Published online Sep 27, 2020. doi: 10.4331/wjbc.v11.i2.14
Peer-review started: June 23, 2020
First decision: July 25, 2020
Revised: August 1, 2020
Accepted: September 8, 2020
Article in press: September 8, 2020
Published online: September 27, 2020
Processing time: 92 Days and 17 Hours
The newly emerged coronavirus (severe acute respiratory syndrome coronavirus 2 SARS-CoV-2) and the disease that it causes coronavirus disease 2019 (COVID-19) have changed the world we know. Yet, the origin and evolution of SARS-CoV-2 remain mostly vague. Many virulence factors and immune mechanisms contribute to the deteriorating effects on the organism during SARS-CoV-2 infection. Both humoral and cellular immune responses are involved in the pathophysiology of the disease, where the principal and effective immune response towards viral infection is the cell-mediated immunity. The clinical picture of COVID-19, which includes immune memory and reinfection, remains unclear and unpredictable. However, many hopes are put in developing an effective vaccine against the virus, and different therapeutic options have been implemented to find effective, even though not specific, treatment to the disease. We can assume that the interaction between the SARS-CoV-2 virus and the individual's immune system determines the onset and development of the disease significantly.
Core Tip: Since the coronavirus disease 2019 (COVID-19) results from the interaction between the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus and the individual′s immune system, we can assume that its onset and development significantly depend on this communication. Immunological aspects of the disease reflect the importance of the immune system to inhibit the viral factors and to control and regulate the pathophysiological processes during SARS-CoV-2 infection. Moreover, immune-mediated and humoral immune responses, immune memory, the cytokine storm, and neuroendocrine-immune regulation are critical factors that can determine the prognosis and outcome for patients. Now, the science is directed to acquiring new data on the immunology, including immune memory against the virus, the development of new technologies for the detection of infection and effective vaccines. However, much more information remains unclear than verified knowledge of the SARS-CoV-2 virus and COVID-19.
- Citation: Velikova TV, Kotsev SV, Georgiev DS, Batselova HM. Immunological aspects of COVID-19: What do we know? World J Biol Chem 2020; 11(2): 14-29
- URL: https://www.wjgnet.com/1949-8454/full/v11/i2/14.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v11.i2.14
The novel coronavirus (formerly called HCoV-19) is a new coronavirus in humans that emerged at the end of 2019 (December) in Wuhan, China. Later, it received the name the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that stands for severe acute respiratory syndrome coronavirus 2. Now it is the cause of the current pandemic[1]. Not surprisingly, coronaviruses (CoV) were intensively studied for the last decade, especially during the last months of the current pandemic, but not enough information is elucidated for them. It is known that CoV are zoonotic, single-stranded ribonucleic acid (RNA) viruses that cause a wide range of symptoms. The latter include those from common cold to more severe respiratory complaints as well as gastroenteric, hepatic, and neurological symptoms[2]. Except SARS-CoV-2, there are six other reported human coronavirus members. These are HCoV-OC43, HCoV229E, SARS-CoV, HCoV-HKU1, Middle East respiratory syndrome corona virus (MERS-CoV), and HCoVNL63[3,4]. Over the last twenty years, CoV have caused two significant epidemics: SARS[5] and MERS[6].
The positive-sense single-stranded RNA of SARS-CoV-2 is enveloped in a lipid bilayer. The virus belongs to the genus Betacoronavirus and family Coronaviridae[7]. Yet, the origin and evolution of SARS-CoV-2 remain vague. Furthermore, SARS-CoV-2-related viruses were found in Malayan pangolins (Manis javanica), as several recent studies showed. These data provided new insights into the evolution and host distribution of these SARS-CoV-2-related viruses[8,9].
Coronavirus disease 2019 (COVID-19) is the illness associated with SARS-CoV-2 infection. The clinical syndrome is characterized by variable symptoms, ranging from mild upper respiratory symptoms to severe interstitial pneumonia and acute respiratory distress syndrome (ARDS)[10,11].
SARS-CoV-2 and SARS-CoV and MERS-CoV belong to the same Betacoronavirus genus, and they share about 80% nucleotide identity. However, despite the close relation between SARS-CoV and SARS-CoV-2, the latter seems to cause milder infections[7]. Moreover, SARS and MERS were characterized mainly with nosocomial spread, whereas SARS-CoV-2 has community transmission[12].
Regarding clinical features, COVID-19 seems similar to SARS; however, it is considered to be less lethal than MERS, which differs from the other two CoV in terms of both phylogenetic and pathogenetic features. Due to the less severe clinical picture, COVID-19 can spread in the community more easily than MERS and SARS, which is reported in the nosocomial settings[13-15].
The spread of COVID-19 is rapid, which is somehow expected because the transfer is carried out by close contact and droplets[16]. However, there is scarce evidence to suggest airborne transmission, as very minimal to no viral RNA was detected in airborne samples, and no viral RNA was found in urine or serum samples of positive patients[17].
Under the experimental circumstances tested, the stability of SARS-CoV-2 is similar to that of SARS-CoV-1. This indicates that their different epidemiologic features are probably due to other factors, such as high viral concentrations in the upper airways and the potential asymptomatic spread of SARS-CoV-2[18,19]. Also, the aerosol and fomite transmission of SARS-CoV-2 is likely since the virus can remain infectious depending on the inoculum shed. It was shown that the virus is viable in aerosols for hours and on surfaces up to days. These findings mirror those of SARS-CoV-1, in which both the nosocomial and super-spreading transmissions were observed. All of these characteristics provide information for pandemic mitigation efforts[20]. In summary, current evidence state that the COVID-19 virus is primarily transferred between people through respiratory droplets and contact routes[21]. Transmission due to droplets is documented in close contact (considered within 1-1.5 m) with a symptomatic person (e.g., coughing or sneezing). The risk is estimated by the exposition of mucosae (mouth and nose) or conjunctiva (eyes) to potentially infective respiratory droplets. Fomites in the immediate environment around the infected person can also lead to transmission[22]. Therefore, the virus can be spread directly by infected people and indirectly through surfaces or objects used by the infected person[22].
Coronavirus is one of the significant pathogens that targets the human respiratory system primarily[10]. SARS-CoV-2 was found to have higher rates of transmissibility and therefore pandemic risk than SARS-CoV, as the current effective reproductive number (R) is 2.9, which is much higher than the R of SARS (1.77). Several COVID-19 studies have estimated the basic reproduction number ranges between 2.6 to 4.71[23]. Nevertheless, the Chinese health authorities have documented average incubation of 7 d (2-14 d)[24]. However, the case fatality rate (CFR) of any disease is not fixed and varies with location and time. It was shown that CFR of COVID-19 varies widely between countries ranging from 0.2% in Germany to 7.7 % in Italy[25]. The differences in the CFR might be due to various factors. On one hand, the greater the number of performed polymerase chain reaction tests, the less the CFR will be. On the other hand, underlying conditions and comorbidities of the patients should also be taken into account - has a patient died because of COVID-19 or as a result of an exacerbation of a chronic disease/or another sudden acute medical condition, such as stroke and heart failure, but with a positive polymerase chain reaction for SARS-CoV-2 RNA.
SARS-CoV-2 proved to be a great challenge. Adapted to the human body, it spread around the world in a short time. The air-drop mechanism is straightforward to implement. The natural spread of SARS-CoV-2 is due to the mechanism and the fact that in about 85% of patients, COVID-19 occurs in a mild form. Patients with a mild form do not seek medical help. Usually, they treat themselves and infect their contacts[26]. They do not limit themselves to their home, but travel, work, and are in contact with many people. It has been shown that asymptomatic individuals can be contagious and secrete SARS-CoV-2. This further facilitates the spread of COVID-19. From an epidemiological point of view, mild and asymptomatic infections are hazardous because people have no idea that they are contagious. They do not seek medical help, they are not examined, and by contact, infect others. This requires a strict search and examination of the contacts of proven individuals with COVID-19 for the detection of asymptomatic infectious.
