Published online Sep 26, 2010. doi: 10.4331/wjbc.v1.i9.281
Revised: June 22, 2010
Accepted: June 29, 2010
Published online: September 26, 2010
AIM: To explore the possibility that PDX-1 gene is reactivated as a consequence of molecular events that occur during liver regeneration.
METHODS: Rat hepatocytes were maintained in DMEM-F12, 10% fetal bovine serum (FBS), penicillin/streptomycin and geneticin when applicable. Rat insulinoma RIN 1046-38 cells were maintained in M-199-10% FBS and penicillin/streptomycin. The final concentration of glucose was 11.1 mmol/L. During regeneration, lateral and medial liver lobes of adult male Wistar rats were surgically removed, with up 70% loss of liver mass. In methylation experiments, 5-aza-deoxycytidine (5-aza-dC) was used. Primer3 software was used for polymerase chain reaction (PCR). Quantitative real time PCR (qRT-PCR) was performed using SYBR Green technology; primers were designed by Beacon Designer 6 software. Western blotting and SDS-PAGE were performed according to standard procedures. Antibodies were purchased from commercial suppliers.
RESULTS: We explored the possibility that liver regeneration could trigger PDX-1 expression, and hence insulin production. Twenty-four hours after surgical liver removal, regeneration was active as demonstrated by the increased proliferating cell nuclear antigen; however, all the other checked genes (involved in insulin gene expression): PC-1, Ngn3, NeuroD1, Btc, PDX-1 and Ins-1, were not related to the molecular events caused by this process. The only marker detected in regenerating liver was E47: a transcription factor of the the basic helix-loop-helix family known to be expressed ubiquitously in mammalian cells. In the rat pancreas, almost all of the tested genes were expressed as shown by RT-PCR, except for Ngn3, which was silenced 2 d after birth. Therefore, the molecular events in liver regeneration are not sufficient to promote PDX-1 expression. DNA methylation is a known mechanism to achieve stable repression of gene expression in mammals: Hxk 2 gene is silenced through this mechanism in normal hepatocytes. The administration of 5-aza-dC to cultured cells is in fact able to upregulate Hxk 2 mRNA. We investigated whether PDX-1 silencing in liver cells could be exerted through methylation of CpG islands in both the promoter and the gene coding regions. The results show that the drug increased the expression level of the Hxk 2 control gene but failed to rescue the expression of PDX-1, thus DNA demethylation is not sufficient to override repression of the PDX-1 gene.
CONCLUSION: During liver regeneration, PDX-1 gene is not reactivated. Demethylation does not de-repress PDX-1 gene expression. Therefore gene silencing is not achieved through this epigenetic mechanism.
- Citation: Pillich RT, Scarsella G, Risuleo G. Regeneration and DNA demethylation do not trigger PDX-1 expression in rat hepatocytes. World J Biol Chem 2010; 1(9): 281-285
- URL: https://www.wjgnet.com/1949-8454/full/v1/i9/281.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v1.i9.281
Diabetes mellitus is a metabolic disease that is characterized by persistent hyperglycemia resulting from defects in insulin secretion and/or action[1,2]. When the amount of glucose in the blood increases, the release of insulin from the pancreas is triggered. This hormone removes glucose from the blood and stimulates the liver to metabolize glucose, thus controlling the level of sugar in the organism. In diabetic patients, the blood sugar levels remain high. This might derive from lack of insulin production, from insufficient levels of the hormone or from its diminished effectiveness. This peptide hormone is primarily involved in the glucose metabolism, and is produced by the pancreatic β cells that, also due to autoimmune responses, are destroyed in type 1 diabetes[3]. Insulin is a very potent regulator because it can exert its specific action at a blood concentration as low as 10-8 mol/L[4].
