, EOCRC is often detected at advanced stages, reducing the chances of long-term survival[6]. Many studies have shown that EOCRC is molecularly distinct from later-onset CRC (LOCRC), or CRC diagnosed after the age of 50-year-old. Compared with LOCRC, EOCRC has a differing frequency of oncogenic mutations[7], increased prevalence of mucinous and signet (poorly differentiated) histology[8], a more distal location[2], and exhibits a distinct DNA methylation profile[9]. Despite aggressive treatment of EOCRC patients, their overall survival is worse compared to those with LOCRC[5,10].
There is no clear cause for most EOCRC cases, although environmental risk factors are likely key contributors to cancer development. Lifestyle factors such as smoking, unhealthy diet, obesity, and alcohol consumption increase the risk of developing CRC early on[1]. In the United States, EOCRC has a strong birth cohort effect, implicating generational lifestyle changes in the development of EOCRC[11].
Several recent studies have demonstrated the association between obesity and metabolic disorders with the development of EOCRC[12,13]. Tang et al[14] found that EOCRC patients had a worse metabolic profile, with higher levels of triglycerides and lower levels of high-density lipoprotein cholesterol compared with LOCRC patients[14]. Molecular links between obesity, metabolic disorders, and CRC have been suggested, including the promotion of intestinal stem cell populations[15,16], increased insulin resistance, adipocyte levels, and inflammation[17]. How EOCRC risk factors affect clinical presentation is still under investigation. One aspect of EOCRC clinical presentation of particular interest is tumor location[18].
Over half of pre-malignant polyps in EOCRC are found in the distal colon and rectum[18], and this has prompted calls for screening sigmoidoscopy at an earlier age than current guidelines, which were recently changed from 50 to 45[19]. While left-sided colon cancer is more predominant in EOCRC, right-sided EOCRC is associated with lower overall survival compared to left-sided EOCRC (44% vs 61%)[20]. Several factors have been implicated in the difference in survival between right-sided and left-sided CRC. During embryonic development, the proximal colon originates from the midgut while the distal colon originates from the hindgut. This developmental difference may impact cancer cell origins as well as the metastatic potential of tumors due to differences in vascularization. Additionally, several microbiota changes have been characterized between the proximal and distal colon which may play a role in oncogenesis[21,22]. Proximal colonic tumors also have distinct histopathological features as they tend to be more mucinous with microsatellite instability and mismatch repair (MMR) deficiency compared to distal tumors[23,24]. These distinctions may indicate unique molecular drivers of distal and proximal EOCRC tumors.
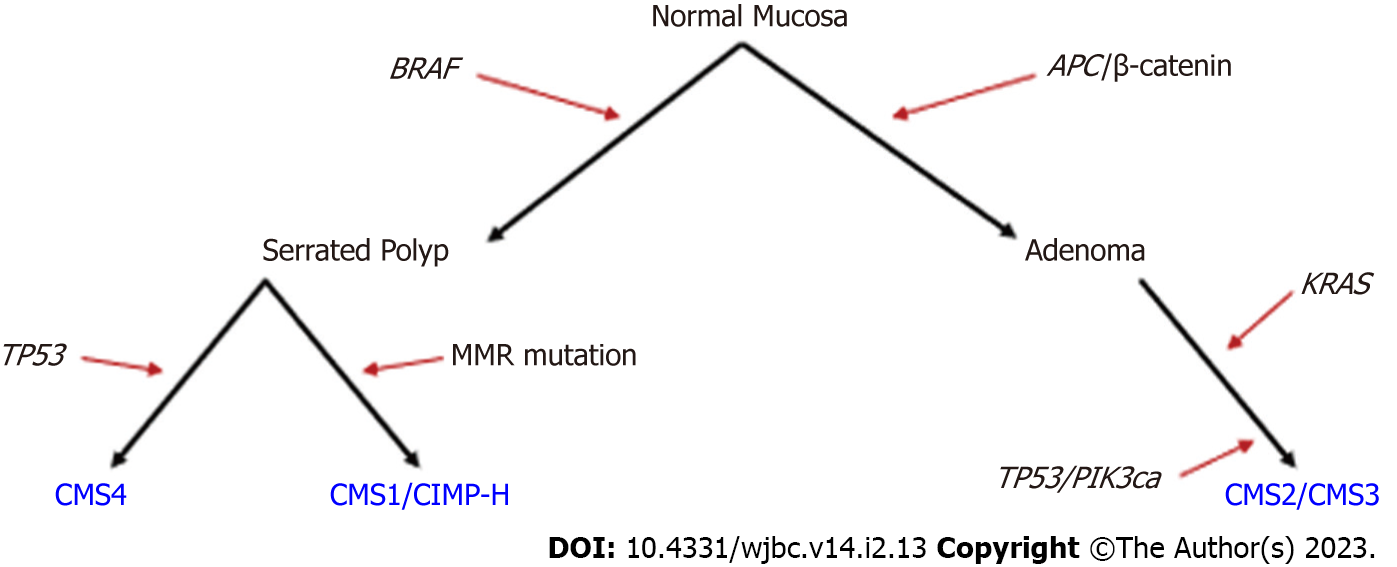
The multi-step progression from normal colonic mucosa to adenoma to CRC was first described in 1990 by Fearon and Vogelstein[25]. In this model, an APC-inactivating mutation is an initiating event, followed by KRAS-activating mutations driving adenoma development[25]. Further studies found that the malignant transformation of adenomas was driven by additional mutations in the tumor growth factor beta, PIK3CA, and TP53 pathways (Figure 1)[26-28]. Sessile serrated polyps act as a precursor to up to one-third of CRCs and are thought to arise through mechanisms distinct from canonical APC mutation-driven polyps[29-31]. Instead, serrated polyps are thought to develop from a BRAF mutation and are also often characterized by DNA hypermethylation[29]. Mutations to BRAF, KRAS, and TP53 are also found in other cancers and the multi-step progression to adenocarcinoma may not strictly follow the canonical or serrated pathways described. While most sporadic LOCRCs can be categorized by deregulation of canonical Wnt/β-catenin/APC signaling or serrated BRAF mutation pathways[30] (Figure 1), it is less clear how sporadic EOCRCs develop.
CRC is often categorized into consensus molecular subtypes (CMS). At the genomic level, CRC can be categorized as microsatellite instable (MSI, often caused by a defect in MMR genes) or chromosome instable. At the transcriptomic level, there are four main CMS. CMS1 is associated with immune JAK-STAT activation, microsatellite instability, and hypermutated tumor DNA[32]. CMS2 is associated with canonical Wnt/MYC activation, CMS3 is characterized by metabolic alterations, and CMS4 is associated with epithelial-mesenchymal transition and immunosuppression[32]. CMS1 tumors are more often considered CpG island methylator phenotype-high (CIMP-high, characterized by genome-wide hypermethylation). These CMS1 tumors also often have BRAF(V600E) mutations and are associated with sessile serrated adenomas[32]. RSPO fusions and RNF43 mutations are often seen in hypermutated CMS1 CRC[32], and Yan et al[33] also identified these alterations in a subset of EOCRC tumoroids[33].
Differing frequencies of CMS in EOCRCs compared with LOCRCs have been identified. Willauer et al[34] showed that patients under 40 were more likely to exhibit CMS1 or CMS2, with CMS1 being considerably more prevalent in EOCRC compared with the average of most CRCs, though no differentiation between sporadic and hereditary mutations was made[34]. Therefore, the increased prevalence of Lynch syndrome, a hereditary MMR deficiency syndrome, in younger patients[7,35] could contribute to this observation. In fact, MSI/CIMP-high tumors were associated with Lynch syndrome in young patients, whereas in older patients, they were associated with BRAF mutations[36]. Sporadic EOCRC patients are less likely to have Lynch syndrome[37] and are less likely to have tumors with a CpG island methylator phenotype[38], which are associated with CMS1[38]. Limitations to sample size necessitate future studies to examine the CMS of sporadic EOCRCs compared with LOCRCs.
Many excellent reviews explain the molecular subtypes and mutations in CRC[30,32,39]. Likewise, many excellent reviews have focused on the different clinical presentations and outcomes of EOCRC and LOCRC patients, with some mention of prevalent molecular distinctions of EOCRC[2,4,40]. However, reviews that focus primarily on the molecular characterizations of EOCRCs are limited. There has been a rise in EOCRC literature over the past 5 years that we aimed to summarize here. In this review, we searched the literature for early- and young-onset CRC, along with specific searches for molecular genetics, DNA methylation, histone alterations, transcriptomics, splicing, proteomics, ethnic disparities, and biomarkers. Relevant papers were selected for discussion. Here, we summarize key findings of the molecular genetic underpinnings of EOCRC thus far.