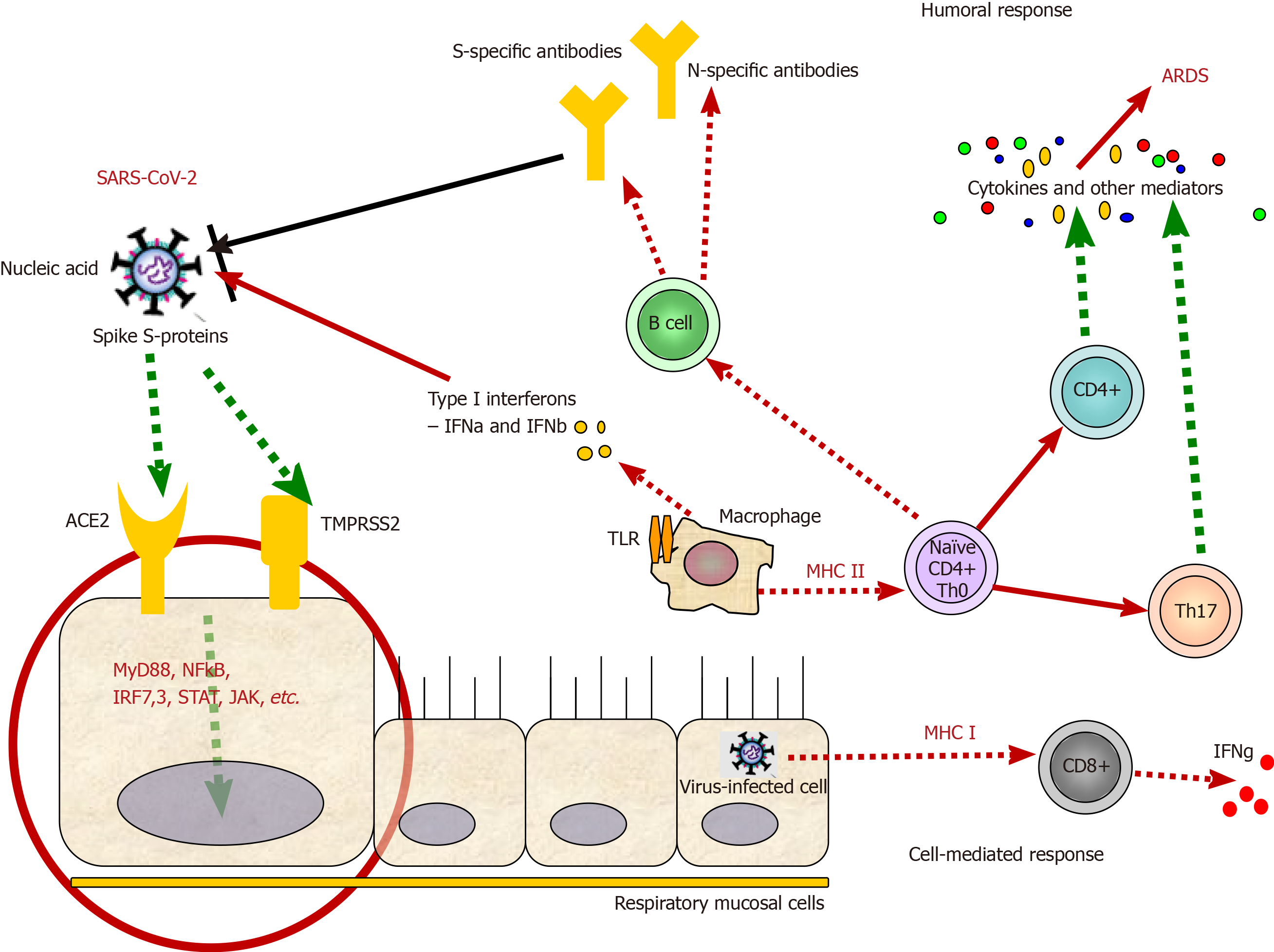
As already mentioned above, COVID-19 is an infection that has shortly become a health problem of global concern. Although COVID-19 is not the first outbreak of a coronaviral disease, neither SARS nor MERS led to such a high number of cases worldwide. This suggests that SARS-CoV-2 is highly contagious and much more virulent than both SARS-CoV and MERS-CoV. The continuous expansion of COVID-19 could be explained with the viral-specific characteristics and virulence factors[27]. CoV are enveloped viruses, named for the spikes on their surface that resemble a crown. Their genome is organized in a positive single-stranded RNA. CoV are divided into four genera: α, β, γ, and δ. Along with SARS-CoV and MERS-CoV, the current coronavirus SARS-CoV-2 is classified as a β-coronavirus. Its genome sequence is about 88% identical to that of two bat-derived SARS-like viruses, and about 80% and 50% identical to the genome sequences of SARS-CoV and MERS-CoV, respectively[28]. Mutations and recombination of the viral genome frequently occur due to error-prone RNA-dependent-RNA polymerases of the CoV. These events are closely related to viral adaptation[27]. CoV proteins include structural proteins: Spike (S), envelope (E), nucleocapsid (N), and membrane (M) proteins and some proteins with unknown function. S protein is a glycoprotein essential for viral entry by attachment and fusion to the cellular membrane (Figure 1). It is the main presented antigen on the viral surface and a target of neutralizing antibodies formed during the humoral immune response to the virus[28].
SARS-CoV and SARS-CoV-2 enter the host cell by binding to the angiotensin-converting enzyme 2 (ACE-2) receptor, while the dipeptidyl peptidase 4 receptor is required for MERS-CoV entry. After the virus enters the cell, viral RNA is released and involved in consecutive processes of new viral particle formation, which are then released[29]. ACE-2 receptors are mainly expressed in the vascular epithelium, renal tubular epithelium, and Leydig cells in the testes. For this reason, SARS-CoV might lead to hypogonadism and harm male fertility. In the respiratory system, SARS-CoV-2 enters the alveoli by binding to the ACE-2 receptors, predominantly expanded on the type II pneumocytes[30]. Once infected, type II pneumocytes are destroyed, and surfactant production is reduced. Macrophages are then recruited to destroy the damaged tissue of the lungs. Macrophages secrete interleukin (IL)-1, IL-6, and tumor necrosis factor alpha (TNFα), which cause vasodilatation and high temperature and enhance neutrophils and lymphocytes migration to the affected area. Alveoli edema causes respiratory failure due to blood-gas exchange disturbance. Nonstructural proteins might affect the innate immune response of the host and play a crucial role in the viral virulence and pathophysiology of SARS-CoV-2 infection[30].
In vitro and ex vivo examinations compared the viral tropism of SARS-CoV-2 with that of SARS-CoV, MERS-CoV, and 2009 pandemic influenza H1N1. It showed more extensive infection of bronchial epithelium, ciliated cells, and goblet cells with SARS-CoV-2 than with the other viruses[31]. A robust replication in human bronchus was observed, although ACE-2 receptor expression was relatively low in comparison with the lung parenchyma. However, the endothelium of the blood vessels was not found to be infected[32].
SARS-CoV-2 could also be detected in tears, anal swabs, and stool specimens. Moreover, infection and productive replication of the virus were observed in the conjunctiva and colorectal carcinoma cell lines. These findings suggest that conjunctiva and the bronchus epithelium might also be portals of infection and also poses the possibility of fecal-oral transmission. Various routes of viral transmission would explain the extensive spread of the SARS-CoV-2 throughout the whole world, causing a pandemic[32].
CoV have developed many mechanisms to avoid the immune system, which allow them to better survive in host cells. One of the multiple strategies is the forming of double-membrane vesicles that lack pattern recognition receptors when shedding, thus avoiding the recognition of their evolutionary old and conservative pathogen-associated molecular patterns, such as double-stranded RNA[33]. Moreover, studies in mice showed that SARS-CoV and MERS-CoV infection might inhibit the most potent anti-viral molecules, such as interferons (IFN)-I (IFN-α and IFN-β)[34,35]. The described mechanisms behind this inhibition in MERS-CoV include blocking of melanoma differentiation-associated protein 5, inhibiting the nuclear transport of IFN regulatory factor 3, suppressing the antigen presentation[29], etc. Therefore, the ability of SARS-CoV-2 to avoid the action of the immune system is a critical factor in the treatment of current infection and the development of specific drugs.