A number of different transcription factors are involved in the control of insulin expression and/or regulation[5]. The better studied is possibly PDX-1[6], which acts synergistically with E47, a member of the basic helix-loop-helix (bHLH) family of transcription factors: synergism requires DNA binding and activation domains of both PDX-1 and bHLH proteins. It has also been shown that synergistic transactivation results from the co-expression of E47, PDX-1 and NeuroD1[7]. The PDX-1 protein contains 283 amino acids with a predicted molecular weight of 31 kDa. Like most other transcription factors, PDX-1 is characterized by a modular architecture with separate functional domains. However, in spite of the vast body of literature on the function of PDX-1, its actual role in the activation/regulation of the insulin gene is controversial. In any case, it is clear that post-translational modification of the PDX-1 gene products is necessary for its full function. In particular, sumoylation has been reported to mediate PDX-1 translocation to the nucleus and to stimulate insulin gene expression. The SUMO modification also accounts for the heterogeneity of PDX-1 molecular weight: this post-translational modification in fact shifts PDX-1 molecular mass from 31 to 46 kDa[8].
The effects of PDX-1 on each gene under particular conditions is dependent on a subtle interaction between transcription factors (positively-acting and negatively-acting) that could also be activated under similar conditions. It is therefore difficult to correlate specific effects of PDX-1 expression with more general multi-faceted effects on gene expression.
In a recent study, we demonstrated that the PDX-1 transcription factor can significantly alter hepatocyte glucose metabolism by transcriptional regulation of at least one important gene of the glycolytic pathway[9]. These results should therefore be taken into consideration when using PDX-1 as a key factor for approaches based on gene therapy. Hence, Pdx-1 overexpression is likely to affect negatively the metabolic function also in animal models. The liver is considered a good candidate for the expression of insulin in patients with type 1 diabetes. However, this transcription factor is not expressed in the liver, and therefore, transformation of hepatocytes with exogenous PDX-1 gene copies is necessary. The rationale of this work was to assess whether endogenous PDX-1 expression could be spontaneously reactivated during liver regeneration in partially hepatectomized rats, and to verify whether PDX-1 silencing in normal hepatocytes might be achieved through DNA methylation.
Clone-9 rat hepatocytes (American Type Culture Collection, ATCC No. CRL-1439) were maintained in DMEM-F12 (Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin/streptomicin and geneticin (100 μg/mL) when applicable. Cells were split every 3-4 d at a 1:10 ratio for no more than 10-15 times. Rat insulinoma RIN 1046-38 cells were maintained in M-199 supplemented with 10% FBS, penicillin and streptomycin. Glucose was added to a final concentration of 11.1 mmol/L. Cells were split every 4-5 d at a ratio of 1:5. In regeneration studies, the lateral and medial liver lobes of adult male Wistar rats (200-250 g) were surgically removed, which caused a 70% loss of liver mass[10]. In methylation experiments, 5-aza-deoxycytidine (5-aza-dC) was used at a final concentration of 5 μmol/L and treatment was performed as described by Goel and collaborators[11]. Total RNA purification was achieved using the Nucleospin RNA extraction kit (Macherey-Nagel). cDNA synthesis was performed using random primers, 1 μg total RNA as template and 200 U Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen). Polymerase chain reaction (PCR) was performed using a Geneamp 2400 (Applied Biosystems) and ExTaq DNA polymerase (Takara). PCR primers were designed using the Primer3 online software. Quantitative real time PCR (qRT-PCR) was performed using a Biorad iCycler and the SYBR Green technology; primers for qRT-PCR were designed using the Beacon Designer 6 software. All PCR primers have already been validated and used in a previous study[9]. SDS-PAGE and western blotting were performed according to standard procedures. Antibodies were purchased from Sigma (β-actin, cat. No. A-4700) and Santa Cruz Biotechnology (proliferating cell nuclear antigen; PCNA, cat No. SC-25280).