Hereditary mutations often lead to CRC at a younger age in comparison with sporadic mutations. Compared with LOCRC, EOCRC patients have an increased polygenic risk score based on profiling single nucleotide polymorphisms[41]. Although approximately 30% of EOCRC cases report a family history of related cancers, only an estimated 10%-20% have known genetic risk factors like familial adenomatous polyposis, Lynch syndrome, or inflammatory bowel disease[7,10,42,43]. Therefore, our understanding of the genetic and molecular pathways that drive carcinogenesis in most patients is far from complete.
Sporadic EOCRCs, patients with no family history, have been shown to have different mutational profiles compared with LOCRCs (Table 1). Notably, many studies have found a significant decrease in the prevalence of APC and Wnt pathway mutations in EOCRC compared with LOCRC[33,43-45], with the exception of the β-catenin gene, CTNNB1, which is mutated in more EOCRCs compared with LOCRCs (Figure 2)[33,34]. Interestingly, a recent study by Yan et al[33] used organoids to demonstrate the heterogeneity between EOCRC patients, identifying some with APC mutations and others with RSPO fusions, which render the cultures hypersensitive to Wnt withdrawal[33].
| Gene | Prevalence in EOCRC vs LOCRC1 | Role in cancer |
| APC | Decreased[33,34,44,45] | Blocks β-catenin, tumor suppressor |
| CTNNB1 | Increased[33,34] | β-catenin, potentiates Wnt signaling, proliferation, and stemness |
| RNF43 | Increased[43]/NS[33] | E3 ligase, negative regulator of Wnt signaling |
| BRCA2 | Increased[54] | Double stranded DNA repair, tumor suppressor |
| PHLPP1 | Increased[54] | Promotes apoptosis, inhibits AKT |
| TOPORS | Increased[54] | Regulates TP53 stability, likely tumor suppressor |
| ATR | Increased[54] | PI3/PI4 kinase, activates checkpoint proteins |
| MYCBP2 | Increased[54] | MYC binding protein, activates MYC |
| FBXW7 | Increased[44,54] | Ubiquitin ligase component, ubiquitinates MYC |
| POLE | Increased[44,54] | DNA polymerase E subunit, proofreading and DNA repair |
| BRAF | Decreased[34,38,57]/increased[43] | Proto-oncogene, activates MAPK signaling |
| TP53 | Decreased[38,52] | Cell cycle inhibitor, tumor suppressor |
| NOMO1 | Increased[53,126] | Inhibits nodal signaling. Deletion increases CRC cell migration |
| MYC | Increased[50,51]/NS[43,47] | Proto-oncogenic transcription factor, promotes proliferation and stemness |
| DNMT3B | Decreased[43] | De-novo DNA methyltransferase |
| MET | Decreased[43] | Proto-oncogene, promotes cell growth and survival |
| PTEN | Increased[57] | Tumor suppressor, negatively regulates AKT signaling |
| KRAS | Decreased[99,120] | Proto-oncogene, activates oncogenic signaling pathways |
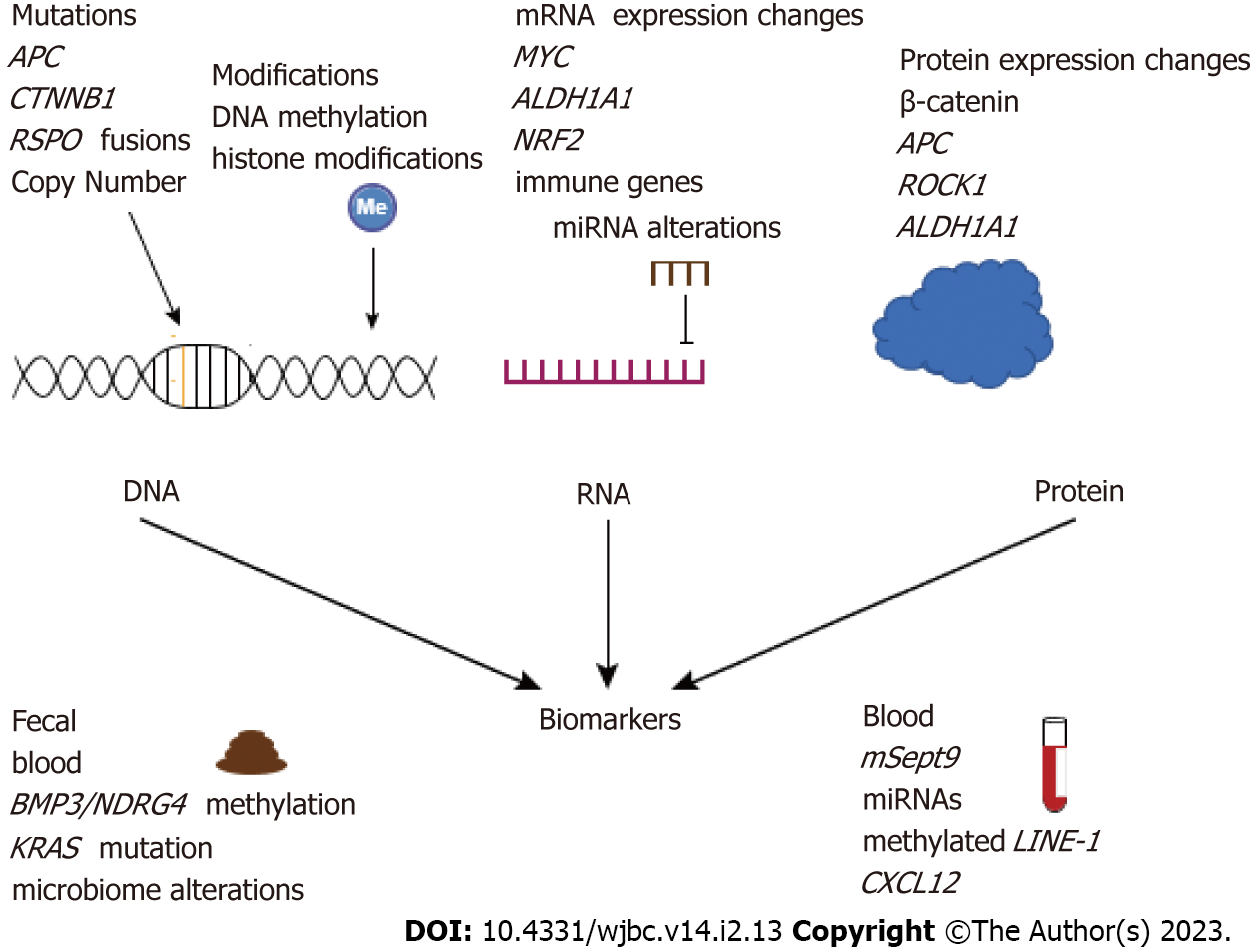
MYC is a key oncogenic target of the Wnt/β-catenin pathway that is often deregulated in CRC[46]. Copy number variations of MYC are seen in 8%-15% of CRCs[47-49], however, we recently reported that 35% of EOCRC tumors from a 21-patient cohort had an increase in MYC copy number[50]. In addition, Pan et al[51] reported increased MYC copy number in younger CRC patients[51], however, another study found no association between MYC copy number and age[47]. Overall, chromosomal deletions and copy number variations have been shown to be more common in EOCRC tumors compared with LOCRC tumors[52,53]. Alterations to MYC regulatory genes have also been identified in EOCRC, including MYCBP2[54], an E3 ubiquitin ligase that may regulate MYC transcription[55], and FBXW7[44,54], a tumor suppressive ubiquitin ligase that mediates degradation of MYC, among other oncoproteins, though more work is needed to determine the functional effect of these mutations[56]. Together, these findings provide evidence for a Wnt-independent increase in MYC activity in EOCRC.
In addition to differences in the prevalence of Wnt pathway/MYC mutations, many studies reported a decrease in BRAF (V600E) mutations (Table 1)[34,38,57], although Xu et al[43] noted an increase in BRAF (V600E) in EOCRC compared with LOCRC tumors[43]. This finding has clinical implications, as BRAF (V600E) mutant tumors are associated with CIMP-high status, have a worse prognosis, and respond differently to cancer treatments[58]. The decrease in APC and BRAF mutations in EOCRC indicates that a higher percentage of EOCRC tumors may not follow the canonical or serrated carcinogenesis pathways commonly observed in LOCRC (Figure 1)[34]. Overall, many key oncogenes and tumor suppressors are differentially mutated in EOCRC compared with LOCRC, which may impact cancer progression and prognosis (Table 1, Figure 1). As new mutations unique to EOCRC are uncovered, more work is needed to determine how such mutations affect gene activity.
In addition to DNA mutations, studies have suggested that EOCRC has a distinct DNA methylation profile from LOCRC[9,59]. DNA methylation regulates gene expression and has been implicated in CRC[60]. Methylation of the long non-coding RNA LINE-1 is often thought to represent global DNA methylation[61]. Studies suggest that DNA is overall hypomethylated in EOCRC compared with intermediate or LOCRC[42,59]. Epigenetic modifications may also be detectable in the blood, and a study by Walters et al[62] found hypermethylation of DNA repetitive elements, including LINE-1, in white blood cells from EOCRC patients[62]. While previous studies examined global DNA methylation, a recent high-throughput study by Joo et al[9] identified 234 differentially methylated regions unique to EOCRC tumors[9]. The authors then compared EOCRC DNA methylation patterns to those which occur upon age-related methylomic drift in the normal mucosa. They suggest that EOCRC tumors more rapidly accumulate cancer-related methylomic drift compared to intermediate or LOCRC tumors, though it remains unclear when this drift occurs during cancer progression[9]. More work is needed to assess DNA methylation over time and within patient-matched tumors and normal mucosa to better understand how age-related DNA methylation changes contribute to EOCRC.