The presentation of virus antigens stimulates the humoral and cellular immune response, which are exerted by virus-specific B and T lymphocytes, respectively. Like common acute viral infections, the antibody production against SARS-CoV viruses has a typical immunoglobulin (Ig)M and IgG pattern. SARS-specific IgM antibodies may disappear at the end of the 3rd mo. In contrast, the IgG antibody may persist longer, indicating that IgG antibodies are likely to be protective[29]. SARS-specific IgG antibodies are predominantly against S- and N-proteins.
Immunological follow-up revealed a progressive increase in plasma SARS-CoV-2-specific IgM and IgG antibodies from 1st to 3rd wk[36].
Patients with COVID-19 may worsen clinically after some days of illness (especially around day 9), which is accompanied by an increase in the pro-inflammatory response, which often leads to admission to intensive care units and the need for supportive mechanical ventilation. This secondary deterioration is reminiscent of SARS-CoV, in which 80% of patients with SARS-CoV develop acute respiratory disease at the time of anti-viral IgG seroconversion[37]. IgG seroconversion is associated with two points in the immune response - the disappearance of the virus and the appearance of IgG antibodies. IgM antibodies may persist for some time together with IgG. Moreover, patients who produce anti-S-neutralizing antibodies at the onset of the disease have a higher risk of death. The mechanisms behind these observations are not clear, but it is assumed that excessive complement activation may play a role. In addition, a phenomenon called antibody-mediated enhancement of viral infection may be responsible for persistent viral load and subsequently cause a direct or indirect effect on ACE-2 activity in the lung[37] and eventually death.
Compared to humoral responses, there are more studies on cellular immunity in coronavirus infection, which is not surprising, taking into account that the principal and effective immune response towards viral infection is cell-mediated immunity.
A clinical case of mild SARS-2-CoV infection and favorable outcome for the patient revealed that the antibody-producing immune cells, defined by the expression of CD3-CD19+CD27hiCD38hi and CD4+CXCR5+ICOS+PD-1+ Th follicular cells, appear in the blood during viral clearance (day 7; 1.48%) and peak on day 8 (6.91%); and on day 7 (1.98%), increasing on day 8 (3.25%) and day 9 (4.46%), respectively. The peak of both cell subpopulations was significantly higher in the COVID-19 patient than in healthy controls and persisted during convalescence[38]. Cytotoxic T cells also increased rapidly from day 7 (3.57%) to day 8 (5.32%) and day 9 (11.8%), followed by a decrease on day 20, respectively. Besides, the incidence of CD38+human leukocyte antigen (HLA)-DR+CD8+ T cells was significantly higher in this patient than in healthy subjects (1.47% ± 0.50%)[37]. We have to emphasize once again that activated cytotoxic T cells, representatives of adaptive or so-called specific immunity, along with natural killer (NK) cells, part of innate immunity, are critical players in the cell-mediated immune response that accompanies any viral infection.
Helper CD4+ T cells simultaneously expressing CD38 and HLA-DR increased between day 7 (0.55%) and day 9 (3.33%) in this patient compared to healthy donors (0,63% ± 0.28%), although being in lower percentages than for CD8+ T cells. CD38+HLA-DR+CD8+ T cells produce about 34%-54% more granzymes A and B and perforin. The appearance and rapid increase of activated CD38+HLA-DR+ T cells, especially CD8+ T cells, on day 7-9 precede the disappearance of symptoms[37].
Analysis of CD16+CD14+ mononuclear cells, which are associated with the immunopathology of COVID-19, showed a lower frequency of these cells in the blood of this patient on days 7, 8, and 9 (1.29%, 0.43%, and 1.47%, respectively), compared to healthy control donors (9.03% ± 4.39%), probably indicative of their entry from the blood to the site of infection. No differences were found in activated HLA-DR+CD3-CD56+ NK cells during infection and compared to healthy levels[37].
However, recent data conclude that even reduced in numbers, Th and Tc cells are overactivated in patients infected with SARS-CoV-2[38]. This significant reduction is also observed in the acute-phase response in COVID-19 patients. However, once presented in the organism, CD4+ and CD8 + anti-virus memory T cells persist in the bloodstream of recovered patients for up to 4 years, even in the absence of viral antigens[39]. Moreover, other studies detected SARS-CoV-S protein-specific memory T memory cells 4 years after the patients` infection[40] and MERS-CoV-specific CD8+ T cells in mice, mainly involved in viral clearance[41]. These findings are a reasonable basis for designing effective vaccines against SARS-CoV-2.
Th17 cell responses are also involved in the immune pathogenesis of COVID-19, mainly with the secretion of various cytokines, such as IL-17, granulocyte-macrophage colony-stimulating factor, IL-21, and IL-22[42]. IL-1b and TNFα, as promoters of human Th17 cell differentiation, are also involved in stimulating vascular permeability and leakage. Nevertheless, it was shown that patients with a severe form of COVID-19 had increased levels of CCR+ Th17 cells[39]. This may suggest that Th17 cells and their cytokines are also involved in the cytokine storm. Besides, elevated Th17 and Th1 cell responses were also described in MERS-CoV and SARS-CoV patients[43,44]. These results support the hypothesis that enhanced IL-17-related pathways, including higher IL-17 but lower IFNγ and IFNα, are associated with worse outcomes for the patients[39].
As we have some proof of the contributing role of Th17 cells to the cytokine storm, Th17 cells may likely promote pulmonary edema, tissue damage, and lung failure. In line with this, targeting Th17 cells may be beneficial for some patients with COVID, especially in those with a dominant Th17 immune profile[43,45].
Some of the immunological responses that are accompanying the SARS-CoV-2 infection are presented in Figure 1.
A symptomatic COVID-19 case is defined as an infected person with a clinical picture suggestive of COVID-19. On the other hand, an asymptomatic case is an infected person who has not developed any signs or symptoms of COVID-19[46].
The incubation period of the disease is 5-6 d on average but can be up to 14 d. Critical epidemiological and immunological aspects of the disease are that an infected person may be contagious 1-3 d before the onset of the symptoms[47]. About 40% of COVID-19 patients experience mild clinical course, and another 40% present with moderate disease. Severe disease that requires oxygen support is observed in about 15% of the patients. Five percent of the infected ones develop a severe disease that progresses to respiratory failure (ARDS) but also sepsis and septic shock, thromboembolism, multiorgan failure, including acute kidney, and cardiac injury[48]. Advanced age, smoking, as well as comorbidities such as diabetes, hypertension, cardiac and chronic lung disease, cerebrovascular diseases, immunosuppression, and cancer have been reported to predispose to a severe course of COVID-19. Children and infants usually experience mild disease or asymptomatic infection[49].
There are no specific clinical symptoms of COVID-19 that can be taken as a reliable pathognomonic sign. Patients most commonly present with fever, cough (usually dry), fatigue, and anorexia. Dyspnea is usually seen in severe cases. Other nonspecific symptoms include sore throat, nasal congestion, headache, nausea, as well as diarrhea and vomiting. Loss of smell and taste have also been reported. In older people, some atypical symptoms such as reduced alertness and/or mobility and delirium might be seen[50,51].
COVID-19 may also present with neurological and mental manifestations, including delirium or encephalopathy, agitation, stroke, meningoencephalitis, anxiety, depression, and sleep problems. In many cases, some of these neurological manifestations have been documented without respiratory symptoms[52,53].
A retrospective study of SARS-CoV-2 infected patients showed mild leucopenia in mild cases. Mild to moderate leukocytosis was observed in severe cases with significantly high neutrophil and significantly low lymphocyte count. In patients with a severe course of the disease, significantly high alanine aminotransferase and aspartate aminotransferase were observed, as well as hypoalbuminemia, elevated concentration of C-reactive protein, lactate dehydrogenase, ferritin, and D-dimer. These laboratory changes are a result of an acute and severe inflammatory response[50].