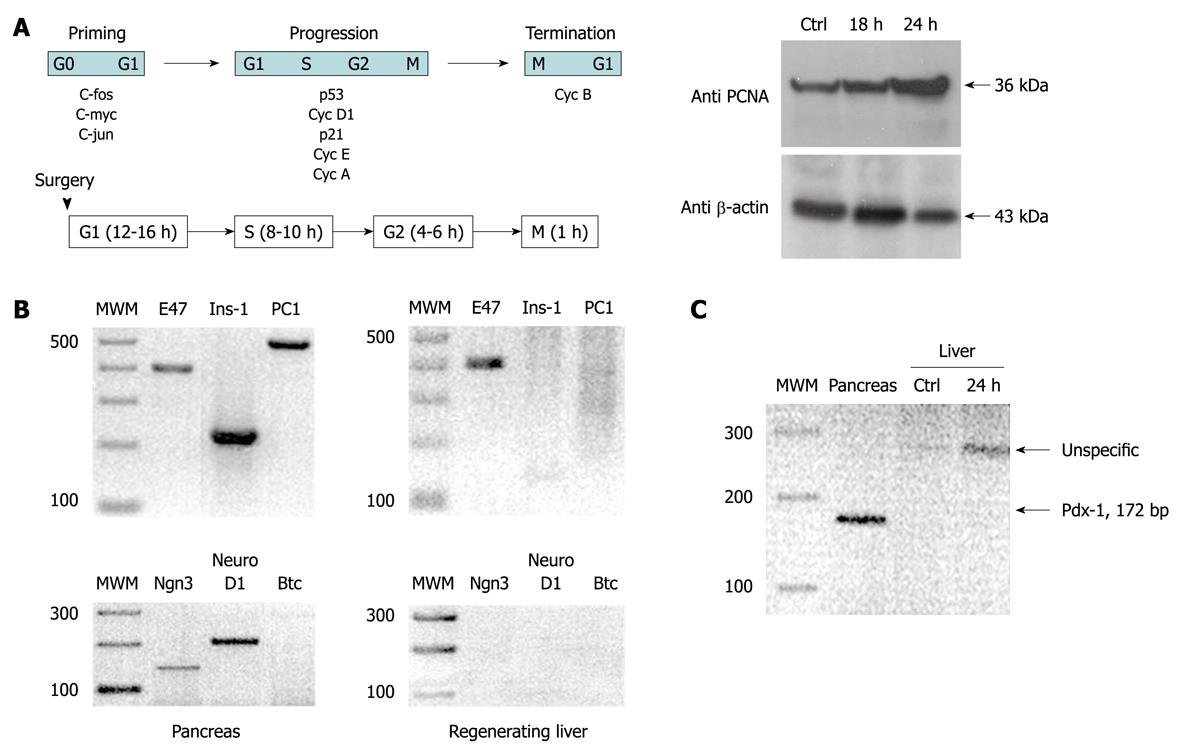
Figure 1 panel A shows that 24 h after surgical resection of the liver, regeneration was fully operational. This was demonstrated by the increased levels of PCNA shown in panels B and C. All the other genes involved in insulin gene expression that were checked (PC-1, Ngn3, NeuroD1, Btc, PDX-1 and Ins-1) did not show any relationships with the molecular events caused by this process. In fact, the only marker that could be detected in the regenerating liver was E47; a transcription factor that belongs to the bHLH family that is known to be expressed ubiquitously in mammalian cells. As a control, in the pancreas, almost all of the tested genes were expressed and could be identified by a band after RT-PCR; the only exception was Ngn3 that was silenced 2 d after birth in rats.