In addition to DNA methylation, histone methylation and acetylation are associated with both aging and CRC[60], however, there have been few studies on histone modifications in EOCRC. A study in 2015 found that high levels of H3K27me3 were associated with a better prognosis in younger CRC patients and a worse prognosis in older CRC patients[63]. DNA and histone modifications are a natural part of aging, however, how they impact gene expression, cancer progression, and drug response remains to be elucidated.
Transcriptome analysis is a comprehensive tool to identify deregulated signaling pathways in cancer[64,65]. When applied to human tissues, this approach considers both genetic and environmental factors that contribute to the profile of deregulated gene expression on a per-patient basis. Several studies have analyzed the transcriptomic profile of EOCRC; however, many studies are limited by sample size and availability of patient-matched control samples.
Deregulation of mRNA targets of the Wnt/β-catenin pathway has been demonstrated in EOCRC, though at a lower frequency compared with LOCRC[33]. We have recently published transcriptome analyses implicating deregulated MYC, and its downstream targets, in EOCRC[50]. The proto-oncogene MYC is upregulated in the intestines of obese individuals[66] and has been suggested to control obesity-mediated metabolic dysfunction in the intestines[67], though Ellegaard et al[68] found no relationship between MYC expression and body mass index[68]. Interestingly, our recent transcriptomic study found increased MYC expression in the EOCRC tumors of a subset of patients who were obese, suggesting a distinct tumor gene expression profile in obese and non-obese patients[50]. We did not find significant deregulation of the Wnt/β-catenin hallmarks of cancer in our EOCRC tumors compared with adjacent normal tissue. The age-associated role of MYC in CRC is supported by another recent study that implicated overexpression of MYC, along with the lncRNA WiNTRLINC1 and the gene ASCL2, in younger colon cancer patients[69].
In addition to MYC, studies comparing EOCRC and LOCRC have found enrichment of cell signaling, apoptosis/inflammation, proliferation, adhesion, and development[38,70]. The recent success in cancer immunotherapies has prompted an interest in examining the immune profiles of CRC[71], however, few studies have interrogated the immune response in EOCRC. A recent study highlights the importance of aging and tumor immune response, showing that aging-related gene ontology sets were enriched in CRC tissues compared with normal tissues and this signature was higher in tumors with high immune infiltration[72]. Profiling approximately 40 tumors from both late- and early-onset CRC patients, Gardner et al[73] found that three immune genes SAA1, C7, and CFD, have deregulated expression in EOCRC primary tumors compared with LOCRC tumors[73]. Changes in the expression of these genes were shown to alter the tumor immune microenvironment and are associated with intestinal inflammation[73]. Another study identified age-associated changes in tumors compared with normal tissues and found enrichment of the nuclear factor erythroid 2-like 2 oxidative stress response in the tumors of younger patients compared with older patients (Figure 2)[74]. The tumor immune microenvironment is a complex system that involves many different cell types. Immune studies in EOCRC are limited by using a homogenized tumor population for bulk RNA sequencing instead of examining alterations at a single cell level.
Most transcriptomic studies of EOCRC have focused on mRNA, however, there is increasing evidence for the relevance of microRNAs (miRNAs) in cancer. miRNAs are short RNA transcripts that generally function to bind and repress a specific target mRNA. Two notable miRNA studies have been performed for EOCRC, the first by Nakamura et al[75] examined miRNAs from tumors and normal samples and found a seven-miRNA panel that was upregulated in EOCRC (n = 42), but not LOCRC (n = 370), in tumor vs normal tissues (Table 2)[75]. An earlier study using microarray analyses of a Turkish EOCRC cohort identified downregulation of miR-143, miR-125b, and upregulation of miR-106a in tumors vs normal tissues, although no comparison with LOCRC was performed[76]. While these miRNAs have been suggested as biomarkers for EOCRC, limited sample sizes, lack of patient-matched controls, and a lack of functional studies leave their roles in cancer progression unclear.
| Gene(s) | Description |
| MYC | Proto-oncogenic transcription factor, increased in EOCRC tumors vs normal samples[50,69] |
| ASCL2 | Transcription factor that promotes intestinal stem cells, increased expression in younger CRC[69] |
| ALDH1A1 | Protein involved in cancer cell stemness, expressed higher in EOCRC tumors[88] |
| PEG10 | Promotes proliferation and invasion, increased in EOCRC tumor vs normal and EOCRC vs LOCRC[70] |
| miR-143, miR-125b | miRNAs, under-expressed in EOCRC tumor vs normal[76] |
| miR-106a | miRNA, overexpressed in EOCRC tumor vs normal[76] |
| hsa-miR-4304, hsa-miR-513a-5p, hsa-miR-628-3p, hsa-miR-194-3p, hsa-miR-193a-5p, hsa-miR-210, and hsa-miR-4453 | miRNAs uniquely overexpressed in EOCRC compared with LOCRC and normal tissues[75] |
| SAA1, C7, CFD | Immune genes differentially expressed in EOCRC vs LOCRC tumors[73] |
| NRF2 | Protein involved in oxidative stress and inflammation, expressed higher in EOCRC vs LOCRC[74] |
As with most EOCRC studies, transcriptomic analyses of EOCRC are limited by sample size and availability of quality sequencing data from tumors and patient-matched normal control samples. In addition to changes in transcript abundance, RNA sequencing can provide information about alternative polyadenylation and splicing events, which can alter protein structure and function. Alternative polyadenylation is associated with cellular proliferation and cancer[77]. It serves to alter the 3’UTR length, which can affect miRNA regulation in many cancers including CRC[78]. Unfortunately, to our knowledge, there have been no studies on alternative splicing or polyadenylation events in EOCRC. However, one study did find a POLE mutation that may be associated with aberrant splicing in EOCRC[79]. As aberrant alternative splicing has been implicated in both CRC[80] and aging[81], we propose that examining EOCRC-specific splicing events would uncover novel insight into disease pathogenesis. Additional post-transcriptional modifications to mRNA, lncRNA, tRNA, and rRNA, such as methy
In addition to post-translational modifications and miRNA analysis, another type of transcriptomic analysis that is gaining popularity is single-cell RNA-sequencing (scRNA-seq), which can be used to identify gene regulation in the different cell types involved in cancer. Single-cell transcriptomics has been used to analyze the age-associated transcriptome in cancers including CRC[84]. Saul and Kosinsky[84] found that many aging- and senescence-associated genes were generally upregulated in cancers, including CRC[84]. These authors also found that CRC displayed distinct populations of epithelial cells with elevated age-related gene expression, underscoring the importance of examining age-related differences in CRC at a single-cell level[84]. Understanding immune infiltration and stem cell populations is crucial for developing cancer treatments that reduce the risk of relapse. Yan et al[33] performed scRNA-seq of EOCRC organoids and showed differing stem cell populations in response to Wnt media supplementation for six different EOCRC and LOCRC tumoroids with different underlying mutations[33]. Expanding scRNA-seq of EOCRCs would further elucidate information about disease progression that is specific to distinct cell populations in younger patients.
Proteomic studies have advanced cancer treatments by identifying therapeutic targets[85]. While the proteomic signature of CRC has been established[85,86], few studies have profiled the proteome of EOCRC[74,87]. Gong et al[87] recently published a paper using mass spectrometry to identify age-associated differential expression of proteins in tumors compared to adjacent normal tissues. The authors found an age-associated proteomic signature in CRC tumors, which included MYC, E2F, and mTORC1 targets, and proteins controlling the G2M checkpoint, DNA repair, and unfolded protein response (UPR) pathways expressed at higher levels in older CRC patients. Overall, 208 proteins were found to positively correlate with age, and only 20 negatively correlated with age. Many of these proteins reside in pathways that are targetable with known cancer drugs, supporting the potential use of specific cancer treatments for different ages of CRC patients[87]. For example, the proteins PIN1, ROCK1, and ANXA5 are expressed higher in EOCRC and are targetable by Food and Drug Administration (FDA)-approved drugs or drugs in clinical trials[87]. While this study demonstrated the difference in proteomic signatures in younger and older CRC tumors, it was limited by the sample size of approximately 50 total patients with young, intermediate, or older onset CRC[87].