Research on cellular immunity in SARS-CoV-2 positive patients showed a significant reduction in CD4+ and CD8+ T lymphocytes[29]. A more significant decrease in T cells was observed in severe cases as well as an elevation of the concentration of serum cytokines (IL-2, IL-6, IL-10, and TNFα). Elevation of IL-6 concentrations was also found in moderate cases[50]. On that basis, the cytokine storm is a crucial factor for the clinical course of COVID-19 and the disease severity.
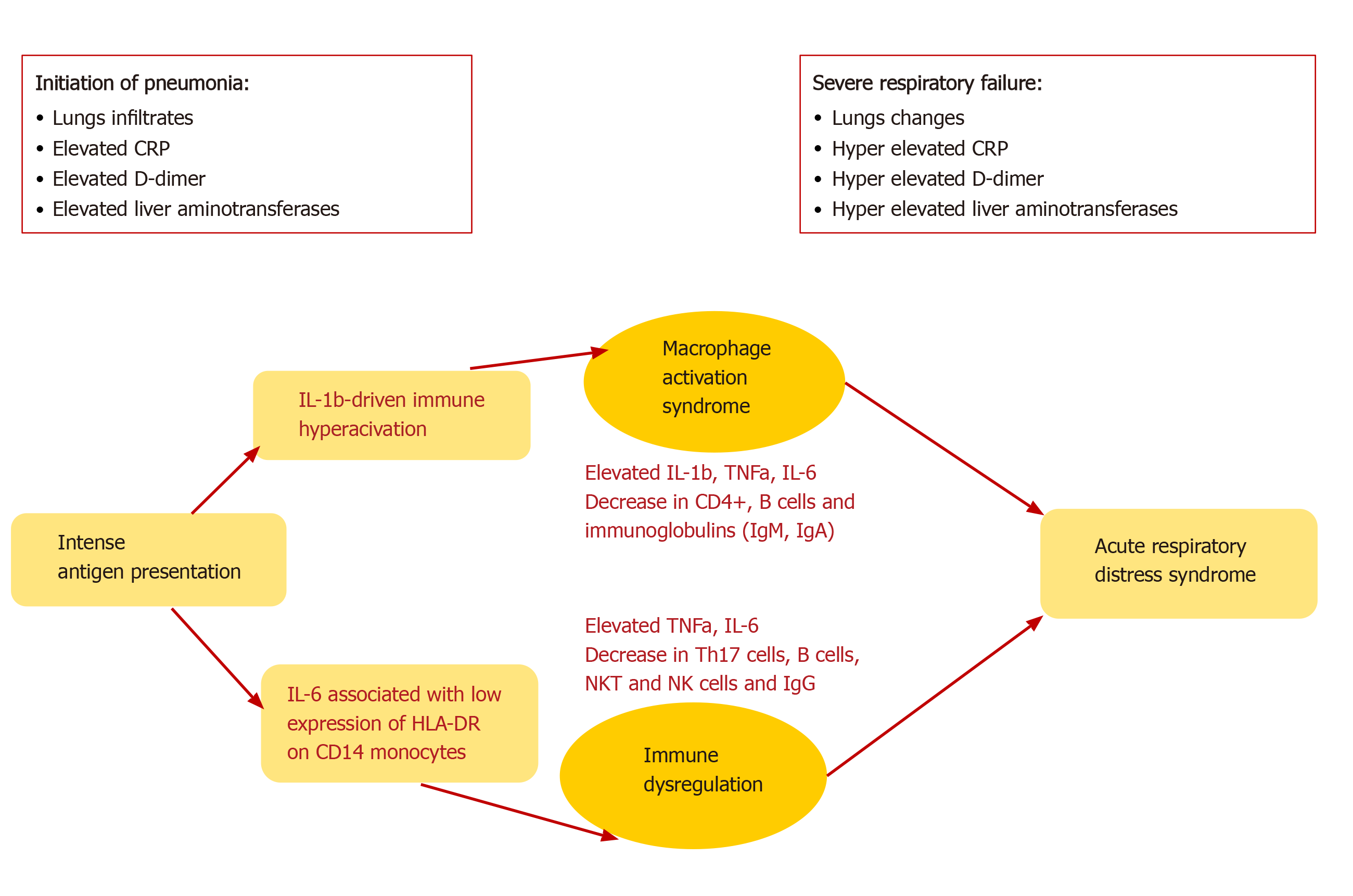
A common complication of SARS-CoV-2 infection is the development of ARDS. The latter is assumed as the leading cause of death in patients with COVID-19, especially among those with underlying diseases and conditions, evaluated as risk factors, smokers, and older age. ARDS is a result of the cytokine storm - an immunopa-thological event that leads to an uncontrolled systemic inflammatory response from the release of pro-inflammatory cytokines IFNα, IFNγ, IL-1β IL-3, IL-6, IL-12, IL-18, TNFα, and chemokines by the immune cells during SARS-CoV infection[29]. These biologically active substances seriously damage the lung parenchyma and lead to respiratory failure. This severe acute hypoxic status is accompanied by increased pulmonary capillary permeability and alveolar epithelial cell damage[51]. TNFα and IL-6 production in COVID-19 follows a different pattern than the pattern in bacterial sepsis or influenza. Furthermore, it was shown that blocking IL-6 by Tocilizumab restored partially HLA-DR expression and increased the number of circulating lymphocytes[51]. The immune mechanisms in COVID-19 are characterized by two main pathways, IL-6 or IL-1b-driven immune hyperactivation, leading to macrophages activations syndrome (MAS) or immune dysregulation (Figure 2).
The initiation of pneumonia in COVID-19 includes intense antigen presentation, accompanied by elevated C-reactive protein (CRP), D-dimer, and liver amino-transferases plus infiltrates in the lungs, whereas severe respiratory failure displays either MAS or deficient HLA-DR expression and profound depletion of Th, B cells, and NK cells[51]. In such a way, during ARDS, CRP, D-Dimer, and liver transaminases are further increased, leading to permanent pathological changes.
The described cytokine storm violently attacks not only the lungs but the organs of every system in the organism, causing multiple organ failure that leads to death in severe cases of SARS-CoV-2, SARS-CoV, and MERS-CoV infection[29].
Affecting the whole organism, the cytokine storm is the primary mechanism that induces disseminated intravascular coagulation (DIC). Pro-inflammatory cytokines TNFα and IL-1 suppress the endogenous anticoagulation. Inflammation damages the endothelium and leads to the release of tissue plasminogen activator, which could explain the elevation of the D-dimer and fibrin degradation products[54]. In summary, the accumulated data showed that COVID-19 is associated with a hypercoagulable state with increased risk of thromboembolic complications.
However, ARDS is assumed as the leading cause of death in COVID-19. During the early stages of the outbreak, it was reported that of the 41 SARS-CoV-2 infected patients admitted, six died of ARDS[48]. ARDS remains the most common immunopathological event for SARS-CoV-2, SARS-CoV, and MERS-CoV infections. One of the primary mechanisms for ARDS is the cytokine storm. The latter is a uncontrolled systemic inflammatory reaction resulting from the release of large amounts of pro-inflammatory cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10,
Another hypothesis on the severity of COVID-19 disease includes the problem associated with the activation of bradykinin B1 receptors on the lung endothelial cells. It is assumed that the enzymatic activity of the ACE-2 receptor inactivates des-Arg9 bradykinin, which is a ligand for B1. Furthermore, unlike B2, B1 is regulated by pro-inflammatory cytokines[59]. Interestingly, without the inactivation of B1 ligands, increased local vascular permeability is observed in lung mucosa, leading to angioedema. Probably, angioedema is a typical manifestation of the severe disease onset and the cause of typical changes visible on the computed tomography scan. The feeling of choking is observed mainly around day 9, denoted the worsening of the patients[60]. As any common viral infection, here, we also observed a progressive inflammatory condition with elevated IL-6, CRP, and ferritin but not procalcitonin or erythrocyte sedimentation rate, which indicates the second stage of the disease[60]. In line with this, the renin-angiotensin system could also contribute to the lung injuries along with sepsis-associated DIC. However, the levels of platelets, prothrombin time, and fibrinogen may remain normal[61].