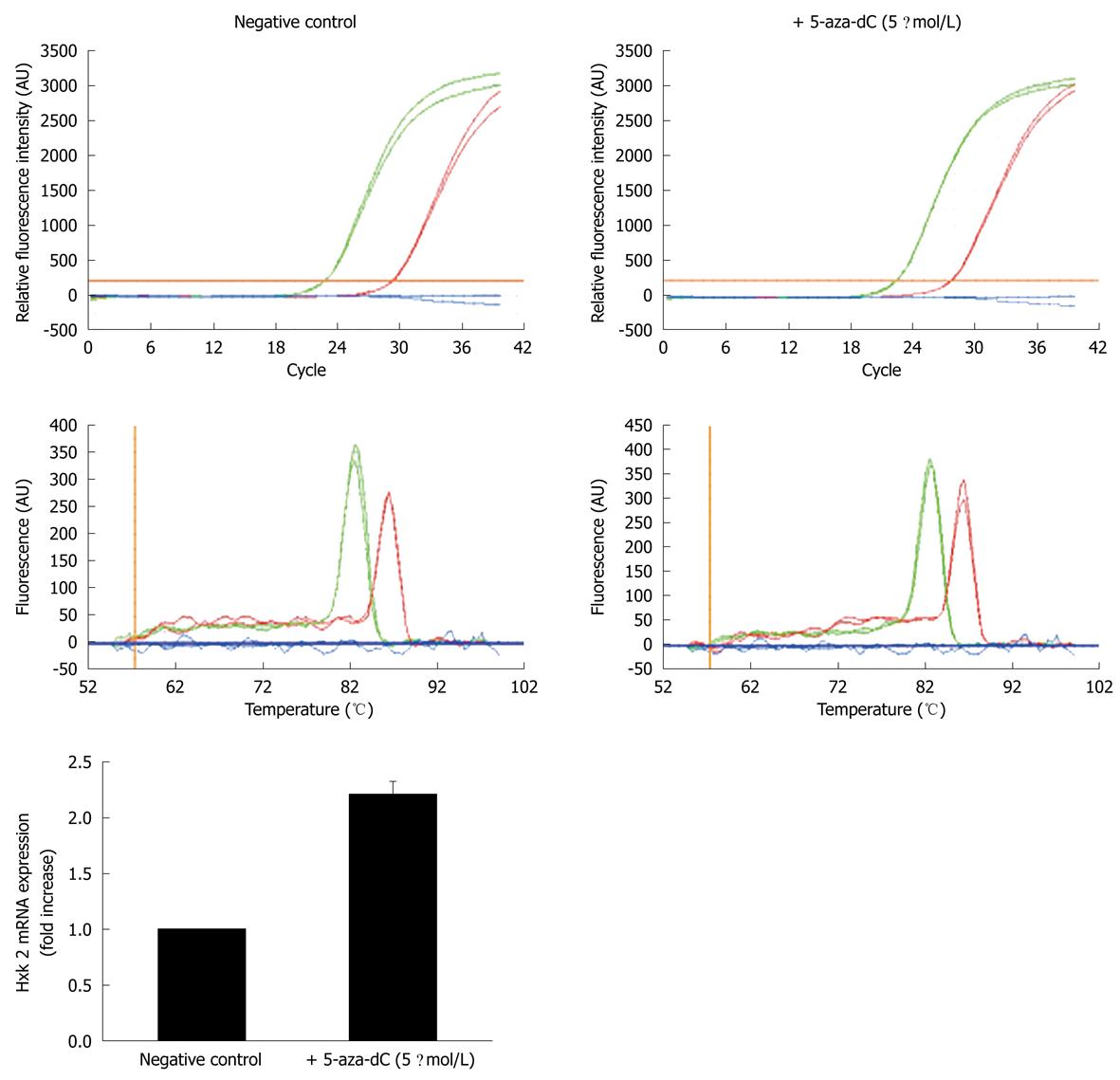
As reported by Goel and collaborators[11], DNA methylation is known to be a mechanism that is commonly used to achieve stable repression of gene expression in mammalian cells: the Hxk 2 gene is silenced through this mechanism in the normal hepatocyte cell line Clone-9 (K-9). The administration of 5 μmol/L 5-aza-dC to K-9 cells in culture is in fact able to upregulate Hxk 2 mRNA transcription by about threefold compared to untreated cells. This drug can be incorporated into nascent DNA but cannot be methylated by cellular methylases; therefore, the gene expression pattern is altered and the expression of silenced genes is reactivated. As a result, we decided to check whether PDX-1 silencing in liver cells could be exerted through methylation of CpG islands in both the promoter and the gene coding regions. We cultured wild-type K-9 cells in the presence or absence of 5 μmol/L 5-aza-dC for 4 d and prepared cDNA from total RNA to be used for QRT-PCR; the Hxk 2 gene was used as a positive control and GAPDH as a reference gene. Figure 2 shows that, as expected, the drug treatment increased the expression level of the Hxk 2 control gene, but failed to rescue the expression of PDX-1, which indicated that the DNA demethylation status induced by the drug was not sufficient to override repression of the PDX-1 gene.