Another recent study by Holowatyj et al[74] found no significant differences (FDR q-value < 0.05) in the plasma proteome of younger-onset (n = 11) compared with older-onset (n = 45) CRCs using an antibody microarray platform to detect 206 inflammatory proteins. An increased sample size may shed light on interesting targets, as the authors found that the cancer-related proteins BRCA2, PTEN, WNT5B, and WNT7A, among others, had a fold change around two (P < 0.05) in EOCRC vs LOCRC serum[74]. While to our knowledge, no other proteome-wide studies have assessed EOCRC, some studies have identified individual proteins that are uniquely expressed in EOCRC tumors. For example, overexpression of the ALDH1/ALDH1A1 protein has been identified in most EOCRC tumors compared with LOCRCs (Figure 2)[88], and an increase of β-catenin in the nucleus and cytoplasm of EOCRC compared with more membrane staining in LOCRC was shown via immunostaining[38].
Protein modifications such as glycosylation[89], ubiquitination[90], phosphorylation, and acetylation[91] are associated with CRC, but age-related characterizations remain limited. A recent study found that an increase in glycosylated hemoglobin in the serum of younger non-diabetic adults correlated with an increased risk for CRC[92], though no studies could be found that focused on post-translational modifications within EOCRC tumors. In addition to changes in protein modifications and expression changes, disruptions to protein folding are common in cancers, eliciting the UPR, which promotes cancer cell survival[93]. Indeed, our previous work showed enrichment of the UPR gene set in EOCRC tumors compared with adjacent control samples[50].
While many studies focus on profiling EOCRC in western countries, the incidence of EOCRC is also increasing in many Asian countries or regions such as Korea, Thailand, Japan, India, and Hong Kong[94,95]. While India reports one of the lowest rates of CRC incidence in the world[94], over half of the sporadic rectal cancers in this country are diagnosed in patients under 50-year-old[96-98]. In addition, studies from Indian cohorts found that under half of early-onset sporadic rectal cancer (EOSRC) tumors exhibit a Wnt signature, the most common driver of CRC, indicating distinct tumor drivers in this population[97,99]. Tumors without Wnt signaling showed increased activation of calcium/nuclear factor of activated T-cell signaling compared to EOSRC with high Wnt signaling[97]. Molecular studies in Indian EOCRC patients have also found a decrease in KRAS mutations[99] and deregulation of MAPK and PI3K/AKT pathways[100] compared with LOCRC patients. Whether EOSRC in Indian patients is molecularly distinct from Western or Caucasian patients, from whom most available CRC data were collected, remains unclear.
A recent study by Xu et al[43] compared germline mutations in a Western Caucasian EOCRC cohort to a Chinese EOCRC cohort and found the Chinese cohort had significantly fewer hereditary syndromes, with no germline APC mutations (mutated in 13% of the western cohort) or BRCA1, SMAD4, or CHEK2 mutations, while these genes were mutated in 16% of western cohort patients under 50-year-old (330 Chinese and 430 Caucasian)[43]. Another recent molecular study in China examined clinical information for 947 EOCRC and 3521 LOCRC and found that EOCRCs were more likely to have a family history of cancer, higher TNM stage, and higher 3-year overall survival, but also a lower 3-year disease-free survival[101]. EOCRCs were also more likely to have defective MMR[101], though it is unclear whether this is a product of Lynch syndrome or sporadic mutations.
While data on CRC age-of-onset are available from many European and Asian countries, limited information on CRC epidemiology is available from countries in Africa[94]. New data identified an increased prevalence of CRC, with most African countries where data is available reporting an average age of CRC diagnosis between 43-year-old and 46-year-old[102]. One study compared EOCRC in Nigerians and African Americans (AA) and found that over 60% of Nigerian CRC patients were diagnosed before the age of 50-year-old, compared with 13.2% of AA[103]. The authors identified many differences between the two populations, where Nigerian EOCRCs were younger and had more rectal cancers[103]. Unfortunately, the demographic patterns of EOCRC in black individuals remain severely understudied[103].
In the United States, a clear racial disparity in CRC diagnosis and treatment exists, where the median age of CRC diagnosis is 68 for whites and 64 for blacks[1]. A study by Galadima et al[104] found that young AA had higher CRC incidence compared with young individuals of other races[104]. In addition, EOCRC rates in the United States are highest among Indigenous and black Americans[105]. Unfortunately, non-Hispanic blacks with EOCRC have a significantly worse 5-year survival than their white counterparts[106,107]. Previous studies have demonstrated ethnicity-specific differences in underlying CRC mutations[108], but few have focused on EOCRC, especially in people of African descent. One study did find a decrease in the prevalence of APC mutations and an increase in gene methylation in an AA EOCRC cohort compared to the mostly white CRC dataset provided by The Cancer Genome Atlas[109].
Overall, the incidence of CRC is increasing in young patients on a global scale, likely due to dietary and lifestyle changes across the world[94]. EOCRC may present differently and have different mutations in different populations around the world, likely owing to both lifestyle and genetic differences[43,97,103,108]. Therefore, it is crucial to increase our understanding of unique EOCRC drivers and to translate this knowledge to improve clinical outcomes for patients worldwide.
The majority of EOCRCs are diagnosed between the ages of 40-49[110], leading to the American Cancer Society lowering the recommended CRC screening age from 50 to 45 in 2018[19]. However, several concerns remain on the cost/benefit analysis of this decision[111], indicating a crucial need for cost-effective early screening options.
While colonoscopy remains the gold standard of CRC screenings, blood and fecal tests are cheaper and less invasive options. Blood-based miRNA and DNA methylation biomarkers have been shown to accurately identify EOCRC[112]. There is currently one FDA-approved blood-based CRC screening test, Epi proColon®, which measures methylation of the gene SEPT9 in cell-free DNA in serum. A recent study showed that methylation of SEPT9 could accurately distinguish EOCRC patients from healthy controls, indicating that this test is effective for younger patients[112]. Another study suggested that the DNA repetitive elements LINE-1, Sat2, and Alu are hypermethylated in the white blood cells isolated from EOCRC patients, providing an additional potential methylation biomarker signature[62]. In addition to DNA methylation, miRNA expression is gaining popularity as a potential blood-based cancer biomarker. A recent study identified a miRNA signature of four miRNAs that could distinguish both EOCRC and LOCRC serum from healthy controls (Table 3)[75]. Serum expression of inflammatory genes has also been suggested to identify EOCRC patients, and one study found that the chemokine CXCL12 has lower expression in younger compared to older patients[74].
| Name | Type | Description |
| mSEPT9 | Methylation, DNA | Blood-based biomarker used in Epi proColon® or both EOCRC and LOCRC[112] |
| miR-193a-5p, miR-210, miR-513a-5p, miR-628-3p | miRNAs | miRNA in serum, panel works for both EOCRC and LOCRC[75] |
| Sat2, LINE-1, Alu | Methylation, DNA | DNA repetitive elements with increased methylation in EOCRC in white blood cells[62] |
| miR-31-5p, DMD | miRNA, mRNA | Transcripts uniquely overexpressed in sporadic EOCRC tumor vs normal and not in LOCRC. miR-3105p targets DMD and it's downregulated in EOCRC[116] |
| MYC | mRNA | Transcription factor with increased tumor expression in EOCRCs may subset patients into distinct groups[50,69] |
Another minimally invasive screening option is a fecal test, such as Cologuard™, which measures methylation of the genes BMP3 and NDRG4 and assesses samples for the KRAS mutation. Cologuard also includes a fecal immunohistochemical test (FIT), which measures human globin, or blood, in the stool (Figure 2). Studies of whether these biomarkers can detect EOCRC are limited, though recent studies have found no significant difference in these markers within CRC tumors in younger and older patients[113-115]. Therefore, while Cologuard and FIT may be effective to detect EOCRC in fecal samples, additional studies are required before such recommendations can be made.
Gene expression biomarkers within tumors have also been suggested to serve as prognostic indicators. miR-31-5p was found to be uniquely overexpressed in sporadic EOCRC tumors vs normal samples, while it was not overexpressed in LOCRC. Moreover, the miR-31-5p target, DMD, was also shown to be decreased in EOCRC tumors, and this change in expression correlated with a worse prognosis (Table 3)[116]. In addition to genetic, transcriptomic, and proteomic alterations, biomarkers of the gut microbiome have also been suggested to identify EOCRC, as EOCRC has been shown to have a distinct microbiome compared with LOCRC[117]. Microbiota studies in CRC and EOCRC are outside the scope of this review but are nicely summarized by Abdullah et al[118]. Overall, a limited number of studies have shown that common CRC screening options may apply to EOCRC as well. miRNA and DNA methylation biomarkers have been proposed to help identify EOCRC (Table 3), however additional studies with larger sample sizes and clinical validations are required.
Currently, clinical practice guidelines do not differentiate the treatment of EOCRC vs LOCRC[119]. However, CRC treatment is dependent on tumor mutational profiling, and thus, the lower frequency of BRAF[34,38,57] and KRAS[99,120] mutations in EOCRC means the mutation-specific treatments will be less commonly used in EOCRC patients. The efficacy of these drugs in EOCRC has not been directly studied but the mechanism is likely very similar to LOCRC.