The pathophysiology of the disease may be a combination of injury of pulmonary type II pneumocyte, viral pneumonia, ARDS, DIC, sepsis, cytokine storm, MAS, and overall immune dysregulation[62,63]. Some of these aspects are shown in Figure 2.
One of the main immunological features of COVID-19 is the exhaustion of lymphocytes. This, along with the reduced functional diversity of immune cells, is associated with a severe course of the disease[64]. The impaired immune response makes the possibility of adequate immune memory questionable. However, recovered patients displayed some SARS-CoV-specific antibodies up to 2-6 years after infection, unless they experienced ARDS, which led to undetectable peripheral memory B cell responses[40,65]. It is not yet known whether the T cell response is enough for the protection of reinfection[66]. To develop its own specific protective anti-viral immune response during the incubation period and in mild cases, the host must be in good general health and have an appropriate genetic terrain (e.g., certain HLA antigens). Moreover, genetic variants are known to contribute to individual differences in the immune response to pathogens.
But this is especially important when designing a vaccination strategy because an effective and protective vaccine depends desperately on the possibility of having immune memory after virus encountering[66].
Usually, COVID-19 survivors developed antibodies from 2-15 d after developing symptoms, as the immune response is broadly reminiscent of the typical anti-viral reaction[67]. The majority of patients produced antibodies against the spike protein[68]. Moreover, it was shown that the levels of antibodies correlated with patients age and the severity of the disease. The drawback of testing antibodies is that in mild cases, it is more likely not to produce detectable antibodies. Besides, it is not known whether these antibodies are protective or not. Some in vitro experiments with anti-SARS-CoV-2 antibodies showed that they are neutralizing and prevent the virus from entering the host cells[7]. However, we always have in mind that cellular immunity against the virus is more critical for viral clearance than the humoral. Thus, the role of Th, T cytotoxic, and NK cells should not be neglected. Studies that included survivors from the 2003 SARS epidemic have recently shown that neutralizing antibodies were found, 17 years after the infection[69]. Similar results were reported for the MERS epidemic, although the levels of neutralizing antibodies faded significantly after 5 years.
Furthermore, it was shown that people exposed for the second time to the virus developed much milder symptoms, even though reinfection can occur[70]. The reason for the decline in the antibodies levels remains unclear, even though some hypotheses are related to the short-life of memory B cells, particularly to coronavirus, hiding in the secondary lymphoid organs, such as lymph nodes and the spleen as well as in the bone marrow and the lungs. Interestingly, except for the antibodies levels, memory B cells can also be isolated and investigated as a marker of previous encountering the virus. Furthermore, some of the memory B cells of some patients reacted to the proteins from the live virus in vitro[71].
Here, we come to the myth of immune passports for COVID-19. The decision to make immune passports for people traveling from one country to another for tourism and recreation is not meaningless, but it exists only in theory because the real situation during the COVID-19 pandemic has been identified as more complicated where the “immune passport” cannot be a solution.
Let us look at the facts; if you go somewhere outside your country and carry the immune passport, you would guarantee that you are healthy and have antibodies to SARS-CoV-2. Especially in pandemics, the security of a society is the number one goal, where every breakthrough exposed the society to severe consequences. False-positive and negative results give some discrepancies and difficulties in interpretations. Moreover, the probability of false peace of mind that “I′m not infected, I can fly/travel safely” and not to keep the protective measures, immune passports are more valuable not as a medical document but as a business card. In such a way, the immune passports not only resolve the problem but create another, the more rapid spread of the infection.
It is scientifically challenging to claim that any given person will be protected from the infection if they reach a specific level of antibodies on testing. Any predictions based on the antibody presence carry legal and ethical issues. This false feeling of protection could also encourage dangerous behavior, such as refusal to wear masks and keep social distancing. Moreover, there will be people who would intentionally try to get sick to re-enter normal life.
Another aspect of the unclear immune response and immune memory regarding SARS-CoV-2 infection is the desire of the world to have an effective vaccine against the virus to be invented. This vaccine is essential to reduce the severity of the disease and its spread and clear the virus, thus helping to control current and future coronavirus outbreaks. There are several strategies for developing vaccines against SARS-CoV and MERS-CoV tested in animals. Among them are: Using live attenuated virus, viral vectors, inactivated virus, vaccine subunits, recombinant DNA, and protein vaccines[71]. There may currently be other promising targets to use in the creation of vaccines against SARS-CoV-2 infection and therapy, but additional laboratory and clinical evidence are still needed. Some new pharmaceutical drugs, including anti-human immunodeficiency virus and stem cell drugs, have been shown in these clinical trials. These studies are ongoing, but still it will take months to years to develop vaccines for SARS-CoV-2.
In summary, it is worrying that after discharge from the hospital, some patients remain positive for the virus, while others return with a relapse. This suggests that the immune response to SARS-CoV-2 intended to neutralize and eliminate the virus may be insufficient in at least some of the patients. Furthermore, this observation can predict that vaccines may not be effective in these individuals. Recovered patients who have not reached a severe stage should be monitored for the presence of the virus along with the measurement of T/B cell responses. All of these scenarios need to be considered when defining vaccine development strategies. Besides, since there are many types or subtypes of coronavirus, vaccines aimed directly at SARS-CoV-2 seems to be challenging to develop. Therefore, Edward Jenner's approach should be considered. In other words, the development of a vaccine against the whole family of CoV or against those representatives that cause disease only in animals.
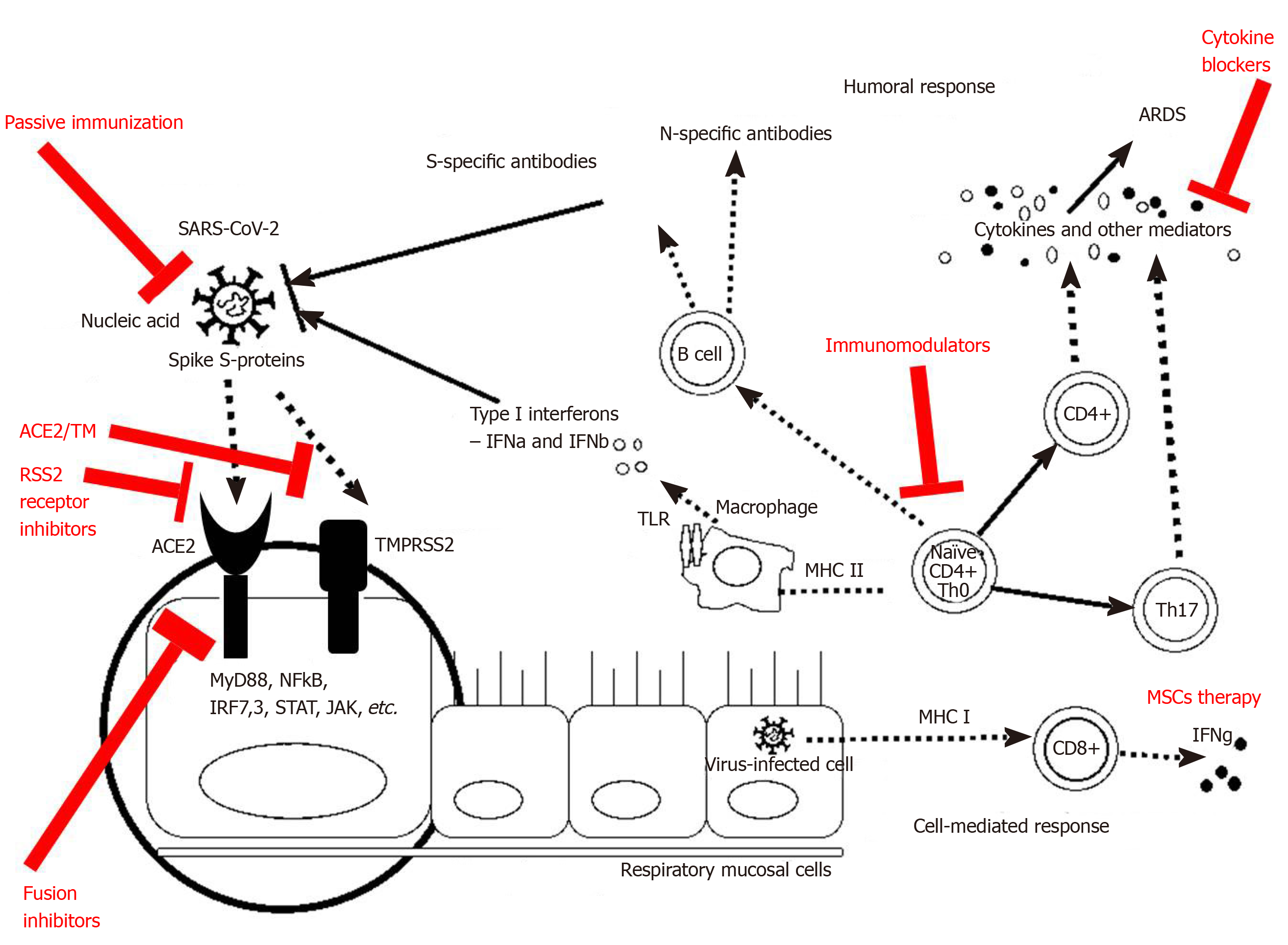
COVID-19 infection revealed some treatment options related to the immune responses. If we accept the division of SARS-CoV-2 infection into three stages, at the beginning of the infection (stage I, an asymptomatic incubation period with or without detectable virus), some of the mechanisms of innate immunity play a role, including NK cells, interferon production, and some cytokines. Therefore, strategies to boost immunity, such as administration of anti-serums (ready-made antibodies from survivors) or pegylated IFNα, are undoubtedly crucial at this stage.