Liver regeneration is a complex process that involves reactivation of cell proliferation and expression of fetal markers such as α-fetoproteins that are normally silent in adult tissue[12-15]. The protein PDX-1 is an important transcription factor that is expressed during gut endoderm differentiation and organ formation. This was the rationale to explore the possibility that liver regeneration could also trigger PDX-1 expression, and hopefully, insulin production.
Regeneration was fully operational after partial surgical ablation of the liver; this was demonstrated by the increased levels of PCNA. Other genes involved in insulin expression did not show any correlation with the molecular events that derived from the enhancement of tissue proliferation. In fact, the only marker that could be detected in the regenerating liver was E47; a transcription factor that belongs to the bHLH family, which is known to be expressed ubiquitously in mammalian cells. In the pancreas of the same animals used as controls, on the contrary, almost all tested genes were expressed and could be identified by RT-PCR. The only gene that was not expressed is Ngn3 but it is known that this gene is silenced 2 d after birth in rats. Therefore, in the light of the results reported in this brief communication we can confidently state that the molecular events that occur during liver regeneration are not per se sufficient to promote PDX-1 expression; a key transcription factor in insulin regulation.
DNA methylation is a commonly accepted mechanism for the stable repression of gene expression in mammalian cells. The Hxk 2 gene is silenced through this mechanism in the normal hepatocytes. To explore the possibility that the expression of PDX-1 might also be blocked through DNA methylation, we grew K-9 cells in the presence of 5-aza-dC; a de-methylating agent that is incorporated into nascent DNA but cannot be methylated by cellular methylases. Treatment with this drug upregulates Hxk 2 mRNA transcription, thus, the gene is actually repressed by methylation, but its function can be rescued after exposure to 5-aza-dC. We checked whether PDX-1 silencing in liver cells could be exerted through methylation of CpG islands in both the promoter and the gene coding regions. We cultured wild-type K-9 cells in the presence or absence of 5 μmol/L 5-aza-dC; the Hxk 2 gene was used as positive control and GAPDH as a reference gene. Treatment with 5-asa-dC increased expression of the Hxk 2 control gene but failed to rescue the expression/function of PDX-1. The overall meaning of this result is that DNA demethylation induced by the drug is not sufficient to override repression of the PDX-1 gene.
The data presented in this short paper clearly indicate that the molecular events that occur during liver regeneration are not sufficient to reactivate spontaneously PDX-1 gene expression. In addition, the use of a demethylating agent in cultured rat hepatocytes is not able to de-repress PDX-1 gene expression, which indicates that silencing of the gene is unlikely to be achieved through this epigenetic mechanism. In conclusion, the results presented here clearly suggest that, because neither molecular mechanism is able to re-activate PDX-1 gene function, the repression of this gene must be ascribed to a different and possibly more complex phenomenon. The peptide hormone insulin is primarily involved in glucose metabolism, and is produced by pancreatic β cells that, also due to autoimmune responses, are destroyed in type 1 diabetes. However, the reactivation at liver level of the enzyme PDX-1 for therapeutic purposes does not seem to be an immediately attainable target.
Diabetes mellitus derives from an insulin deficiency; this hormone is produced by the pancreatic β cells, which are destroyed in type 1 diabetes. A number of different transcription factors are involved in the control of insulin expression and/or regulation and the best studied is possibly PDX-1. The actual role of this transcription factor in the activation/regulation of the insulin gene is controversial and the effects of PDX-1 on each gene depend on a fine interaction between transcription factors. We recently demonstrated that the PDX-1 transcription factor can significantly alter hepatocyte glucose metabolism. In this study, we assessed whether PDX-1 expression could be spontaneously reactivated during liver regeneration in partially hepatectomized rats; also, we verified whether PDX-1 silencing in the normal hepatocytes might be achieved through DNA demethylation.