Several clinical features have been associated with EOCRC. Approximately 75% of sporadic cases occur in the 40–year-old to 49-year-old age group, with 55%-80% of EOCRCs occurring in the distal colon or rectum[121,122]. The increasing rate of EOCRC has been predominated by an increasing rate of distal colon and rectal cancer, with individuals born circa 1990 having double and quadruple the risk of colon and rectal cancer, respectively, compared to those born circa 1950[123]. While many strides have been made to understand alterations at the DNA, RNA, and protein levels that contribute to EOCRC, questions remain on how EOCRC patients should be treated compared with their older counterparts. Currently, young patients are more likely to be treated, or overtreated, with systemic chemotherapy, but have similar clinical outcomes compared with older patients[10,104,124].
As the number of EOCRC cases continues to rise globally, there is a critical need to optimize cancer treatment strategies, as well as to further develop non-invasive screening options to identify people at risk for EOCRC. Currently, scientific studies are limited by low sampling size, especially in non-white patients. Researchers are addressing this limitation by continuing to grow biobanks with younger and non-diseased samples. Furthermore, machine learning approaches have been suggested to increase the statistical power of limited sample sizes[125], which could be applied to identify EOCRC risk genes or genetic loci in under-represented minorities. Another limitation is the lack of a clear model system to test hypotheses on EOCRC. While models for EOCRC exist, as APCmin mice develop CRC at a young age, and HCT-116 cells are from a young patient[126], these systems fail to recapitulate the diversity in EOCRC subtypes that are observed in patient samples. The development of stable cell lines from CRC samples generally requires transformation, altering the cellular profiles, and limiting normal controls. Organoid models are gaining popularity due to their ability to recapitulate the colonic crypt structure from both normal and tumor cells[33,127]. Future work will tease out the molecular mechanisms unique to EOCRC with growing biobanks and organoids as well as other innovative model systems.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Han J, China; Zheng T, China; S-Editor: Li L L-Editor: Filipodia P-Editor: Li L
| 1. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3261] [Article Influence: 652.2] [Reference Citation Analysis (2)] |
| 2. | Hofseth LJ, Hebert JR, Chanda A, Chen H, Love BL, Pena MM, Murphy EA, Sajish M, Sheth A, Buckhaults PJ, Berger FG. Early-onset colorectal cancer: initial clues and current views. Nat Rev Gastroenterol Hepatol. 2020;17:352-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 254] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 3. | Patel SG, Ahnen DJ. Colorectal Cancer in the Young. Curr Gastroenterol Rep. 2018;20:15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 4. | Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, Siena S. Early-onset colorectal cancer in young individuals. Mol Oncol. 2019;13:109-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 407] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 5. | Connell LC, Mota JM, Braghiroli MI, Hoff PM. The Rising Incidence of Younger Patients With Colorectal Cancer: Questions About Screening, Biology, and Treatment. Curr Treat Options Oncol. 2017;18:23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 6. | Eng C, Jácome AA, Agarwal R, Hayat MH, Byndloss MX, Holowatyj AN, Bailey C, Lieu CH. A comprehensive framework for early-onset colorectal cancer research. Lancet Oncol. 2022;23:e116-e128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 104] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 7. | Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, Goldberg RM, Paskett E, Shields PG, Freudenheim JL, Stanich PP, Lattimer I, Arnold M, Liyanarachchi S, Kalady M, Heald B, Greenwood C, Paquette I, Prues M, Draper DJ, Lindeman C, Kuebler JP, Reynolds K, Brell JM, Shaper AA, Mahesh S, Buie N, Weeman K, Shine K, Haut M, Edwards J, Bastola S, Wickham K, Khanduja KS, Zacks R, Pritchard CC, Shirts BH, Jacobson A, Allen B, de la Chapelle A, Hampel H; Ohio Colorectal Cancer Prevention Initiative Study Group. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol. 2017;3:464-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 515] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 8. | Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, Welton M, Shelton A, Ma L, Arber DA. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25:1128-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 260] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 9. | Joo JE, Clendenning M, Wong EM, Rosty C, Mahmood K, Georgeson P, Winship IM, Preston SG, Win AK, Dugué PA, Jayasekara H, English D, Macrae FA, Hopper JL, Jenkins MA, Milne RL, Giles GG, Southey MC, Buchanan DD. DNA Methylation Signatures and the Contribution of Age-Associated Methylomic Drift to Carcinogenesis in Early-Onset Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Strum WB, Boland CR. Clinical and Genetic Characteristics of Colorectal Cancer in Persons under 50 Years of Age: A Review. Dig Dis Sci. 2019;64:3059-3065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Siegel RL, Jakubowski CD, Fedewa SA, Davis A, Azad NS. Colorectal Cancer in the Young: Epidemiology, Prevention, Management. Am Soc Clin Oncol Educ Book. 2020;40:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 12. | Liu PH, Wu K, Ng K, Zauber AG, Nguyen LH, Song M, He X, Fuchs CS, Ogino S, Willett WC, Chan AT, Giovannucci EL, Cao Y. Association of Obesity With Risk of Early-Onset Colorectal Cancer Among Women. JAMA Oncol. 2019;5:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 351] [Article Influence: 58.5] [Reference Citation Analysis (2)] |
| 13. | Sanford NN, Giovannucci EL, Ahn C, Dee EC, Mahal BA. Obesity and younger versus older onset colorectal cancer in the United States, 1998-2017. J Gastrointest Oncol. 2020;11:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Tang CT, Li J, Yang Z, Zeng C, Chen Y. Comparison of some biochemical markers between early-onset and late-onset colorectal precancerous lesions: A single-center retrospective study. J Clin Lab Anal. 2022;36:e24637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | DeClercq V, McMurray DN, Chapkin RS. Obesity promotes colonic stem cell expansion during cancer initiation. Cancer Lett. 2015;369:336-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, Pinello L, Katz Y, Shinagare S, Abu-Remaileh M, Mihaylova MM, Lamming DW, Dogum R, Guo G, Bell GW, Selig M, Nielsen GP, Gupta N, Ferrone CR, Deshpande V, Yuan GC, Orkin SH, Sabatini DM, Yilmaz ÖH. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 592] [Cited by in RCA: 611] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 17. | Cirillo F, Catellani C, Sartori C, Lazzeroni P, Amarri S, Street ME. Obesity, Insulin Resistance, and Colorectal Cancer: Could miRNA Dysregulation Play A Role? Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 18. | Segev L, Kalady MF, Church JM. Left-Sided Dominance of Early-Onset Colorectal Cancers: A Rationale for Screening Flexible Sigmoidoscopy in the Young. Dis Colon Rectum. 2018;61:897-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 19. | Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC, Shih YT, Walter LC, Andrews KS, Brawley OW, Brooks D, Fedewa SA, Manassaram-Baptiste D, Siegel RL, Wender RC, Smith RA. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68:250-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 1309] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 20. | Tom CM, Mankarious M, Jeganathan NA, Deutsch M, Koltun WA, Berg AS, Scow JS. Characteristics and Outcomes of Right- Versus Left-Sided Early Onset Colorectal Cancer. Dis Colon Rectum. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 21. | Zhong M, Xiong Y, Ye Z, Zhao J, Zhong L, Liu Y, Zhu Y, Tian L, Qiu X, Hong X. Microbial Community Profiling Distinguishes Left-Sided and Right-Sided Colon Cancer. Front Cell Infect Microbiol. 2020;10:498502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Phipps O, Quraishi MN, Dickson EA, Steed H, Kumar A, Acheson AG, Beggs AD, Brookes MJ, Al-Hassi HO. Differences in the On- and Off-Tumor Microbiota between Right- and Left-Sided Colorectal Cancer. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Hyngstrom JR, Hu CY, Xing Y, You YN, Feig BW, Skibber JM, Rodriguez-Bigas MA, Cormier JN, Chang GJ. Clinicopathology and outcomes for mucinous and signet ring colorectal adenocarcinoma: analysis from the National Cancer Data Base. Ann Surg Oncol. 2012;19:2814-2821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 265] [Article Influence: 20.4] [Reference Citation Analysis (1)] |