During the incubation period, as well as the non-severe stage (stage II, a mild symptomatic period with the presence of a virus in the body), a specific adaptive immune response is required to neutralize and eliminate the virus, which will eventually prevent disease progression to severe stages. The adaptive response, however, occurs more slowly and is activated at a later stage. When the protective immune response is compromised, the virus will spread, and massive damage of the affected tissues and organs will occur, especially in those that highly express ACE-2 receptors, such as the gut and kidneys. This leads to lung inflammation, mostly mediated by pro-inflammatory immune cells. Inflammation of the lungs is the leading cause of life-threatening respiratory failure in the severe stage of the disease (stage III, severe respiratory symptomatic stage with high viral load). Therefore, once severe lung damage occurs, attention should be moved to the inflammation and the efforts to suppress the immune responses and control the symptoms[72]. Some of the treatment options related to the immunological mechanisms could be seen in Figure 3.
Antibody and plasma therapy are the next therapy option, closely related to the immune mechanisms. It has been reported that many cured patients donate plasma against SARS-CoV-2, as there were clinical trials for SARS-CoV[73] and MERS-CoV[74]. Preliminary data indicate favorable results for use in patients with acute and severe SARS-CoV-2 infection. In addition, the development of a recombinant human monoclonal antibody, such as CR3022, is a reasonably easy way to neutralize the virus by attachment to the receptor-binding domain of SARS-CoV-2. This SARS-specific human monoclonal antibody has the potential to be developed as a drug for SARS-CoV-2 infection[75]. Other monoclonal antibodies that neutralize SARS-CoV, such as m396 and CR3014, may be alternatives for the treatment of SARS-CoV-2[76].
Passive antibody therapy is the administration of ready-made antibodies against an infectious agent to a susceptible individual to prevent or treat an infectious disease caused by that microorganism. Thus, the passive application of antibodies is the only means of ensuring the immediate and ready immunity of endangered persons. Experience with previous epidemics with other CoV, such as SARS-CoV-1, has shown that survivors' sera may contain neutralizing antibodies to the virus, thus the expected mechanism of action of passive antibody therapy is viral neutralization and elimination. However, other mechanisms, such as antibody-dependent cellular cytotoxicity and/or phagocytosis, are also possible[72].
Currently, the only ready-to-use antibodies are those obtained from survived patients. As more people become ill with COVID-19 and recover, the number of potential donors will continue to increase. The serum of recovered individuals can be used prophylactically to prevent infection in high-risk individuals, such as those with chronic diseases, medical personnel, and those who have been in contact with patients. The efficacy of this approach is unknown, but historical experience has shown that products containing passive antibodies are more effective in preventing than in treating existing disease.
If used for therapy, antibodies are most effective when administered soon after the onset of symptoms. The antibody acts by altering the inflammatory responses, which is also more easily achieved during the initial immune response, or the asymptomatic stage. The reason for the differences in efficacy is not well understood. Still, it may reflect the fact that the antibody works by neutralizing the initial inoculum from the infectious agent, which is probably much less than that in an already developed infection and a large number of viral copy. In line with this, to be effective, a sufficient amount of antibodies must be administered. When given to a person at risk of infection, this antibody will reach the tissues via blood and can provide protection against the infection. Depending on the amount and composition of immunoglobulins, passive protection can last from weeks to months (for IgG). The challenges, however, are related to the difficulties in that, as we have mentioned above, some patients do not possess high antibody titers after the illness. Therefore, due to the individual variation of the immune responses, the insufficiently active immune response in some people will be a reason for them to be prone to reinfection.
In a prophylactic regimen of the use of passive immunotherapy in persons at risk, the aim is to prevent disease. Used therapeutically, the passive serum is administered to patients with clinical manifestations of the disease to reduce the severity of symptoms and mortality. The efficacy of these approaches cannot be measured without conducting a controlled clinical trial[72].
The risks of passive application of sera products fall into two categories - known and theoretical. The known risks are those associated with the use of blood products - infectious diseases and allergic reactions to the components of the serum, including serum sickness. With modern blood banking techniques, these risks are low. For sera used for therapy, there are also theoretical risks for transfusion-related acute lung injury and antibody-dependent enhancement (ADE) of infection. Several mechanisms of ADE have been described for CoV, and there are concerns that antibodies against one type of coronavirus may exacerbate infection to another virus strain. It is possible to experimentally predict the risk of ADE in SARS-CoV-2, as suggested for MERS. It is assumed that when using sera rich in virus-neutralizing SARS-CoV-2 antibodies, the risk of developing ADE is minimal[72]. However, it is good to test these hypotheses in clinical trials. Another theoretical risk is that the use of antibodies to individuals exposed to SARS-CoV-2 may weaken the immune response so that not enough of their own antibodies would form. This puts these people at risk for subsequent reinfection. However, if the risk proves to be real, these individuals can be vaccinated against COVID-19 when the vaccine becomes available.
The preparation of highly purified and enriched neutralizing antibodies against SARS2-CoV-2 is preferable because they are safer and have higher activity. However, such preparations will not be available in the coming months, while locally derived serums may be available much sooner[77].
Other therapy strategies are oriented against the cytokine storm. Because lymphocytopenia is commonly seen in severe cases of COVID-19, the cytokine storm caused by the SARS-CoV-2 virus may be mediated by leukocytes other than T cells. High white blood cell counts are common, which, along with lymphocytopenia, is used as a differential diagnostic criterion for COVID-19. In any case, the blocking of IL-6 can be effective, as well as the blocking of IL-1 and TNFα. The approach to blocking these pro-inflammatory cytokines has been adopted in various autoimmune diseases. We also proposed anti-IL-6 therapy in IBD patients as a way of limiting the inflammation and development of colorectal carcinoma[78].
Anti-inflammatory strategies, including the blockade of specific cytokines that increase B1 expression on endothelial cells locally at the site of inflammation in combination with B1 and or B2 receptor blockade, should be considered. IL-1 (consisting of IL-1α and IL-1β) and TNF are potent inducers of the B1 receptor. Blocking the translocation of nuclear factor kappa B, TNF-α, or IL-1 prevents the regulation of B1 receptors both functional and molecular by lipopolysaccharide. Therefore, one strategy could be the treatment with anakinra, a monoclonal antibody that blocks not only IL-1α but also IL-1β, and has an excellent safety profile. IL-1a is probably in extremely high concentrations locally due to its release from damaged cells. TNF blockade is an option but is associated with many more infectious complications. Also, complement activity has been described and may play a role in this stage of the disease. Moreover, it may be affected by blockade of C5 component with eculizumab, a monoclonal antibody that was randomized to COVID-19 (NCT04288713)[60]. The use of antibodies against cytokines is a general approach in several autoimmune and autoinflammatory diseases in which one or more cytokines play a direct role in the pathogenesis. However, this approach is not without any side effects, so the benefit should always be considered.