The transcription factor PDX-1 is a key molecule for approaches based on gene therapy of type 1 diabetes. The liver is considered a good candidate for the expression of insulin in patients affected by this disease. However, this transcription factor is not expressed in the liver, therefore, transformation of hepatocytes with exogenous PDX-1 gene copies is necessary. Thus, we explored the possibility that PDX-1 might be re-activated in regenerating liver, and if its silencing is caused by DNA methylation.
The molecular events that occur during liver regeneration are not sufficient to reactivate spontaneously PDX-1 gene expression. In addition, the use of a demethylating agent in cultured rat hepatocytes is not able to de-repress PDX-1 gene expression, which indicates that silencing of the gene is unlikely to be achieved through this epigenetic mechanism. These are new significant data that might have an important consequence on the activation of PFX-1 gene at the liver level for clinical/therapeutic purposes.
The results of this work suggest that neither addition of exogenous PDX-1 genes nor demethylation of the resident gene function can be used as an approach to gene therapy of type 1 diabetes. Therefore, reactivation PDX-1 enzyme at liver level for therapeutic purposes does not seem to be an immediately attainable target.
This short paper describes some interesting findings that PDX gene is not involved in liver regeneration of rats. However,major revision is needed for publication.
Peer reviewers: Veerapol Kukongviriyapan, PhD, Associate Professor, Department of Pharmacology, Faculty of Medicine, Khon Kaen University, 40002 Khon Kaen, Thailand; Takuji Tanaka, MD, PhD, Professor, FIAC, Director of The Tohkai Cytopathology Institute: Cancer Research and Prevention, 5-1-2 Minami-Uzura, Gifu 500-8285, Japan
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM
| 1. | Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220-223. |
| 2. | Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005;435:224-228. |
| 3. | Nichols J, Cooke A. Overcoming self-destruction in the pancreas. Curr Opin Biotechnol. 2009;20:511-515. |
| 4. | Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular cell biology. 4th ed. New York: WH Freeman and Co 2000; . |
| 5. | Shao S, Fang Z, Yu X, Zhang M. Transcription factors involved in glucose-stimulated insulin secretion of pancreatic beta cells. Biochem Biophys Res Commun. 2009;384:401-404. |
| 6. | Kaneto H, Matsuoka TA, Miyatsuka T, Kawamori D, Katakami N, Yamasaki Y, Matsuhisa M. PDX-1 functions as a master factor in the pancreas. Front Biosci. 2008;13:6406-6420. |
| 7. | Glick E, Leshkowitz D, Walker MD. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J Biol Chem. 2000;275:2199-2204. |
| 8. | Kishi A, Nakamura T, Nishio Y, Maegawa H, Kashiwagi A. Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am J Physiol Endocrinol Metab. 2003;284:E830-E840. |
| 9. | Pillich RT, Scarsella G, Risuleo G. Overexpression of the Pdx-1 homeodomain transcription factor impairs glucose metabolism in cultured rat hepatocytes. Molecules. 2008;13:2659-2673. |
| 10. | Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186-202. |
| 11. | Goel A, Mathupala SP, Pedersen PL. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J Biol Chem. 2003;278:15333-15340. |
| 12. | Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413-427. |
| 13. | Taub R, Greenbaum LE, Peng Y. Transcriptional regulatory signals define cytokine-dependent and -independent pathways in liver regeneration. Semin Liver Dis. 1999;19:117-127. |
| 14. | Spear BT. Alpha-fetoprotein gene regulation: lessons from transgenic mice. Semin Cancer Biol. 1999;9:109-116. |
| 15. | Kountouras J, Boura P, Lygidakis NJ. Liver regeneration after hepatectomy. Hepatogastroenterology. 2001;48:556-562. |