| 24. | Kim YH, Min BH, Kim SJ, Choi HK, Kim KM, Chun HK, Lee H, Kim JY, Chang DK, Son HJ, Rhee PL, Rhee JC, Kim JJ. Difference between proximal and distal microsatellite-unstable sporadic colorectal cancers: analysis of clinicopathological and molecular features and prognoses. Ann Surg Oncol. 2010;17:1435-1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8003] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 26. | Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1648] [Cited by in RCA: 1588] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 27. | Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 371] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 28. | Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1354] [Cited by in RCA: 1421] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 29. | Langner C. Serrated and non-serrated precursor lesions of colorectal cancer. Dig Dis. 2015;33:28-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Müller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 31. | Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology. 2020;158:291-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 311] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 32. | Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 590] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 33. | Yan HHN, Siu HC, Ho SL, Yue SSK, Gao Y, Tsui WY, Chan D, Chan AS, Wong JWH, Man AHY, Lee BCH, Chan ASY, Chan AKW, Hui HS, Cheung AKL, Law WL, Lo OSH, Yuen ST, Clevers H, Leung SY. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut. 2020;69:2165-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 34. | Willauer AN, Liu Y, Pereira AAL, Lam M, Morris JS, Raghav KPS, Morris VK, Menter D, Broaddus R, Meric-Bernstam F, Hayes-Jordan A, Huh W, Overman MJ, Kopetz S, Loree JM. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 2019;125:2002-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 241] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 35. | Mork ME, You YN, Ying J, Bannon SA, Lynch PM, Rodriguez-Bigas MA, Vilar E. High Prevalence of Hereditary Cancer Syndromes in Adolescents and Young Adults With Colorectal Cancer. J Clin Oncol. 2015;33:3544-3549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 175] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 36. | Perea J, Rueda D, Canal A, Rodríguez Y, Álvaro E, Osorio I, Alegre C, Rivera B, Martínez J, Benítez J, Urioste M. Age at onset should be a major criterion for subclassification of colorectal cancer. J Mol Diagn. 2014;16:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 37. | Goel A, Nagasaka T, Spiegel J, Meyer R, Lichliter WE, Boland CR. Low frequency of Lynch syndrome among young patients with non-familial colorectal cancer. Clin Gastroenterol Hepatol. 2010;8:966-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Kirzin S, Marisa L, Guimbaud R, De Reynies A, Legrain M, Laurent-Puig P, Cordelier P, Pradère B, Bonnet D, Meggetto F, Portier G, Brousset P, Selves J. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One. 2014;9:e103159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 39. | Li J, Ma X, Chakravarti D, Shalapour S, DePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021;35:787-820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 303] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 40. | Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, Wu K, Cao Y, Ng K, Ogino S. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. 2021;18:230-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 401] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 41. | Archambault AN, Jeon J, Lin Y, Thomas M, Harrison TA, Bishop DT, Brenner H, Casey G, Chan AT, Chang-Claude J, Figueiredo JC, Gallinger S, Gruber SB, Gunter MJ, Guo F, Hoffmeister M, Jenkins MA, Keku TO, Le Marchand L, Li L, Moreno V, Newcomb PA, Pai R, Parfrey PS, Rennert G, Sakoda LC, Lee JK, Slattery ML, Song M, Win AK, Woods MO, Murphy N, Campbell PT, Su YR, Lansdorp-Vogelaar I, Peterse EFP, Cao Y, Zeleniuch-Jacquotte A, Liang PS, Du M, Corley DA, Hsu L, Peters U, Hayes RB. Risk Stratification for Early-Onset Colorectal Cancer Using a Combination of Genetic and Environmental Risk Scores: An International Multi-Center Study. J Natl Cancer Inst. 2022;114:528-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 42. | Magnani G, Furlan D, Sahnane N, Reggiani Bonetti L, Domati F, Pedroni M. Molecular Features and Methylation Status in Early Onset (≤40 Years) Colorectal Cancer: A Population Based, Case-Control Study. Gastroenterol Res Pract. 2015;2015:132190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 43. | Xu T, Zhang Y, Zhang J, Qi C, Liu D, Wang Z, Li Y, Ji C, Li J, Lin X, Hou T, Liu H, Zhang L, Han-Zhang H, Shen L, Wang X. Germline Profiling and Molecular Characterization of Early Onset Metastatic Colorectal Cancer. Front Oncol. 2020;10:568911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 44. | Kothari N, Teer JK, Abbott AM, Srikumar T, Zhang Y, Yoder SJ, Brohl AS, Kim RD, Reed DR, Shibata D. Increased incidence of FBXW7 and POLE proofreading domain mutations in young adult colorectal cancers. Cancer. 2016;122:2828-2835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 45. | Zhunussova G, Afonin G, Abdikerim S, Jumanov A, Perfilyeva A, Kaidarova D, Djansugurova L. Mutation Spectrum of Cancer-Associated Genes in Patients With Early Onset of Colorectal Cancer. Front Oncol. 2019;9:673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 46. | Rennoll S, Yochum G. Regulation of MYC gene expression by aberrant Wnt/β-catenin signaling in colorectal cancer. World J Biol Chem. 2015;6:290-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 101] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 47. | Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, Kim WH, Lee HS. c-MYC Copy-Number Gain Is an Independent Prognostic Factor in Patients with Colorectal Cancer. PLoS One. 2015;10:e0139727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 48. | Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, Mirlacher M, Tapia C, Simon R, Sauter G, Terracciano L, Tornillo L. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60:768-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 49. | Kwak Y, Yun S, Nam SK, Seo AN, Lee KS, Shin E, Oh HK, Kim DW, Kang SB, Kim WH, Lee HS. Comparative analysis of the EGFR, HER2, c-MYC, and MET variations in colorectal cancer determined by three different measures: gene copy number gain, amplification status and the 2013 ASCO/CAP guideline criterion for HER2 testing of breast cancer. J Transl Med. 2017;15:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 50. | Marx OM, Mankarious MM, Eshelman MA, Ding W, Koltun WA, Yochum GS. Transcriptome Analyses Identify Deregulated MYC in Early Onset Colorectal Cancer. Biomolecules. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 51. | Pan W, Wang W, Huang J, Lu K, Huang S, Jiang D, Bu D, Liu J, Jing H, Yao J, Hou Y. The prognostic role of c-MYC amplification in schistosomiasis-associated colorectal cancer. Jpn J Clin Oncol. 2020;50:446-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 52. | Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I, Fetveit T, Mathisen Ø, Husby A, Sandvik O, Nesbakken A, Thiis-Evensen E, Lothe RA. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 53. | Perea J, García JL, Pérez J, Rueda D, Arriba M, Rodríguez Y, Urioste M, González-Sarmiento R. NOMO-1 gene is deleted in early-onset colorectal cancer. Oncotarget. 2017;8:24429-24436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Tricoli JV, Boardman LA, Patidar R, Sindiri S, Jang JS, Walsh WD, McGregor PM 3rd, Camalier CE, Mehaffey MG, Furman WL, Bahrami A, Williams PM, Lih CJ, Conley BA, Khan J. A mutational comparison of adult and adolescent and young adult (AYA) colon cancer. Cancer. 2018;124:1070-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 55. | Guo Q, Xie J, Dang CV, Liu ET, Bishop JM. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci U S A. 1998;95:9172-9177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 116] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O, Spruck C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006-9012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 392] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 57. | Lee W, Wang Z, Saffern M, Jun T, Huang KL. Genomic and molecular features distinguish young adult cancer from later-onset cancer. Cell Rep. 2021;37:110005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 58. | Morris VK, Bekaii-Saab T. Improvements in Clinical Outcomes for BRAF(V600E) -Mutant Metastatic Colorectal Cancer. Clin Cancer Res. 2020;26:4435-4441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 59. | Akimoto N, Zhao M, Ugai T, Zhong R, Lau MC, Fujiyoshi K, Kishikawa J, Haruki K, Arima K, Twombly TS, Zhang X, Giovannucci EL, Wu K, Song M, Chan AT, Cao Y, Meyerhardt JA, Ng K, Giannakis M, Väyrynen JP, Nowak JA, Ogino S. Tumor Long Interspersed Nucleotide Element-1 (LINE-1) Hypomethylation in Relation to Age of Colorectal Cancer Diagnosis and Prognosis. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 60. | Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 516] [Article Influence: 103.2] [Reference Citation Analysis (0)] |
