Published online Nov 27, 2020. doi: 10.4331/wjbc.v11.i3.99
Peer-review started: June 4, 2020
First decision: July 4, 2020
Revised: July 10, 2020
Accepted: September 18, 2020
Article in press: September 18, 2020
Published online: November 27, 2020
Processing time: 160 Days and 2.3 Hours
X-linked adrenoleukodystrophy (X-ALD), an inborn error of peroxisomal β-oxidation, is caused by defects in the ATP Binding Cassette Subfamily D Member 1 (ABCD1) gene. X-ALD patients may be asymptomatic or present with several clinical phenotypes varying from severe to mild, severe cerebral adrenoleuko-dystrophy to mild adrenomyeloneuropathy (AMN). Although most female heterozygotes present with AMN-like symptoms after 60 years of age, occasional cases of females with the cerebral form have been reported. Phenotypic variability has been described within the same kindreds and even among monozygotic twins. There is no association between the nature of ABCD1 mutation and the clinical phenotypes, and the molecular basis of phenotypic variability in X-ALD is yet to be resolved. Various genetic, epigenetic, and environmental influences are speculated to modify the disease onset and severity. In this review, we summarize the observations made in various studies investigating the potential modifying factors regulating the clinical manifestation of X-ALD, which could help understand the pathogenesis of the disease and develop suitable therapeutic strategies.
Core Tip: The monogenic peroxisomal disorder, X-linked adrenoleukodystrophy (X-ALD), presents with different clinical phenotypes. The molecular basis for the phenotypic variation has yet to be resolved and is considered to be influenced by genetic, epigenetic, cellular, or environmental factors. We herein discuss the various modifying factors, which can potentially alter the phenotypic presentation of X-ALD.
- Citation: Palakuzhiyil SV, Christopher R, Chandra SR. Deciphering the modifiers for phenotypic variability of X-linked adrenoleukodystrophy. World J Biol Chem 2020; 11(3): 99-111
- URL: https://www.wjgnet.com/1949-8454/full/v11/i3/99.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v11.i3.99
Monogenic disorders are primarily caused by a single defective gene, but mutations in a single gene can result in a disease with varying clinical phenotypes. X-linked adrenoleukodystrophy (X-ALD), caused by mutations in the ABCD1 gene, is one such monogenic disorder affecting peroxisomal β-oxidation. ABCD1, mapped on Xq28, comprises of 10 exons[1] and codes for a 75kDa peroxisomal membrane protein called the ABCD1 protein or adrenoleukodystrophy protein (ALDP)[2]. ALDP is highly expressed in specific cell types like oligodendrocytes, astrocytes, microglial cells, adrenocortical cells, and endothelial cells in the brain, adrenal glands, testis, and kidney, liver, lung, and placenta[1,3,4]. ALDP transports very-long-chain fatty acids (VLCFAs), activated by coenzyme-A, into the peroxisomes, for β oxidation. A defect in the ABCD1 gene results in the synthesis of a dysfunctional ALDP protein, unable to transport VLCFA across the peroxisomal membrane. This leads to the buildup of VLCFA, mainly hexacosanoic and tetracosanoic acids, in various body tissues, primarily the brain, spinal cord, adrenal cortex, testis, and plasma[1]. The elevated plasma concentration of VLCFA acts as a diagnostic marker for this disorder.
The exact role of VLCFA in the pathogenesis of X-ALD remains unclear, and no correlation has been established between the concentration of VLCFA and the different phenotypes of X-ALD. The abnormally accumulated VLCFA can disrupt the integrity of the plasma membranes through interdigitating between the leaflets of the lipid bilayer and can induce lipotoxicity, endoplasmic reticulum stress, mitochondrial dysfunction, and oxidative stress leading to apoptosis favoring the process of cerebral demyelination in the brain[5-8].
X-ALD patients have a diverse clinical presentation. They may be asymptomatic or present with the rapidly progressive forms after 3 years of age[1]. The main types of presentation in male patients are: (1) Cerebral ALD (CALD), the cerebral demyelinating form; (2) Adrenomyeloneuropathy (AMN), with spinal cord demyelination and axonal degeneration; and (3) Addison-like phenotype due to adrenocortical insufficiency.
Cerebral ALD affects males and is typified by progressive inflammatory cerebral demyelination leading to neurodegeneration. It includes the childhood cerebral form (CCALD) - appearing in mid-childhood (4-8 years), adolescent cerebral form or adolescent CALD (10-20 years), and adult cerebral form or adult CALD (> 20 years). Children with CCALD present with behavioral problems and a decline in school performance due to impairment of auditory discrimination and spatial orientation, thereby affecting writing and speech. Rarely, seizures may be the initial manifestation. As the disease progresses, there are further signs of damage to the brain white matter, including spastic quadriparesis, dysphagia, and visual loss leading to a vegetative condition. Adolescent CALD manifests between 10 and 20 years of age with clinical features of cerebral involvement. In adult CALD, psychiatric symptoms, seizures, spastic paraparesis, and dementia develop in males over the age of 20.
The second most common phenotype, AMN, is usually characterized as a gradually developing, non-inflammatory axonopathy, mainly affecting males over 20 years of age. AMN is sub-divided as “pure AMN” and “AMN-cerebral.” In patients with pure AMN, there is spinal cord involvement resulting in gait disturbances and bladder dysfunction, whereas patients with AMN-cerebral form show clinical features of cerebral inflammation besides the symptoms of pure AMN[9]. The transformation of pure AMN to the cerebral form of AMN is not clearly understood.
A significant proportion of male patients with X-ALD develop adrenocortical insufficiency, which may occur either after the appearance of neurological symptoms or decades ahead. A majority of males present with adrenocortical insufficiency in association with features of CALD or AMN[1]. Rare cases have shown to manifest adrenocortical insufficiency without cerebral demyelination and are characterized as “Addison only” type of X-ALD[1,10].
In 20%-50% of heterozygotes or female carriers of X-ALD, symptoms similar to AMN, typically consisting of gait disturbance, dysuria, and urgency, occur after 40 years. There are also reports of female carriers with CALD and adrenal insufficiency[11]. For instance, Hershkovitz et al[12] reported a case of CALD in a girl of age, 8.9 years, where the genetic analysis was indicative of heterozygosity with a deletion at Xq27.2-tel. Similarly, Chen et al[13] reported CALD and adrenal insufficiency in a 38-year-old Chinese woman. The possible explanation for the symptomatic state observed in certain heterozygotes could be skewed X-chromosome inactivation, resulting in the expression of the chromosome carrying the faulty ABCD1 gene[14]. Studies have found a significant association between the degree of skewness and the severity of neurological deficits[14,15]. However, factors favoring this event remain unidentified.
The various clinical types of X-ALD frequently appear in the same kindreds and nuclear families carrying the same mutation in the ABCD1 gene. In half of the kindreds, both CALD and AMN are found[16]. Diverse phenotypes and clinical features have been seen in mother and son, monozygotic twins, heterozygous siblings, and affected members of several generations of families[17-19]. For instance, a study reported a family with X-ALD where the proband was diagnosed with the CCALD. In contrast, his two other siblings and maternal uncle were diagnosed with the adolescent form of CALD, Addison’s only phenotype of X-ALD, and AMN. Mutational analysis found a hemizygous mutation of c.1780C>G in the ABCD1 gene in all three siblings[20].
A Brazilian study reports two siblings with CCALD presenting with different clinical features at diagnosis. Both parents had the p.Trp132Ter mutation in ABCD1. Addison's disease phenotype was found in their maternal grandfather[21]. Similarly, different clinical phenotypes have been reported in a Tunisian family with p.Gln316Pro mutation in ABCD1[22].
In an early study of 15 Dutch kindreds, van Geel and coworkers[23] found only CALD in 20%, only AMN in 40%, and both CALD and AMN in 40%. Another large study of 178 kindreds found CALD in 30%, AMN in 20%, and both CALD and AMN in 50%[1].
Korenke et al[19] describe phenotypic variation in monozygotic twins with the same mutation (C2203T) in exon8 of ABCD1, where neuroimaging studies were found normal for the first twin, and parietooccipital demyelination was found in the second twin at ten years of age. Sobue et al[24] also report genetically confirmed monozygotic twins who presented with different clinical types of X-ALD. Although myeloneuropathy was present in both twins, widespread brain demyelination with cognitive dysfunction and behavioral symptoms was pronounced in the older twin, while the younger twin presented with adrenal insufficiency.
Despite various studies, the exact cause of the phenotypic variability is not clear. Over > 800 mutations have been characterized in the ABCD1 gene, but, based on the observations of various studies, it is clear that there is no association between the genotype and the different phenotypes of X-ALD. Identical defects in the ABCD1 gene have been found in cases with different types of X-ALD (CCALD, adult CALD, AMN, Addison only, and asymptomatic)[25,26]. Mutations that can cause deleterious damage to the protein, such as large deletions are reported in severe cerebral forms and milder types such as AMN and asymptomatic cases[27]. These data support the assumption that factors other than the X-linked locus participate in deciding the phenotype. It is possible that the specific mutation, along with the influence of individual genetic and environmental factors commonly referred to as “modifiers”, could play a crucial role in penetrance and the disease severity. Exploring these potential modifiers and understanding their roles in defining the phenotype in X-ALD associated with a specific mutation in the ABCD1 gene is crucial in predicting the disease phenotype.
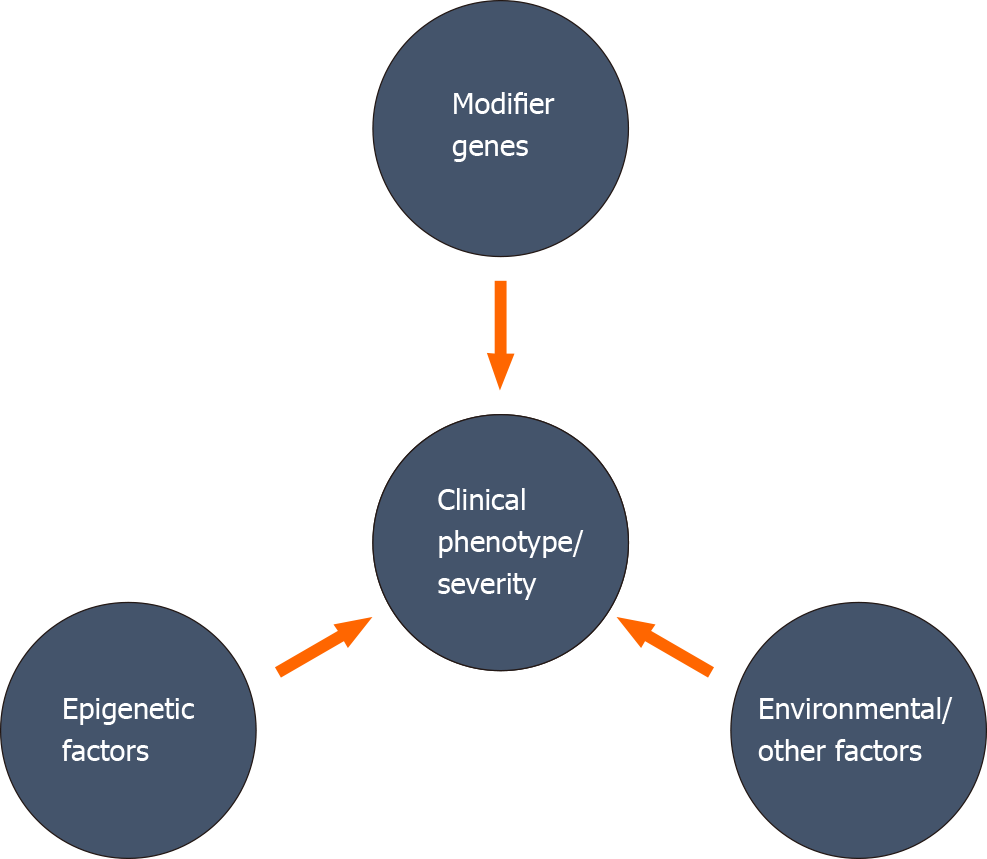
A variant at a particular genetic locus may not be adequate to determine the clinical phenotype, severity, and progression in human diseases[28]. Direct or indirect association of different genetic, epigenetic factors, and environmental factors can change the expressivity, penetrance, and severity of a disease progression (Figure 1). The relative involvement of multiple modifiers to the disease phenotype may generate a combined impact on the phenotypic expression, and the combination of these modifiers may differ among individuals. Identifying these modifying factors and establishing a collective association with different clinical phenotypes is very challenging, but maybe crucial for appropriate management of the disease.