On the other hand, a significant drawback is the fact that blocking of a cytokine rarely has the desired effect because the cytokines are connected in a network. Corticosteroids are also an option for therapy in these cases. Anti-inflammatory strategies may provide time, but will not cure the disease on their own if the virus is present or bradykinin-associated angioedema is not controlled[60].
Although various clinics in China have proclaimed the use of mesenchymal stromal/stem cells (MSCs) in severe cases of COVID-19 infection, no reliable results have yet been published. One of the advantages of this therapy is that MSCs must be activated by IFNγ. After that, MSCs are capable of exerting their anti-inflammatory effects. By producing various growth factors, MSCs can help repair damaged lung tissue. However, severely affected patients may lack these MSCs properties because T cells do not activate well in SARS-CoV-2 infection. The use of a “licensing approach” is being considered: Pre-treatment of MSCs with IFNγ with or without TNFα or IL-1. Thus, cytokine-treated MSCs may be more effective in suppressing the hyperactive immune response and promoting tissue repair, as such MSCs are useful in acute lung damage caused by lipopolysaccharide[72]. In any case, the use of MSCs in clinical practice should be limited to strict indications, taking into account the benefit-risk for each patient. Their oncogenic potential should not be neglected.
Based on the COVID-19 results from the interaction between the SARS-CoV-2 virus and the individual′s immune system, we can assume that the onset and development of COVID-19 significantly depend on this communication.
Viral factors, such as viral type, load, titer, in vitro viability, mutations, as well as individuals’ factors like genetics, age, gender, overall health, nutritional status, neuroendocrine-immune regulation, etc., can determine the duration and severity of the disease, probably the risk of reinfection.
Now, the science is directed to acquiring new data on the immunology, including the immune memory against the virus, the development of new technologies for the detection of infection, and effective vaccines. However, the unknown is much greater than verified knowledge of the SARS-CoV-2 virus and COVID-19.
Being a global health issue, COVID-19 urges scientists to put a lot of effort into developing effective therapeutic strategies for treating and saving lives. Little is known about the histopathological changes in the lungs and the damaged organs, as there are few reports of postmortem examinations. Thus, further research would be of great significance for the detailed understanding of SARS-CoV-2 infection.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Bulgaria
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ciotti M, Manenti A S-Editor: Huang P L-Editor: Filipodia P-Editor: Li JH
| 1. | World Health Organization. Coronavirus disease 2019 (COVID-19) situation report-48. 2020. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200308-sitrep-48-covid-19.pdf?sfvrsn¼16f7ccef_4. |
| 2. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17613] [Article Influence: 3522.6] [Reference Citation Analysis (0)] |
| 3. | Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24:490-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1725] [Cited by in RCA: 1874] [Article Influence: 208.2] [Reference Citation Analysis (0)] |
| 4. | Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1873] [Cited by in RCA: 1872] [Article Influence: 374.4] [Reference Citation Analysis (1)] |
| 5. | Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88-S97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 740] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 6. | Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4030] [Cited by in RCA: 4017] [Article Influence: 309.0] [Reference Citation Analysis (0)] |
| 7. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14097] [Article Influence: 2819.4] [Reference Citation Analysis (1)] |
| 8. | Zhang T, Wu Q, Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol. 2020;30:1346-1351.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 870] [Cited by in RCA: 749] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 9. | Liu P, Chen W, Chen JP. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 10. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14869] [Cited by in RCA: 12964] [Article Influence: 2592.8] [Reference Citation Analysis (1)] |
| 11. | Liu K, Fang YY, Deng Y, Liu W, Wang MF, Ma JP, Xiao W, Wang YN, Zhong MH, Li CH, Li GC, Liu HG. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl). 2020;133:1025-1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 856] [Article Influence: 171.2] [Reference Citation Analysis (0)] |
| 12. | Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A Novel Coronavirus Emerging in China - Key Questions for Impact Assessment. N Engl J Med. 2020;382:692-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 834] [Article Influence: 166.8] [Reference Citation Analysis (0)] |
| 13. | Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26:729-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 727] [Cited by in RCA: 689] [Article Influence: 137.8] [Reference Citation Analysis (0)] |
| 14. | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6893] [Cited by in RCA: 7479] [Article Influence: 1495.8] [Reference Citation Analysis (0)] |
| 15. | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8473] [Cited by in RCA: 7587] [Article Influence: 1517.4] [Reference Citation Analysis (0)] |
| 16. | Paules CI, Marston HD, Fauci AS. Coronavirus Infections-More Than Just the Common Cold. JAMA. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1204] [Cited by in RCA: 1133] [Article Influence: 226.6] [Reference Citation Analysis (0)] |
| 17. | Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA, Niemeyer D, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Bruenink S, Schneider J, Ehmann R, Zwirglmaier M, Drosten C, Wendtner C. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. MedRxiv. 2020;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 18. | Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, Wang M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323:1406-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3006] [Cited by in RCA: 2686] [Article Influence: 537.2] [Reference Citation Analysis (0)] |
| 19. | Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3727] [Cited by in RCA: 3401] [Article Influence: 680.2] [Reference Citation Analysis (0)] |
| 20. | van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med. 2020;382:1564-1567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5894] [Cited by in RCA: 5655] [Article Influence: 1131.0] [Reference Citation Analysis (0)] |
| 21. | Liu J, Liao X, Qian S, Yuan J, Wang F, Liu Y, Wang Z, Wang FS, Liu L, Zhang Z. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020;26:1320-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 415] [Cited by in RCA: 379] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 22. | Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA. 2020;323:1610-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1390] [Article Influence: 278.0] [Reference Citation Analysis (0)] |
| 23. | Liu T, Hu J, Kang M, Lin L, Zhong H, Xiao J, He G, Song T, Huang Q, Rong Z, Deng A, Zeng W, Tan X, Zeng S, Zhu Z, Li J, Wan D, Lu J, Deng H, He J, Ma W. Transmission dynamics of 2019 novel coronavirus (2019-nCoV). bioRxiv. 2020;. [DOI] [Full Text] |
| 24. | Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S. Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med. 2020;201:7-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 389] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 25. | Hamid S, Mir MY, Rohela GK. Novel coronavirus disease (COVID-19): a pandemic (epidemiology, pathogenesis and potential therapeutics). New Microbes New Infect. 2020;35:100679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 26. | Meng L, Hua F, Bian Z. Coronavirus Disease 2019 (COVID-19): Emerging and Future Challenges for Dental and Oral Medicine. J Dent Res. 2020;99:481-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1067] [Cited by in RCA: 887] [Article Influence: 177.4] [Reference Citation Analysis (0)] |
| 27. | De Soto J, Hakim S, Boyd F. The Pathophysiology of Virulence of the COVID-19 Virus. 2020 2020040077. Preprints. [DOI] [Full Text] |
| 28. | Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 417] [Cited by in RCA: 507] [Article Influence: 101.4] [Reference Citation Analysis (0)] |
| 29. | Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1000] [Cited by in RCA: 935] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 30. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14234] [Article Influence: 2846.8] [Reference Citation Analysis (0)] |
| 31. | Belser JA. Assessment of SARS-CoV-2 replication in the context of other respiratory viruses. Lancet Respir Med. 2020;8:651-652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Hui KPY, Cheung MC, Perera RAPM, Ng KC, Bui CHT, Ho JCW, Ng MMT, Kuok DIT, Shih KC, Tsao SW, Poon LLM, Peiris M, Nicholls JM, Chan MCW. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8:687-695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 388] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 33. | Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927-5940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 407] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 34. | Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1056] [Cited by in RCA: 1164] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 35. | Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB, Meyerholz DK, Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;129:3625-3639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 430] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 36. | de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2157] [Cited by in RCA: 2277] [Article Influence: 253.0] [Reference Citation Analysis (0)] |
| 37. | Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, Jia X, Nicholson S, Catton M, Cowie B, Tong SYC, Lewin SR, Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26:453-455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 812] [Cited by in RCA: 757] [Article Influence: 151.4] [Reference Citation Analysis (0)] |
| 38. | Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 230] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 39. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5776] [Article Influence: 1155.2] [Reference Citation Analysis (2)] |
| 40. | Fan YY, Huang ZT, Li L, Wu MH, Yu T, Koup RA, Bailer RT, Wu CY. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154:1093-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 41. | Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, Lv H, Wang TB, Yang H, Richardus JH, Liu W, Cao WC. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186:7264-7268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 381] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 42. | Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Baric RS, Enjuanes L, Gallagher T, McCray PB, Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111:4970-4975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 386] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 43. | Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53:368-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 581] [Article Influence: 116.2] [Reference Citation Analysis (0)] |
| 44. | Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, Bortolotti P, Martinez L, Dubucquoi S, Dessein R, Gosset P, Mathieu D, Guery B. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014;9:e88716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 193] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 45. | Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4:e00165-e00113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 46. | Wei WE, Li Z, Chiew CJ, Yong SE, Toh MP, Lee VJ. Presymptomatic Transmission of SARS-CoV-2 - Singapore, January 23-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:411-415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 653] [Cited by in RCA: 621] [Article Influence: 124.2] [Reference Citation Analysis (0)] |
| 47. | Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172:577-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4286] [Cited by in RCA: 3362] [Article Influence: 672.4] [Reference Citation Analysis (1)] |
| 48. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30071] [Article Influence: 6014.2] [Reference Citation Analysis (3)] |
| 49. | Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054-1062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17476] [Cited by in RCA: 18179] [Article Influence: 3635.8] [Reference Citation Analysis (0)] |
| 50. | Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2835] [Cited by in RCA: 3410] [Article Influence: 682.0] [Reference Citation Analysis (0)] |
| 51. | Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, Ntaganou M, Kyriakopoulou M, Dimopoulos G, Koutsodimitropoulos I, Velissaris D, Koufargyris P, Karageorgos A, Katrini K, Lekakis V, Lupse M, Kotsaki A, Renieris G, Theodoulou D, Panou V, Koukaki E, Koulouris N, Gogos C, Koutsoukou A. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992-1000.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1306] [Cited by in RCA: 1553] [Article Influence: 310.6] [Reference Citation Analysis (0)] |
| 52. | Spinato G, Fabbris C, Polesel J, Cazzador D, Borsetto D, Hopkins C, Boscolo-Rizzo P. Alterations in Smell or Taste in Mildly Symptomatic Outpatients With SARS-CoV-2 Infection. JAMA. 2020;323:2089-2090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 518] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 53. | Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4761] [Cited by in RCA: 4694] [Article Influence: 938.8] [Reference Citation Analysis (0)] |
| 54. | Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7:e438-e440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 878] [Cited by in RCA: 1058] [Article Influence: 211.6] [Reference Citation Analysis (0)] |
| 55. | Williams AE, Chambers RC. The mercurial nature of neutrophils: still an enigma in ARDS? Am J Physiol Lung Cell Mol Physiol. 2014;306:L217-L230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 309] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 56. | Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008;133:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 273] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 57. | Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1895] [Cited by in RCA: 1789] [Article Influence: 223.6] [Reference Citation Analysis (0)] |
| 58. | Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, Shin HM, Choi JY, Inn KS, Kim JH, Moon JY, Choi MS, Cho NH, Kim YS. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep. 2016;6:25359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 275] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 59. | Marceau F, Bawolak MT, Fortin JP, Morissette G, Roy C, Bachelard H, Gera L, Charest-Morin X. Bifunctional ligands of the bradykinin B2 and B1 receptors: An exercise in peptide hormone plasticity. Peptides. 2018;105:37-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 60. | Van de Veerdonk F, Netea MG, van Deuren M, van den Hoogen FHJ, de Mast Q, Bruggemann R, van der Hoeven H. Kinins and Cytokines in COVID-19: A Comprehensive Pathophysiological Approach. 2020 2020040023. Preprints. [DOI] [Full Text] |
| 61. | Belen-Apak FB, Sarıalioğlu F. Pulmonary intravascular coagulation in COVID-19: possible pathogenesis and recommendations on anticoagulant/thrombolytic therapy. J Thromb Thrombolysis. 2020;50:278-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 62. | O'Donnell JS, Sharif K, Emery P, Bridgewood C, McGonagle D. Why the immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia are distinct from macrophage activation syndrome with disseminated Intravascular coagulation. Autoimmun Rev. 2020;. [DOI] [Full Text] |
| 63. | Fox SE, Akmatbekov A, Harbert JL, Li G, Brown Q, Vander Heide RS. Pulmonary and cardiac pathology in Covid-19: the first autopsy series from New Orleans. MedRxiv. 2020;Preprint. [DOI] [Full Text] |
| 64. | Zheng HY, Zhang M, Yang CX, Zhang N, Wang XC, Yang XP, Dong XQ, Zheng YT. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 597] [Cited by in RCA: 705] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 65. | Libraty DH, O'Neil KM, Baker LM, Acosta LP, Olveda RM. Human CD4(+) memory T-lymphocyte responses to SARS coronavirus infection. Virology. 2007;368:317-321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Yang LT, Peng H, Zhu ZL, Li G, Huang ZT, Zhao ZX, Koup RA, Bailer RT, Wu CY. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120:171-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 67. | Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26:1478-1488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1123] [Cited by in RCA: 1118] [Article Influence: 223.6] [Reference Citation Analysis (0)] |
| 68. | Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, Ling Y, Zhang Y, Xun J, Lu L, Jiang S, Lu H, Wen Y, Huang J. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 249] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 69. | Poh CM, Carissimo G, Wang B, Amrun SN, Lee CY, Chee RS, Fong SW, Yeo NK, Lee WH, Torres-Ruesta A, Leo YS, Chen MI, Tan SY, Chai LYA, Kalimuddin S, Kheng SSG, Thien SY, Young BE, Lye DC, Hanson BJ, Wang CI, Renia L, Ng LFP. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 306] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 70. | Payne DC, Iblan I, Rha B, Alqasrawi S, Haddadin A, Al Nsour M, Alsanouri T, Ali SS, Harcourt J, Miao C, Tamin A, Gerber SI, Haynes LM, Al Abdallat MM. Persistence of Antibodies against Middle East Respiratory Syndrome Coronavirus. Emerg Infect Dis. 2016;22:1824-1826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 71. | Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 506] [Cited by in RCA: 483] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 72. | Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451-1454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 920] [Cited by in RCA: 1002] [Article Influence: 200.4] [Reference Citation Analysis (0)] |
| 73. | Mair-Jenkins J, Saavedra-Campos M, Baillie JK, Cleary P, Khaw FM, Lim WS, Makki S, Rooney KD, Nguyen-Van-Tam JS, Beck CR; Convalescent Plasma Study Group. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 689] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 74. | Koenig KL. Identify-Isolate-Inform: A Modified Tool for Initial Detection and Management of Middle East Respiratory Syndrome Patients in the Emergency Department. West J Emerg Med. 2015;16:619-624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 75. | Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 899] [Cited by in RCA: 942] [Article Influence: 188.4] [Reference Citation Analysis (0)] |
| 76. | Zhang L, Liu Y. Potential interventions for novel coronavirus in China: A systematic review. J Med Virol. 2020;92:479-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 674] [Cited by in RCA: 725] [Article Influence: 145.0] [Reference Citation Analysis (1)] |
| 77. | Casadevall A, Pirofski LA. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130:1545-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 636] [Cited by in RCA: 652] [Article Influence: 130.4] [Reference Citation Analysis (0)] |
| 78. | Velikova TV, Miteva L, Stanilov N, Spassova Z, Stanilova SA. Interleukin-6 compared to the other Th17/Treg related cytokines in inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2020;26:1912-1925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (1)] |