| 61. | Baba Y, Yagi T, Sawayama H, Hiyoshi Y, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Baba H. Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers. Digestion. 2018;97:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 62. | Walters RJ, Williamson EJ, English DR, Young JP, Rosty C, Clendenning M, Walsh MD, Parry S, Ahnen DJ, Baron JA, Win AK, Giles GG, Hopper JL, Jenkins MA, Buchanan DD. Association between hypermethylation of DNA repetitive elements in white blood cell DNA and early-onset colorectal cancer. Epigenetics. 2013;8:748-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 63. | Goossens-Beumer IJ, Benard A, van Hoesel AQ, Zeestraten EC, Putter H, Böhringer S, Liefers GJ, Morreau H, van de Velde CJ, Kuppen PJ. Age-dependent clinical prognostic value of histone modifications in colorectal cancer. Transl Res. 2015;165:578-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Pira G, Uva P, Scanu AM, Rocca PC, Murgia L, Uleri E, Piu C, Porcu A, Carru C, Manca A, Persico I, Muroni MR, Sanges F, Serra C, Dolei A, Angius A, De Miglio MR. Landscape of transcriptome variations uncovering known and novel driver events in colorectal carcinoma. Sci Rep. 2020;10:432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 65. | Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the juncture of clinical translation. Nat Rev Genet. 2018;19:93-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 175] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 66. | Liu Z, Brooks RS, Ciappio ED, Kim SJ, Crott JW, Bennett G, Greenberg AS, Mason JB. Diet-induced obesity elevates colonic TNF-α in mice and is accompanied by an activation of Wnt signaling: a mechanism for obesity-associated colorectal cancer. J Nutr Biochem. 2012;23:1207-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 67. | Luo Y, Yang S, Wu X, Takahashi S, Sun L, Cai J, Krausz KW, Guo X, Dias HB, Gavrilova O, Xie C, Jiang C, Liu W, Gonzalez FJ. Intestinal MYC modulates obesity-related metabolic dysfunction. Nat Metab. 2021;3:923-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 68. | Ellegaard AM, Knop FK. MYC mRNA expression throughout the intestine is not associated with body mass index or type 2 diabetes. Endocrinol Diabetes Metab. 2022;5:e00327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 69. | Yokota K, Tanaka Y, Harada H, Kaida T, Nakamoto S, Soeno T, Fujiyama Y, Yokota M, Kojo K, Miura H, Yamanashi T, Sato T, Nakamura T, Watanabe M, Yamashita K. WiNTRLINC1/ASCL2/c-Myc Axis Characteristics of Colon Cancer with Differentiated Histology at Young Onset and Essential for Cell Viability. Ann Surg Oncol. 2019;26:4826-4834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 70. | Watson KM, Gardner IH, Byrne RM, Ruhl RR, Lanciault CP, Dewey EN, Anand S, Tsikitis VL. Differential Expression of PEG10 Contributes to Aggressive Disease in Early Versus Late-Onset Colorectal Cancer. Dis Colon Rectum. 2020;63:1610-1620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 71. | Markman JL, Shiao SL. Impact of the immune system and immunotherapy in colorectal cancer. J Gastrointest Oncol. 2015;6:208-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 101] [Reference Citation Analysis (0)] |
| 72. | Yue T, Chen S, Zhu J, Guo S, Huang Z, Wang P, Zuo S, Liu Y. The aging-related risk signature in colorectal cancer. Aging (Albany NY). 2021;13:7330-7349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 73. | Gardner IH, Siddharthan R, Watson K, Dewey E, Ruhl R, Khou S, Guan X, Xia Z, Tsikitis VL, Anand S. A Distinct Innate Immune Signature of Early Onset Colorectal Cancer. Immunohorizons. 2021;5:489-499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 74. | Holowatyj AN, Gigic B, Herpel E, Scalbert A, Schneider M, Ulrich CM; MetaboCCC Consortium; ColoCare Study. Distinct Molecular Phenotype of Sporadic Colorectal Cancers Among Young Patients Based on Multiomics Analysis. Gastroenterology. 2020;158:1155-1158.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 75. | Nakamura K, Hernández G, Sharma GG, Wada Y, Banwait JK, González N, Perea J, Balaguer F, Takamaru H, Saito Y, Toiyama Y, Kodera Y, Boland CR, Bujanda L, Quintero E, Goel A. A Liquid Biopsy Signature for the Detection of Patients With Early-Onset Colorectal Cancer. Gastroenterology. 2022;163:1242-1251.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 76. | Ak S, Tunca B, Tezcan G, Cecener G, Egeli U, Yilmazlar T, Ozturk E, Yerci O. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J Surg Res. 2014;191:113-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 77. | Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 586] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 78. | Mao Z, Zhao H, Qin Y, Wei J, Sun J, Zhang W, Kang Y. Post-Transcriptional Dysregulation of microRNA and Alternative Polyadenylation in Colorectal Cancer. Front Genet. 2020;11:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 79. | Lasabová Z, Kalman M, Holubeková V, Grendár M, Kašubová I, Jašek K, Meršaková S, Malicherová B, Baranenko D, Adamek M, Kruzliak P, Plank L. Mutation analysis of POLE gene in patients with early-onset colorectal cancer revealed a rare silent variant within the endonuclease domain with potential effect on splicing. Clin Exp Med. 2019;19:393-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 80. | Chen Y, Huang M, Liu X, Huang Y, Liu C, Zhu J, Fu G, Lei Z, Chu X. Alternative splicing of mRNA in colorectal cancer: new strategies for tumor diagnosis and treatment. Cell Death Dis. 2021;12:752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Bhadra M, Howell P, Dutta S, Heintz C, Mair WB. Alternative splicing in aging and longevity. Hum Genet. 2020;139:357-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 82. | Gao Y, Wang H, Li H, Ye X, Xia Y, Yuan S, Lu J, Xie X, Wang L, Zhang J. Integrated analyses of m(1)A regulator-mediated modification patterns in tumor microenvironment-infiltrating immune cells in colon cancer. Oncoimmunology. 2021;10:1936758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 83. | Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, Zhang X, Cao Y, Ma D, Zhu X, Zhang Y, Fang JY, Chen H, Hong J. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 292] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 84. | Saul D, Kosinsky RL. Single-Cell Transcriptomics Reveals the Expression of Aging- and Senescence-Associated Genes in Distinct Cancer Cell Populations. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 85. | Vasaikar S, Huang C, Wang X, Petyuk VA, Savage SR, Wen B, Dou Y, Zhang Y, Shi Z, Arshad OA, Gritsenko MA, Zimmerman LJ, McDermott JE, Clauss TR, Moore RJ, Zhao R, Monroe ME, Wang YT, Chambers MC, Slebos RJC, Lau KS, Mo Q, Ding L, Ellis M, Thiagarajan M, Kinsinger CR, Rodriguez H, Smith RD, Rodland KD, Liebler DC, Liu T, Zhang B; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell. 2019;177:1035-1049.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 527] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 86. | Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJ, Carr SA, Tabb DL, Coffey RJ, Slebos RJ, Liebler DC; NCI CPTAC. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 968] [Cited by in RCA: 1116] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 87. | Gong Y, Liu Y, Wang T, Li Z, Gao L, Chen H, Shu Y, Li Y, Xu H, Zhou Z, Dai L. Age-Associated Proteomic Signatures and Potential Clinically Actionable Targets of Colorectal Cancer. Mol Cell Proteomics. 2021;20:100115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 88. | Vermani L, Kumar R, Kannan RR, Deka MK, Talukdar A, Kumar NS. Expression pattern of ALDH1, E-cadherin, Vimentin and Twist in early and late onset sporadic colorectal cancer. Biomark Med. 2020;14:1371-1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 89. | Kirwan A, Utratna M, O'Dwyer ME, Joshi L, Kilcoyne M. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. Biomed Res Int. 2015;2015:490531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 90. | Nag JK, Appasamy P, Sedley S, Malka H, Rudina T, Bar-Shavit R. RNF43 induces the turnover of protease-activated receptor 2 in colon cancer. FASEB J. 2023;37:e22675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 91. | Zhu Y, Gu L, Lin X, Liu C, Lu B, Cui K, Zhou F, Zhao Q, Prochownik EV, Fan C, Li Y. Dynamic Regulation of ME1 Phosphorylation and Acetylation Affects Lipid Metabolism and Colorectal Tumorigenesis. Mol Cell. 2020;77:138-149.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 92. | Yu X, Chen C, Song X, Guo Y, Tong Y, Zhao Y, Song Z. Glycosylated Hemoglobin as an Age-Specific Predictor and Risk Marker of Colorectal Adenomas in Non-Diabetic Adults. Front Endocrinol (Lausanne). 2021;12:774519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 93. | Huang J, Pan H, Wang J, Wang T, Huo X, Ma Y, Lu Z, Sun B, Jiang H. Unfolded protein response in colorectal cancer. Cell Biosci. 2021;11:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 94. | Siegel RL, Torre LA, Soerjomataram I, Hayes RB, Bray F, Weber TK, Jemal A. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68:2179-2185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 537] [Article Influence: 89.5] [Reference Citation Analysis (1)] |