Phenotypic variability of a disease can be explained by the influence of other genes apart from the gene involved in the disease, and these genes are called “modifier genes”[29]. Modifier genes can affect the expression or function of another gene. The final impact of these genes on clinical variability could depend on their collective interaction and the interplay of other epigenetic or environmental factors. Genetic segregation analysis of a considerable number of families with X-ALD and analysis of concordant and discordant siblings indicates that a modifying gene, with an allele frequency of approximately 0.5, could be the main determining factor for phenotypic differences[16,30]. Numerous studies have been directed towards identifying potential modifier genes that control the clinical variability of X-ALD. The foremost challenge for many studies is the small sample size for detecting the genetic association. These studies have attempted to investigate a modifier role in various genes involved in the metabolism of VLCFA, inflammatory pathways, methionine metabolism, and bile acid metabolism (Table 1). However, no studies have attempted to elucidate an interactive association of different genes with different phenotypes associated with the primary mutation in the ABCD1 gene.
| Series No. | Gene name | Variants studied | No. of cases | Inference | Ref. |
| Genes associated with VLCFA metabolism | |||||
| I | ABCD2 | rs11172566 | 117 | No significance | Maier et al[34] |
| rs11172661 | No significance | ||||
| I | ABCD2 | A/T(5’UTR) | 280 | No significance | Matsukawa et al[35] |
| M94V | No significance | ||||
| II | ABCD3 | rs4148058 | 280 | No significance | Matsukawa et al[35] |
| rs2147794 | No significance | ||||
| rs16946 | No significance | ||||
| rs681187 | No significance | ||||
| rs662813 | No significance | ||||
| rs337592 | No significance | ||||
| III | ABCD4 | rs17182959 | 280 | No significance | Matsukawa et al[35] |
| rs17158118 | No significance | ||||
| rs17782508 | No significance | ||||
| rs2301345 | No significance | ||||
| rs4148077 | No significance | ||||
| rs4148078 | No significance | ||||
| rs3742801 | No significance | ||||
| IV | Cytochrome P450 4F subfamily (CYP4F2) | rs21086622 | 152 | Minor allele A associated with CALD (P = 0.036) | van Engen et al[38] |
| rs3093207 | No significance | ||||
| rs1272 | No significance | ||||
| rs3093200 | No significance | ||||
| rs3093194 | No significance | ||||
| rs3093166 | No significance | ||||
| rs4808400 | No significance | ||||
| rs3093153 | No significance | ||||
| rs3093135 | No significance | ||||
| rs3093105 | No significance | ||||
| Genes associated with methionine metabolism | |||||
| I | Cystathionine β-Synthase (CBS) | c.844_845ins68 | 86 | Associated with pure AMN | Linnebank et al[46] |
| I | Cystathionine β-Synthase (CBS) | c.844_845ins68 | 172 | No significance | Semmler et al[48] |
| II | Methionine synthase (MTR) | c.2756A>G | 86 | No significance | Linnebank et al[45] |
| II | Methionine synthase (MTR) | c.2756A>G | 172 | No significance | Semmler et al[48] |
| III | Methylenetetrahydrofolate reductase (MTHFR) | c.677C>T | 86 | No significance | Linnebank et al[45] |
| III | Methylenetetrahydrofolate reductase (MTHFR) | c.677C>T | 172 | No significance | Semmler et al[48] |
| c.1298A>C | No significance | ||||
| IV | Dihydrofolate reductase (DHFR) | c.594+59del19bp | 172 | No significance | Semmler et al[48] |
| V | 5-Methyltetrahydrofolate-Homocysteine Methyltransferase Reductase (MTRR) | c.60A>G | 172 | No significance | Semmler et al[48] |
| VI | Transcobalamin 2 (TC2) | c. 776C>G | 86 | GG genotype prevalent in AMN with demyelination compared to pure AMN (P = 0.001) | Linnebank et al[45] |
| VI | Transcobalamin 2 (TC2) | c. 776C>G (GG) | 172 | GG genotype associated with demyelination (P = 0.036) | Semmler et al[48] |
| VII | Reduced folate carrier 1 (RFC1) | c.80G>A | 172 | No significance | Semmler et al[48] |
| Genes associated with inflammation | |||||
| I | TNF-α | G- 308A | 15 | No significance | McGuinness et al[64] |
| II | Cluster of differentiation (CD1) | CD1A-CD1E | 139 | No significance | Barbier et al[44] |
| III | Human leukocyte antigen (HLA) | HLA-DRB1*16 | 29 | HLA-DRB1*16 associated with X-ALD (P < 0.02) | Berger et al[40] |
| HLA-DRB1*15 | No significance | ||||
| III | Human leukocyte antigen (HLA) | HLA-DBR1* | 70 | No significance | Schmidt et al[41] |
| III | Human leukocyte antigen (HLA) | HLA-DRB1*16 | 106 | No significance | McGuinness et al[65] |
| HLA-DRB1*15 | |||||
| IV | Interleukin 6 (IL6) | 68 | No significance | Schmidt et al[41] | |
| V | Myelin Oligodendrocyte glycoprotein (MOG) | (TAAA)n | 68 | 226bp (TAAA)n polymorphism associated with the presence of Anti-MOG antibody. (P < 0.05). | Schmidt et al[41] |
| V | Myelin oligodendrocyte glycoprotein (MOG) | G15A | 44 | No significance | Gomez-Lira et al[42] |
| CTC 5 repeats | No significance | ||||
| G511C | No significance | ||||
| G520A | No significance | ||||
| 551+68 A→G | No significance | ||||
| 551+77 C→T | No significance | ||||
| Other genes | |||||
| I | Superoxide oxide dismutase (SOD2) | rs4880 | T-allele associated with cerebral involvement in non-CCALD cases | Brose et al[49] | |
| rs2758352 | No significance | ||||
| rs2842980 | No significance | ||||
| rs2758329 | No significance | ||||
| II | Apolipoprotein E (APOE) | rs7412 rs429358 | 83 | APOE4 associated with cerebral involvement | Orchard et al[52] |
| III | Cytochrome P450 family 7 subfamily A member 1 (CYP2A1) | rs3824260 (c.-533T>C) | Study carried out on a patient diagnosed with AMN with c.659T>C mutation in ABCD1 gene in patient and mother | CC allele observed in patient whereas CT in mother | Płatek et al[50] |
| rs3808607 (c.-267C>A) | AA allele observed in patient whereas CA in mother | ||||
| IV | 3 β-hydroxysteroid dehydrogenase type 7 (HSD3B7) | rs9938550 (c.748A>G) | GG allele observed in both | ||
| rs2305880 (c.1068T>C) | CC allele observed in patient | ||||
| V | Bile acyl-CoA synthetase (SLC27A5) | rs4810274 (c.1668-6T>C) | CC observed in patient | ||
| VI | Aldo-keto reductase family 1 member D1 (AKR1D1) | c.-71G>C | GC observed in patient | ||
| VII | Cytochrome P450 Family 27 Subfamily A Member 1 (CYP27A1) | rs397795841 (c.-357dupC) | Homozygous mutation in both | ||
Genes involved in peroxisomal metabolism of VLCFA: The molecular defect in X-ALD is a deficiency of the ALDP protein due to which there is a defective passage of VLCFA into the peroxisome. In the peroxisomal matrix, saturated and unbranched VLCFA are metabolized by enzymes of the β-oxidation pathway[31]. In patients with X-ALD, VLCFA, particularly C26:0, collect in various tissues and are incorporated into different complex lipids. Excess levels of VLCFAs and VLCFA-containing lipids are considered as biochemical triggers playing a central part in the development of X-ALD.
The superfamily of ATP-binding cassette transporters, which ALDP belongs to, also includes ALDRP, PMP70, and ABCD4 coded by ABCD2, ABCD3, and ABCD4 genes. Experimental data suggest that ABCD2 and ABCD3 genes, when over-expressed, can supplement the biochemical defect in ALD fibroblasts[32]. However, Asheuer et al[33] demonstrated that the concentrations of ABCD2 transcripts were similar in the unaffected white brain matter in different ALD phenotypes suggesting that difference in ABCD2-gene expression was not likely to contribute to the vulnerability for cerebral demyelination. In contrast, the expression of ABCD4 genes correlated with the predisposition for brain demyelination and showed a trend of an association with CCALD, AMN-cerebral, and pure AMN phenotypes. Two other independent association studies have reported that ALD phenotypes are not associated with the ABCD2 genotype[34]. A Japanese study found no significant association of SNPs in ABCD2, ABCD3, and ABCD4, and ALD phenotypes, except for five single nucleotide polymorphisms in ABCD4, were less commonly found in AMN patients than in controls, but no significant association with CCALD (Table 1). However, a repetition of this study of five SNPs on another group of French ALD patients found no significant link with CCALD or pure AMN[35].
Accumulation of VLCFA could also result from the excessive lengthening of long-chain fatty acids to VLCFA in the cell[36]. This increased elongation can be due to enhanced expression of elongases and/or imbalance in the degradation and synthesis of VLCFA. Ofman et al[37] reported no change in the expression of ELOVL1 in X-ALD fibroblast, therefore ruling out the possibility of VLCFA accumulation due to increased expression of ELOVL1. However, knockdown of ELOVL1 showed a reduction in C26:0 concentrations in X-ALD fibroblasts, thus indicating that ELOVL1 could be a possible modifier for X-ALD.
The oxidation of VLCFA starts with its activation by coenzyme A and enzymes with very-long-chain acyl-CoA synthetase (VLACS) activity. In a study of the unaffected brain white matter from X-ALD cases, Asheuer et al[33] have found that the expression of VLACS genes did not correlate with the clinical phenotypes (CALD and AMN phenotypes). On the contrary, lower expression of the BG1 gene, which codes for a non-peroxisomal synthetase which activates VLCFA to its coenzyme A derivatives, was found in the white matter of ALD patients which correlated with the presence of cerebral demyelination. Hence BG1 could be considered as a potential modifier gene[33].
VLCFAs can also undergo ω-oxidation, and are further converted to dicarboxylic acids by the cytochrome P450 system. These reactions may present another route for the metabolism of the accumulated VLCFAs. The gene, CYP4F2, codes for a critical enzyme in the ω-oxidation of VLCFA to very long-chain dicarboxylic acids. van Engen et al[38] reported that the CYP4F2 polymorphism (CYP4F2 p.433M) increased the chances of acquiring CALD in male Caucasians (Table 1). They further demonstrated the functional impact of the CYP4F2 p.433M variant on cellular models, which showed reduced CYP4F2 protein level, led to a reduction in the metabolism of VLCFA through ω-oxidation.
Inflammation-related genes: Brain inflammation and the ensuing progressive inflammatory demyelination is characteristically found in the demyelinating forms of X-ALD. Acute inflammation occurs only in the CNS and not in other tissues of the affected cases[39]. Variants in the genes playing a role inflammation have been speculated to influence disease variability. The putative modifying genes could participate in an inflammatory response to the buildup of VLCFA or some other related metabolite in the brain. Since the pathology of the cerebral form is akin to that seen in multiple sclerosis (MS), some of the genetic factors involved in triggering inflammation in multiple sclerosis could also participate in the pathogenesis of X-ALD.
Genetic variants of specific major histocompatibility complex class II antigens (HLA-DRB1) are reported to be associated with the risk for MS and could be suitable candidate modifiers for X-ALD. Berger et al[40] described a significant relationship between the HLA-DRB1*16 allele and X-ALD. However, this allele did not show association with CALD, the inflammatory phenotype of X-ALD. The DRB genes are involved in the synthesis of peptides receptors playing a central role in the immune system.
Myelin oligodendrocyte glycoprotein (MOG) is the main
Tumor necrosis factor (TNF-α), a major pro-inflammatory cytokine, is involved in the pathogenesis of many neurological disorders including MS. TNF-α is capable of causing damage to the myelin sheath and oligodendrocytes and has also found to modulate the MBP (Myelin basic protein) gene promoter activity, via activation of NF-κB transcriptional factor in oligodendroglioma cells[43]. However, increased TNF-α bioactivity was not found to be associated with any allelic difference in the TNF-α gene (G- 308A).
Genetic variants of the cytokine, interleukin-6 (IL-6), such as the IL-6 C-allele which is a variable number tandem repeat polymorphism situated on the 3’ flanking region of the IL-6 gene, is reported to be linked to late-onset Alzheimer’s and MS, but no association was found with the different clinical phenotypes of X-ALD[41].
Neuroinflammation in CALD is suspected to be due to the involvement of different classes of lipids enriched in the VLCFA[2]; thus, the participation of CD1, a lipid antigen-presenting molecule, was speculated. Barbier et al[44] assessed the association between the genetic variants of CD1 molecules (CD1A-E) and the presence of neuroinflammation in X-ALD but found no association between them.
Genes associated with methionine metabolism: The pathological characteristic of the cerebral type of X-ALD is CNS demyelination. Demyelination starts in the mid of corpus callosum and advances outwards in both brain hemispheres. This leads to a gradual neurologic decline and death within 3 to 5 years[3].
The sulfur-containing amino acid, methionine, plays a vital metabolic role in providing methyl group required for DNA methylation, brain myelination, and precursors for the generation of glutathione taurine. S-adenosyl methionine (SAM), the active form of methionine, is a methyl donor. Deficiency of SAM can lead to demyelination in the CNS. Studies have reported variants of methionine metabolism as risk factors causing demyelination in X-ALD patients. Linnebank et al[45] studied the combined risk genotype, i.e. the occurrence of a minimum of one distinct genotype of three functional polymorphisms in genes associated with methionine metabolism, 5,10-methylenetetrahydrofolate reductase (MTHFR) c.677CT, methionine synthase (MTR) c.2756AG, and transcobalamin 2 (Tc2) c.776CG, in 86 patients with various phenotypes of X-ALD. These authors reported that CCALD patients tended to have a higher prevalence of the combined risk genotype (46%) in comparison to the group with the benign variant "pure" AMN (33%; P = 0.222) due to a higher prevalence of the MTR (41% vs 22%, P = 0.110) and the Tc2 risk genotype (18% vs 14%, P = 0.675). Moreover, this genotype was overrepresented in patients with AMN with CNS demyelination (AMN-cerebral) when compared to 49 AMN patients without CNS demyelination (“pure” AMN) and suggested that variations in genes associated with methionine metabolism might influence the phenotypic variability in X-ALD. Cystathionine β-synthase (CBS) is another important enzyme in the methionine metabolic pathway, and the CBS c.844_845ins68 variant may affect the availability or concentrations of activated methionine and glutathione. Linnebank and colleagues also found that CBS c.844_845ins68 insertion allele protected X-ALD patients from cerebral demyelination[46].
In another study of CBS c.844_855ins68, MTR c.2756A to G, and TC2 c.776 C to G in 120 Chinese ALD patients, the frequency of only the GG genotype of the TC2 c.776 C/G was more in those with brain demyelination than in controls[47]. TC2 is the transport carrier protein for cobalamin and methylcobalamin, the active form of cobalamin, a crucial cofactor necessary for the enzymatic activity of methionine synthase.
These results were further confirmed by Semmler et al[48] who genotyped eight polymorphisms in methionine metabolism genes, including CBS c.844_855ins68, MTHFR c.677C>T, MTR c.2756A>G and DHFR c.594+59del19bp, and found Tc2 c.776 GG genotype to be more prevalent in X-ALD cases with clinical features of brain demyelination compared to those without demyelination.
Other potential genetic modifiers: Reactive oxygen species (ROS) can trigger oxidative damage to DNA and proteins and ineffective oxidative phosphorylation, and this could result in dying-back axonopathy. Axonal degeneration in the spinal cord is typically observed in the AMN form of X-ALD. The mitochondrial superoxide dismutase (SOD2) is responsible for detoxifying ROS and is considered a modifying factor for the development of demyelination in X-ALD. A study reported that SOD2 variant C47T and GTAC haplotype with reduced activity were associated with adolescent cerebral, adult cerebral X-ALD, and AMN- cerebral patients[49] (Table 1).
Bile acid metabolism occurs in the peroxisomes. An abnormal bile acid profile and mutations in the genes associated with the metabolism of bile acids such as CYP7A1, CYP27A1, CYP7B1, HSD3B7, AKR1D1, and SLC27A52, has been reported in a Polish AMN patient and these genes have been suggested as potential modifiers of X-ALD[50] (Table 1). However, more studies are required to confirm this association
Apolipoprotein E, a protein associating with lipid particles and functioning in lipoprotein-mediated lipid transport between organs, has three isoforms APOE2, APOE3 and APOE4 encoded by three alleles situated on a single gene locus. APOE3 protein maintains the blood-brain-barrier integrity (BBB) through the downregulation of cyclophilin A (CypA), a pro-inflammatory protein[51]. Male X-ALD patients bearing the APOE4 genotype are reported to have greater cerebral involvement as determined by MRI severity score, lesser neurologic function, and elevated concentrations of matrix metalloproteinase-2 (MMP-2) in the cerebrospinal fluid compared to non-carriers[52]. The presence of the APOE4 allele has been suggested to upregulate CypA leading to the activation of MMP-9 and loss of BBB integrity, leading to increased severity of cerebral disease in cerebral ALD[51].
Epigenetic factors, too, can influence the onset of disease by inducing a subtle change in the gene expression without any notable alteration in the DNA sequence. Epigenetic alterations comprise DNA methylation, post-translational modifications of histones such as methylation, phosphorylation, acetylation, and post-transcriptional regulation by non-coding RNA.
DNA methylation acts as a regulatory mechanism for gene expression, and cell differentiation and various studies have demonstrated the association between change in DNA methylation and disease pathogenesis[53]. A study by Schlüter et al[54] compared the genome-wide DNA methylation pattern of unaffected frontal brain white matter of patients with CCALD and AMN with cerebral involvement and found hypermethylation of genes that are majorly involved in differentiation of oligodendrocytes including MBP, CNP, MOG, PLP1 that can result to impaired differentiation of oligodendrocyte precursor cells to remyelinating oligodendrocyte and hypomethylation of genes associated with an immune function such as IFITM1 and CD59. This supports the neuropathological evidence of lack of remyelination and immune activation noted in the cerebral form of X-ALD. This study also showed that combined methylation levels of SPG20, UNC45A, and COL9A3 and combined expression levels of ID4 and MYRF could be useful as biomarkers for differentiating CALD from AMN.
Aberrant expression of microRNAs (miRNAs), a group of small non-coding RNAs regulating post-transcriptional gene expression, has been suggested to play a significant part in the development of neuroinflammation and degeneration[55]. Shah et al[55] found decreased expression of miR-196a and increased expression of ELOVL, IKKα, IKKβ, MAP4K3, and MAP3K2 in cerebral ALD compared to AMN and control fibroblasts, and suggested that the regulation of inflammatory signaling pathway in CALD brain occurs via miR-196a.
Various host or cellular environmental factors may influence disease development in an individual. Oxidative stress is a common phenomenon reported in various neurodegenerative disorders, including X-ALD. Overproduction of free radicles results in lipid peroxidation, whose byproducts can cause deleterious damage to the cells[56]. Nury et al[57] observed reduced plasma levels of oxidative stress markers such as α-tocopherol, GSH, and docosahexaenoic acid (DHA) in different X-ALD phenotypes. These authors also showed that 7α-hydroxycholesterol, 7β-hydroxycholesterol, 7-ketocholesterol, and 9- and 13-hydroxyoctadecadienoic acids were produced as a result of oxidative stress. Increased level of 7-ketocholesterol was found to cause overproduction of free radicles, activation of PRAP-1, and caspase 3 and elevated LC3-II/LC3-I and p62 in BV-12 microglia cells, indicating its ability to induce cell death[53]. A recent study demonstrated 7-ketocholesterol induced activation of PRAP-1 via NF-κB transforms microglial cells from a resting stage to an active stage, ultimately damaging the neurons. As 7-ketocholesterol induces oxidative stress, inflammation, and cell death, high levels could enhance peroxisomal dysfunction in microglial cells, promoting brain damage in the affected patients.
Jang et al[58] demonstrated the abnormal generation of cholesterol 25-hydroxylase and 25-hydroxycholesterol in CCALD patient-derived cell models and showed that 25-hydroxycholesterol aids the aggregation and activation of NLRP3 inflammasome, a caspase-1- activating multi-protein complex, resulting in increased formation of pro-inflammatory cytokines, IL-1β, IL-18[59]. 25-hydroxycholesterol has also been found to induce mitochondrial-dependent apoptosis of cells via the stimulation of glycogen synthesis kinase-3β (GSK-3β)/LXR pathway in the amyotrophic lateral sclerosis cell model[60]. This could also account for severe cerebral inflammatory demyelination, the hallmark of CCALD.
Trauma to the head has been speculated to trigger or worsen symptoms in X-ALD[61], and asymptomatic cases of X-ALD presenting with symptoms after head trauma have been reported. The inflammatory response following a traumatic brain injury, followed by mitochondrial dysfunction, oxidative stress, and disruption of the BBB, has been suggested to activate cerebral inflammatory demyelination resulting in the appearance of symptoms[62].
The major difference between the different X-ALD phenotypes is the presence or absence of neuroinflammation and cerebral demyelination. The inflammatory response in the brain is believed to begin after the abnormal accumulation of VLCFA. ALDP deficiency has been shown to induce alteration in brain endothelial cells favoring the migration of leukocytes by downregulating the expression of c-Myc, leading to a reduction in the expression of cell surface tight junction proteins CLDN5 and ZO1 and increased expression of cell adhesion molecule ICAM-1 and MMP9[63]. We speculate that the migration of immune cells into the brain could be a rate-limiting step in the induction of cerebral demyelination. The onset of the migration across the BBB could be precipitated by various environmental triggers, genetic or epigenetic factors and ABCD1 deficiency acting either alone or in concert, marking the onset of brain inflammation leading to cerebral symptoms (Figure 2). Further, the absence of neuroinflammation in “pure AMN” and “Addison’s only” phenotype could also possibly be due to the involvement of modifiers that are protective against the VLCFA toxicity in the brain. For instance, it is commonly known that MMP2 and MMP9 are required for the migration of the immune cells across the endothelial basal membrane and parenchymal border, respectively. However, the delay in the synthesis and secretion of MMP2 and MMP9 could delay the migration process and, ultimately, the disease onset. Genetic factors such as APOE4, which is shown to be associated with cerebral involvement in young males, are also associated with increased expression of MMP9 via cyclophilin A, leading to BBB leakiness. Thus, a complex interplay between multiple determinants could affect the onset and severity of the disease symptoms.
The role of different modifiers influencing disease phenotypes has been described in various metabolic disorders. Therefore, though X-ALD is a monogenic disorder, other genetic factors, along with the environmental triggers, may be responsible for the severity and penetrance of the disease. Although numerous studies have made efforts to understand different genetic, epigenetic, and environmental factors in X-ALD, the exact cause of phenotypic differences in X-ALD patients with the same genotype is not clear. Improved knowledge of these factors will allow identification of patients prone to developing a particular form or clinical type of X-ALD. Besides, detailed elucidation of the association of different potential modifiers with the clinical heterozygosity in X-ALD is crucial for understanding the disease pathogenesis and for developing novel therapeutic strategies. With the introduction of neonatal screening for X-ALD, X-ALD modifiers will become increasingly essential to categorize patients who are likely to develop cerebral demyelination and plan appropriate management of these patients.
Manuscript source: Invited manuscript
Specialty type: Neurosciences
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Demarquoy J S-Editor: Wang JL L-Editor: Filipodia P-Editor: Li X
| 1. | Moser HW, Mahmood A, Raymond GV. X-linked adrenoleukodystrophy. Nat Clin Pract Neurol. 2007;3:140-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 242] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 2. | Kemp S, Wanders R. Biochemical aspects of X-linked adrenoleukodystrophy. Brain Pathol. 2010;20:831-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Berger J, Forss-Petter S, Eichler FS. Pathophysiology of X-linked adrenoleukodystrophy. Biochimie. 2014;98:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 4. | Kemp S, Pujol A, Waterham HR, van Geel BM, Boehm CD, Raymond GV, Cutting GR, Wanders RJ, Moser HW. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: role in diagnosis and clinical correlations. Hum Mutat. 2001;18:499-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 207] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Parisi LR, Sowlati-Hashjin S, Berhane IA, Galster SL, Carter KA, Lovell JF, Chemler SR, Karttunen M, Atilla-Gokcumen GE. Membrane Disruption by Very Long Chain Fatty Acids during Necroptosis. ACS Chem Biol. 2019;14:2286-2294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Khan M, Singh J, Gilg AG, Uto T, Singh I. Very long-chain fatty acid accumulation causes lipotoxic response via 5-lipoxygenase in cerebral adrenoleukodystrophy. J Lipid Res. 2010;51:1685-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | van de Beek MC, Ofman R, Dijkstra I, Wijburg F, Engelen M, Wanders R, Kemp S. Lipid-induced endoplasmic reticulum stress in X-linked adrenoleukodystrophy. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2255-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Launay N, Ruiz M, Fourcade S, Schlüter A, Guilera C, Ferrer I, Knecht E, Pujol A. Oxidative stress regulates the ubiquitin-proteasome system and immunoproteasome functioning in a mouse model of X-adrenoleukodystrophy. Brain. 2013;136:891-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Powers JM, DeCiero DP, Ito M, Moser AB, Moser HW. Adrenomyeloneuropathy: a neuropathologic review featuring its noninflammatory myelopathy. J Neuropathol Exp Neurol. 2000;59:89-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 125] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | van Geel BM, Assies J, Wanders RJ, Barth PG. X linked adrenoleukodystrophy: clinical presentation, diagnosis, and therapy. J Neurol Neurosurg Psychiatry. 1997;63:4-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Jangouk P, Zackowski KM, Naidu S, Raymond GV. Adrenoleukodystrophy in female heterozygotes: underrecognized and undertreated. Mol Genet Metab. 2012;105:180-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Hershkovitz E, Narkis G, Shorer Z, Moser AB, Watkins PA, Moser HW, Manor E. Cerebral X-linked adrenoleukodystrophy in a girl with Xq27-Ter deletion. Ann Neurol. 2002;52:234-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Chen X, Chen Z, Huang D, Liu X, Gui Q, Yu S. Adult cerebral adrenoleukodystrophy and Addison's disease in a female carrier. Gene. 2014;544:248-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Maier EM, Kammerer S, Muntau AC, Wichers M, Braun A, Roscher AA. Symptoms in carriers of adrenoleukodystrophy relate to skewed X inactivation. Ann Neurol. 2002;52:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Wang Z, Yan A, Lin Y, Xie H, Zhou C, Lan F. Familial skewed x chromosome inactivation in adrenoleukodystrophy manifesting heterozygotes from a Chinese pedigree. PLoS One. 2013;8:e57977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Moser HW, Moser AB, Smith KD, Bergin A, Borel J, Shankroff J, Stine OC, Merette C, Ott J, Krivit W. Adrenoleukodystrophy: phenotypic variability and implications for therapy. J Inherit Metab Dis. 1992;15:645-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 163] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Simpson RH, Rodda J, Reinecke CJ. Adrenoleukodystrophy in a mother and son. J Neurol Neurosurg Psychiatry. 1987;50:1165-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Elrington GM, Bateman DE, Jeffrey MJ, Lawton NF. Adrenoleukodystrophy: heterogeneity in two brothers. J Neurol Neurosurg Psychiatry. 1989;52:310-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Korenke GC, Fuchs S, Krasemann E, Doerr HG, Wilichowski E, Hunneman DH, Hanefeld F. Cerebral adrenoleukodystrophy (ALD) in only one of monozygotic twins with an identical ALD genotype. Ann Neurol. 1996;40:254-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Gosalakkal J, Balky AP. Intra familial phenotypical variations in adrenoleukodystrophy. Neurol India. 2010;58:109-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Soardi FC, Esquiaveto-Aun AM, Guerra-Júnior G, Lemos-Marini SH, Mello MP. Phenotypic variability in a family with x-linked adrenoleukodystrophy caused by the p.Trp132Ter mutation. Arq Bras Endocrinol Metabol. 2010;54:738-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Kallabi F, Ellouz E, Tabebi M, Ben Salah G, Kaabechi N, Keskes L, Triki C, Kamoun H. Phenotypic variability in a Tunisian family with X-linked adrenoleukodystrophy caused by the p.Gln316Pro novel mutation. Clin Chim Acta. 2016;453:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | van Geel BM, Assies J, Weverling GJ, Barth PG. Predominance of the adrenomyeloneuropathy phenotype of X-linked adrenoleukodystrophy in The Netherlands: a survey of 30 kindreds. Neurology. 1994;44:2343-2346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Sobue G, Ueno-Natsukari I, Okamoto H, Connell TA, Aizawa I, Mizoguchi K, Honma M, Ishikawa G, Mitsuma T, Natsukari N. Phenotypic heterogeneity of an adult form of adrenoleukodystrophy in monozygotic twins. Ann Neurol. 1994;36:912-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Kemp S, Ligtenberg MJ, van Geel BM, Barth PG, Wolterman RA, Schoute F, Sarde CO, Mandel JL, van Oost BA, Bolhuis PA. Identification of a two base pair deletion in five unrelated families with adrenoleukodystrophy: a possible hot spot for mutations. Biochem Biophys Res Commun. 1994;202:647-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Kok F, Neumann S, Sarde CO, Zheng S, Wu KH, Wei HM, Bergin J, Watkins PA, Gould S, Sack G. Mutational analysis of patients with X-linked adrenoleukodystrophy. Hum Mutat. 1995;6:104-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature. 1993;361:726-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 817] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 28. | Gallati S. Disease-modifying genes and monogenic disorders: experience in cystic fibrosis. Appl Clin Genet. 2014;7:133-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 29. | Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 393] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 30. | Smith KD, Sack G, Beaty T, Bergin A, Naidu S, Moser A MH. A genetic-basis for the multiple phenotypes of X-linked adrenoleukodystrophy. Am J Hum Genet. 1991;49:165-165. |
| 31. | Sassa T, Kihara A. Metabolism of very long-chain Fatty acids: genes and pathophysiology. Biomol Ther (Seoul). 2014;22:83-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 32. | Smith KD, Kemp S, Braiterman LT, Lu JF, Wei HM, Geraghty M, Stetten G, Bergin JS, Pevsner J, Watkins PA. X-linked adrenoleukodystrophy: genes, mutations, and phenotypes. Neurochem Res. 1999;24:521-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 127] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 33. | Asheuer M, Bieche I, Laurendeau I, Moser A, Hainque B, Vidaud M, Aubourg P. Decreased expression of ABCD4 and BG1 genes early in the pathogenesis of X-linked adrenoleukodystrophy. Hum Mol Genet. 2005;14:1293-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Maier EM, Mayerhofer PU, Asheuer M, Köhler W, Rothe M, Muntau AC, Roscher AA, Holzinger A, Aubourg P, Berger J. X-linked adrenoleukodystrophy phenotype is independent of ABCD2 genotype. Biochem Biophys Res Commun. 2008;377:176-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Matsukawa T, Asheuer M, Takahashi Y, Goto J, Suzuki Y, Shimozawa N, Takano H, Onodera O, Nishizawa M, Aubourg P, Tsuji S. Identification of novel SNPs of ABCD1, ABCD2, ABCD3, and ABCD4 genes in patients with X-linked adrenoleukodystrophy (ALD) based on comprehensive resequencing and association studies with ALD phenotypes. Neurogenetics. 2011;12:41-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Kemp S, Valianpour F, Denis S, Ofman R, Sanders RJ, Mooyer P, Barth PG, Wanders RJ. Elongation of very long-chain fatty acids is enhanced in X-linked adrenoleukodystrophy. Mol Genet Metab. 2005;84:144-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 37. | Ofman R, Dijkstra IM, van Roermund CW, Burger N, Turkenburg M, van Cruchten A, van Engen CE, Wanders RJ, Kemp S. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol Med. 2010;2:90-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 38. | van Engen CE, Ofman R, Dijkstra IM, van Goethem TJ, Verheij E, Varin J, Vidaud M, Wanders RJ, Aubourg P, Kemp S, Barbier M. CYP4F2 affects phenotypic outcome in adrenoleukodystrophy by modulating the clearance of very long-chain fatty acids. Biochim Biophys Acta. 2016;1862:1861-1870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 39. | Moser HW, Smith KD, Watkins PA, Powers J MA. X-linked adrenoleukodystrophy. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease (Vol. 4). New York: McGraw Hill, 2001: 3257-3301. |
| 40. | Berger J, Bernheimer H, Faé I, Braun A, Roscher A, Molzer B, Fischer G. Association of X-linked adrenoleukodystrophy with HLA DRB1 alleles. Biochem Biophys Res Commun. 1995;216:447-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Schmidt S, Marrosu GM, Kölsch H, Haase CG, Ferenczik S, Sokolowski P, Köhler W, Schmidt M, Papassotiropoulos A, Heun R, Grosse-Wilde H, Klockgether T. Genetic variations and humoral immune responses to myelin oligodendroglia glycoprotein in adult phenotypes of X-linked adrenoleukodystrophy. J Neuroimmunol. 2003;135:148-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 42. | Gomez-Lira M, Marzari MG, Uziel G, Pignatti P, Rizzuto N, Salviati A. Myelin oligodendrocyte glycoprotein (MOG) polymorphisms and adrenoleukodystrophy. J Neuroimmunol. 2000;111:245-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Huang CJ, Nazarian R, Lee J, Zhao PM, Espinosa-Jeffrey A, de Vellis J. Tumor necrosis factor modulates transcription of myelin basic protein gene through nuclear factor kappa B in a human oligodendroglioma cell line. Int J Dev Neurosci. 2002;20:289-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 44. | Barbier M, Sabbagh A, Kasper E, Asheuer M, Ahouansou O, Pribill I, Forss-Petter S, Vidaud M, Berger J, Aubourg P. CD1 gene polymorphisms and phenotypic variability in X-linked adrenoleukodystrophy. PLoS One. 2012;7:e29872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Linnebank M, Kemp S, Wanders RJ, Kleijer WJ, van der Sterre ML, Gärtner J, Fliessbach K, Semmler A, Sokolowski P, Köhler W, Schlegel U, Schmidt S, Klockgether T, Wüllner U. Methionine metabolism and phenotypic variability in X-linked adrenoleukodystrophy. Neurology. 2006;66:442-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 46. | Linnebank M, Semmler A, Kleijer WJ, van der Sterre ML, Gärtner J, Fliessbach K, Sokolowski P, Köhler W, Schlegel U, Klockgether T, Wanders RJ, Schmidt S, Wüllner U, Kemp S. The cystathionine beta-synthase variant c.844_845ins68 protects against CNS demyelination in X-linked adrenoleukodystrophy. Hum Mutat. 2006;27:1063-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 47. | Cao GN, Bao XH, Xiong H, Wu Y, Wu XR. [Association of genetic polymorphisms in methionine metabolism genes with X-linked adrenoleukodystrophy]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011;28:279-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 48. | Semmler A, Bao X, Cao G, Köhler W, Weller M, Aubourg P, Linnebank M. Genetic variants of methionine metabolism and X-ALD phenotype generation: results of a new study sample. J Neurol. 2009;256:1277-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 49. | Brose RD, Avramopoulos D, Smith KD. SOD2 as a potential modifier of X-linked adrenoleukodystrophy clinical phenotypes. J Neurol. 2012;259:1440-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Płatek T, Orso E, Zapała B, Polus A, Kieć-Wilk B, Piwowar M, Chojnacka M, Ciałowicz U, Malczewska-Malec M, Schmitz G, Solnica B, Dembińska-Kieć A. Case report of dysregulation of primary bile acid synthesis in a family with X-linked adrenoleukodystrophy. Medicine (Baltimore). 2018;97:e13353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 51. | Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, Berk BC, Zlokovic BV. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 993] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 52. | Orchard PJ, Markowski TW, Higgins L, Raymond GV, Nascene DR, Miller WP, Pierpont EI, Lund TC. Association between APOE4 and biomarkers in cerebral adrenoleukodystrophy. Sci Rep. 2019;9:7858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Fedotova EY, Illarioshkin SN. DNA Methylation in Neurodegenerative Diseases. Russ J Gene. 55:271–277. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 54. | Schlüter A, Sandoval J, Fourcade S, Díaz-Lagares A, Ruiz M, Casaccia P, Esteller M, Pujol A. Epigenomic signature of adrenoleukodystrophy predicts compromised oligodendrocyte differentiation. Brain Pathol. 2018;28:902-919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 55. | Shah N, Singh I. MicroRNA Profiling Identifies miR-196a as Differentially Expressed in Childhood Adrenoleukodystrophy and Adult Adrenomyeloneuropathy. Mol Neurobiol. 2017;54:1392-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Ranea-Robles P, Launay N, Ruiz M, Calingasan NY, Dumont M, Naudí A, Portero-Otín M, Pamplona R, Ferrer I, Beal MF, Fourcade S, Pujol A. Aberrant regulation of the GSK-3β/NRF2 axis unveils a novel therapy for adrenoleukodystrophy. EMBO Mol Med. 2018;10:e8604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 57. | Nury T, Zarrouk A, Ragot K, Debbabi M, Riedinger JM, Vejux A, Aubourg P, Lizard G. 7-Ketocholesterol is increased in the plasma of X-ALD patients and induces peroxisomal modifications in microglial cells: Potential roles of 7-ketocholesterol in the pathophysiology of X-ALD. J Steroid Biochem Mol Biol. 2017;169:123-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 58. | Jang J, Park S, Jin Hur H, Cho HJ, Hwang I, Pyo Kang Y, Im I, Lee H, Lee E, Yang W, Kang HC, Won Kwon S, Yu JW, Kim DW. 25-hydroxycholesterol contributes to cerebral inflammation of X-linked adrenoleukodystrophy through activation of the NLRP3 inflammasome. Nat Commun. 2016;7:13129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 59. | Yang Y, Wang H, Kouadir M, Song H, Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019;10:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 538] [Cited by in RCA: 908] [Article Influence: 151.3] [Reference Citation Analysis (0)] |
| 60. | Vejux A, Lizard G. Cytotoxic effects of oxysterols associated with human diseases: Induction of cell death (apoptosis and/or oncosis), oxidative and inflammatory activities, and phospholipidosis. Mol Aspects Med. 2009;30:153-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 235] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 61. | Budhram A, Pandey SK. Activation of Cerebral X-linked Adrenoleukodystrophy After Head Trauma. Can J Neurol Sci. 2017;44:597-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 62. | Bouquet F, Dehais C, Sanson M, Lubetzki C, Louapre C. Dramatic worsening of adult-onset X-linked adrenoleukodystrophy after head trauma. Neurology. 2015;85:1991-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 63. | Musolino PL, Gong Y, Snyder JM, Jimenez S, Lok J, Lo EH, Moser AB, Grabowski EF, Frosch MP, Eichler FS. Brain endothelial dysfunction in cerebral adrenoleukodystrophy. Brain. 2015;138:3206-3220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 64. | McGuinness MC, Griffin DE, Raymond GV, Washington CA, Moser HW, Smith KD. Tumor necrosis factor-alpha and X-linked adrenoleukodystrophy. J Neuroimmunol. 1995;61:161-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 65. | McGuinness MC, Powers JM, Bias WB, Schmeckpeper BJ, Segal AH, Gowda VC, Wesselingh SL, Berger J, Griffin DE, Smith KD. Human leukocyte antigens and cytokine expression in cerebral inflammatory demyelinative lesions of X-linked adrenoleukodystrophy and multiple sclerosis. J Neuroimmunol. 1997;75:174-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.4] [Reference Citation Analysis (0)] |