| 95. | Sung JJY, Chiu HM, Jung KW, Jun JK, Sekiguchi M, Matsuda T, Kyaw MH. Increasing Trend in Young-Onset Colorectal Cancer in Asia: More Cancers in Men and More Rectal Cancers. Am J Gastroenterol. 2019;114:322-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 96. | Nath J, Wigley C, Keighley MR, Perakath B. Rectal cancer in young adults: a series of 102 patients at a tertiary care centre in India. Colorectal Dis. 2009;11:475-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 97. | Kumar R, Raman R, Kotapalli V, Gowrishankar S, Pyne S, Pollack JR, Bashyam MD. Ca(2+)/nuclear factor of activated T cells signaling is enriched in early-onset rectal tumors devoid of canonical Wnt activation. J Mol Med (Berl). 2018;96:135-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 98. | Gupta S, Bhattacharya D, Acharya AN, Majumdar S, Ranjan P, Das S. Colorectal carcinoma in young adults: a retrospective study on Indian patients: 2000-2008. Colorectal Dis. 2010;12:e182-e189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 99. | Raman R, Kotapalli V, Adduri R, Gowrishankar S, Bashyam L, Chaudhary A, Vamsy M, Patnaik S, Srinivasulu M, Sastry R, Rao S, Vasala A, Kalidindi N, Pollack J, Murthy S, Bashyam M. Evidence for possible non-canonical pathway(s) driven early-onset colorectal cancer in India. Mol Carcinog. 2014;53 Suppl 1:E181-E186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 100. | Singh MP, Rai S, Singh NK, Srivastava S. Transcriptomic landscape of early age onset of colorectal cancer identifies novel genes and pathways in Indian CRC patients. Sci Rep. 2021;11:11765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 101. | Chen Y, Chen Z, Huang J, Hu J, He X, Lan P. Clinicopathological and molecular characteristics of early-onset vs late-onset colorectal cancer according to tumor location. Int J Clin Oncol. 2022;27:749-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 102. | Irabor DO. Emergence of Colorectal Cancer in West Africa: Accepting the Inevitable. Niger Med J. 2017;58:87-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 103. | Holowatyj AN, Maude AS, Musa HS, Adamu A, Ibrahim S, Abdullahi A, Manko M, Aminu SM, Mohammed A, Idoko J, Ukwenya Y, Carpten J, Chandler PD, Hampel H, Faruk M. Patterns of Early-Onset Colorectal Cancer Among Nigerians and African Americans. JCO Glob Oncol. 2020;6:1647-1655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 104. | Galadima HI, Adunlin G, Hughes MS, Cropp CD, Lucero L, Akpinar-Elci M. Racial disparities and treatment trends among young-onset colorectal cancer patients: An analysis of a hospital cancer registry. Cancer Epidemiol. 2021;72:101911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 105. | Petrick JL, Barber LE, Warren Andersen S, Florio AA, Palmer JR, Rosenberg L. Racial Disparities and Sex Differences in Early- and Late-Onset Colorectal Cancer Incidence, 2001-2018. Front Oncol. 2021;11:734998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 106. | Holowatyj AN, Ruterbusch JJ, Rozek LS, Cote ML, Stoffel EM. Racial/Ethnic Disparities in Survival Among Patients With Young-Onset Colorectal Cancer. J Clin Oncol. 2016;34:2148-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (1)] |
| 107. | Kamath SD, Torrejon N, Wei W, Tullio K, Nair KG, Liska D, Krishnamurthi SS, Khorana AA. Racial disparities negatively impact outcomes in early-onset colorectal cancer independent of socioeconomic status. Cancer Med. 2021;10:7542-7550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 108. | Sylvester BE, Huo D, Khramtsov A, Zhang J, Smalling RV, Olugbile S, Polite BN, Olopade OI. Molecular analysis of colorectal tumors within a diverse patient cohort at a single institution. Clin Cancer Res. 2012;18:350-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 109. | Xicola RM, Manojlovic Z, Augustus GJ, Kupfer SS, Emmadi R, Alagiozian-Angelova V, Triche T Jr, Salhia B, Carpten J, Llor X, Ellis NA. Lack of APC somatic mutation is associated with early-onset colorectal cancer in African Americans. Carcinogenesis. 2018;39:1331-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 110. | Cavestro GM, Mannucci A, Zuppardo RA, Di Leo M, Stoffel E, Tonon G. Early onset sporadic colorectal cancer: Worrisome trends and oncogenic features. Dig Liver Dis. 2018;50:521-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 111. | Anderson JC, Samadder JN. To Screen or Not to Screen Adults 45-49 Years of Age: That is the Question. Am J Gastroenterol. 2018;113:1750-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 112. | Loomans-Kropp HA, Song Y, Gala M, Parikh AR, Van Seventer EE, Alvarez R, Hitchins MP, Shoemaker RH, Umar A. Methylated Septin9 (mSEPT9): A promising blood-based biomarker for the detection and screening of early-onset colorectal cancer. Cancer Res Commun. 2022;2:90-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 113. | Limburg PJ, Mahoney DW, Ahlquist DA, Allawi HT, Johnson SC, Kaiser M, Katerov VE, Statz S, Graham RP, Foote PH, Doering KA, Burger KN, Lidgard GP, Kisiel JB. Comparison of Tissue-Based Molecular Markers in Younger versus Older Patients with Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev. 2020;29:1570-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 114. | Chen CH, Tsai MK, Wen CP. Extending Colorectal Cancer Screening to Persons Aged 40 to 49 Years With Immunochemical Fecal Occult Blood Test: A Prospective Cohort Study of 513,283 Individuals. J Clin Gastroenterol. 2016;50:761-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 115. | D'Souza N, Monahan K, Benton SC, Wilde L, Abulafi M; NICE FIT Steering Group. Finding the needle in the haystack: the diagnostic accuracy of the faecal immunochemical test for colorectal cancer in younger symptomatic patients. Colorectal Dis. 2021;23:2539-2549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 116. | Liu C, Wu W, Chang W, Wu R, Sun X, Wu H, Liu Z. miR-31-5p-DMD axis as a novel biomarker for predicting the development and prognosis of sporadic early-onset colorectal cancer. Oncol Lett. 2022;23:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 117. | Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 126] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 118. | Abdullah M, Sukartini N, Nursyirwan SA, Pribadi RR, Maulahela H, Utari AP, Muzellina VN, Wiraatmadja A, Renaldi K. Gut Microbiota Profiles in Early- and Late-Onset Colorectal Cancer: A Potential Diagnostic Biomarker in the Future. Digestion. 2021;102:823-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 119. | Cavestro GM, Mannucci A, Balaguer F, Hampel H, Kupfer SS, Repici A, Sartore-Bianchi A, Seppälä TT, Valentini V, Boland CR, Brand RE, Buffart TE, Burke CA, Caccialanza R, Cannizzaro R, Cascinu S, Cercek A, Crosbie EJ, Danese S, Dekker E, Daca-Alvarez M, Deni F, Dominguez-Valentin M, Eng C, Goel A, Guillem JG, Houwen BBSL, Kahi C, Kalady MF, Kastrinos F, Kühn F, Laghi L, Latchford A, Liska D, Lynch P, Malesci A, Mauri G, Meldolesi E, Møller P, Monahan KJ, Möslein G, Murphy CC, Nass K, Ng K, Oliani C, Papaleo E, Patel SG, Puzzono M, Remo A, Ricciardiello L, Ripamonti CI, Siena S, Singh SK, Stadler ZK, Stanich PP, Syngal S, Turi S, Urso ED, Valle L, Vanni VS, Vilar E, Vitellaro M, You YN, Yurgelun MB, Zuppardo RA, Stoffel EM; Associazione Italiana Familiarità Ereditarietà Tumori; Collaborative Group of the Americas on Inherited Gastrointestinal Cancer; European Hereditary Tumour Group, and the International Society for Gastrointestinal Hereditary Tumours. Delphi Initiative for Early-Onset Colorectal Cancer (DIRECt) International Management Guidelines. Clin Gastroenterol Hepatol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 65] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 120. | Alsop K, Mead L, Smith LD, Royce SG, Tesoriero AA, Young JP, Haydon A, Grubb G, Giles GG, Jenkins MA, Hopper JL, Southey MC. Low somatic K-ras mutation frequency in colorectal cancer diagnosed under the age of 45 years. Eur J Cancer. 2006;42:1357-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 121. | Wang Y, Yang L, Zhou M, Shen L, Zhang J, Deng W, Liang L, Hu R, Yang W, Yao Y, Zhang Z. Disparities in survival for right-sided vs. left-sided colon cancers in young patients: a study based on the Surveillance, Epidemiology, and End Results database (1990-2014). Cancer Manag Res. 2018;10:1735-1747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 122. | Kasi PM, Shahjehan F, Cochuyt JJ, Li Z, Colibaseanu DT, Merchea A. Rising Proportion of Young Individuals With Rectal and Colon Cancer. Clin Colorectal Cancer. 2019;18:e87-e95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 123. | Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, Jemal A. Colorectal Cancer Incidence Patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 832] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 124. | Kneuertz PJ, Chang GJ, Hu CY, Rodriguez-Bigas MA, Eng C, Vilar E, Skibber JM, Feig BW, Cormier JN, You YN. Overtreatment of young adults with colon cancer: more intense treatments with unmatched survival gains. JAMA Surg. 2015;150:402-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 125. | Mägi R, Horikoshi M, Sofer T, Mahajan A, Kitajima H, Franceschini N, McCarthy MI; COGENT-Kidney Consortium, T2D-GENES Consortium, Morris AP. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet. 2017;26:3639-3650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 126. | Pérez-García J, Martel-Martel A, García-Vallés P, Corchete LA, García JL, Gestoso-Uzal N, Vidal-Tocino R, Blanco Ó, Méndez L, Sánchez-Martín M, Fuentes M, Herrero AB, Holowatyj AN, Perea J, González-Sarmiento R. Recurrent NOMO1 Gene Deletion Is a Potential Clinical Marker in Early-Onset Colorectal Cancer and Is Involved in the Regulation of Cell Migration. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 127. | Mizutani T, Clevers H. Primary Intestinal Epithelial Organoid Culture. Methods Mol Biol. 2020;2171:185-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |