Published online Apr 26, 2010. doi: 10.4331/wjbc.v1.i4.41
Revised: April 7, 2010
Accepted: April 19, 2010
Published online: April 26, 2010
Progressive cell loss due to apoptosis is a pathological hallmark implicated in a wide spectrum of degenerative diseases such as heart disease, atherosclerotic arteries and hypertensive vessels, Alzheimer’s disease and other neurodegenerative disorders. Tremendous efforts have been made to improve our understanding of the molecular mechanisms and signaling pathways involved in apoptosistic cell death. Once ignored completely or overlooked as cellular detritus, microRNAs (miRNAs) that were discovered only a decade ago, have recently taken many by surprise. The importance of miRNAs has steadily gained appreciation and miRNA biology has exploded into a massive swell of interest with enormous range and potential in almost every biological discipline because of their widespread expression and diverse functions in both animals and humans. It has been established that miRNAs are critical regulators of apoptosis of various cell types. These small molecules act by repressing the expression of either the proapoptotic or antiapoptotic genes to produce antiapoptotic or proapoptotic effects. Appealing evidence has been accumulating for the involvement of miRNAs in human diseases associated with apoptotic cell death and the potential of miRNAs as novel therapeutic targets for the treatment of the diseases. This editorial aims to convey this message and to boost up the research interest by providing a timely, comprehensive overview on regulation of apoptosis by miRNAs and a synopsis on the pathophysiologic implications of this novel regulatory network based on the currently available data in the literature. It begins with a brief introduction to apoptosis and miRNAs, followed by the description of the fundamental aspects of miRNA biogenesis and action, and the role of miRNAs in regulating apoptosis of cancer cells and cardiovascular cells. Speculations on the development of miRNAs as potential therapeutic targets are also presented. Remarks are also provided to point out the unanswered questions and to outline the new directions for the future research of the field.
- Citation: Wang Z. MicroRNA: A matter of life or death. World J Biol Chem 2010; 1(4): 41-54
- URL: https://www.wjgnet.com/1949-8454/full/v1/i4/41.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v1.i4.41
It has been nearly 40 years since Kerr named the novel death process ‘‘apoptosis,’’ from the Greek word meaning ‘‘falling of the leaves’’, an active process that leads to cell death[1]. The human body destroys approximately 60 × 109 cells/d through an apoptotic process in response to various stresses such as physiological, pathogenic, or cytotoxic stimuli[2]. Unlike necrosis, apoptosis is a complex endogenous gene-controlled event that requires an exogenous signal-stimulated or inhibited by a variety of regulatory factors, such as formation of oxygen free radicals, ischemia, hypoxia, reduced intracellular K+ concentration, and generation of nitric oxide. Progressive cell loss due to apoptosis is a pathological hallmark implicated in a wide spectrum of degenerative diseases such as heart disease, atherosclerotic arteries and hypertensive vessels, and Alzheimer’s disease and other neurodegenerative disorders[3-9]. And facilitating apoptosis is also an appealing therapeutic approach in the treatment of human cancers[10-12]. Understanding the signaling pathways and mechanisms of apoptosis is of paramount importance for developing novel therapeutic strategies for human disease.
Recently, microRNAs (miRNAs), a newly recognized class of small, non-coding RNAs, have emerged as a central player in controlling cell fate: survival, growth, differentiation and death[13-16]. Since their discovery in 1993, the importance of miRNA in gene regulation has steadily gained appreciation and now miRNA biology has exploded into a massive swell of interest with enormous range and potential in almost every biological discipline. An ample amount of information on miRNAs has been accumulated in this rapidly evolving research field. Aberrant miRNA expression has been documented in human disease and in animal models as well, with evidence for a causative role in tumorigenesis and other pathological processes[15,16]. One of the most active fields of miRNA research is miRNA regulation of apoptosis. Thus far, more than 30 of 820 human miRNAs have been experimentally validated to regulate apoptosis, and the number in the list is likely to increase with more future studies. This editorial aims to give a comprehensive summary of basics of miRNAs and analysis of the currently available data regarding miRNAs in controlling apoptosis. For convenience, I term the miRNAs that are involved in regulating apoptosis “apoptosis-regulating miRNAs”. The transcriptional controls, target genes and signaling pathways linking the apoptosis-regulating miRNAs and apoptotic cell death will be discussed. The implication of manipulating the apoptosis-regulating miRNAs as a potential new strategy for molecular therapy of human disease, particularly cancer and heart disease, will be highlighted. Finally, some unanswered questions/unsolved problems and future directions of the research on apoptosis-regulating miRNAs will be pinpointed and speculated.
Genes for miRNAs are located in the chromosomes, and many of them are identified in clusters that can be transcribed as polycistronic primary transcripts. Some miRNAs are encoded by their own genes and others are encoded by the sequences as a part of the host protein-coding genes. Based on the genomic arrangement of miRNA genes, miRNAs can be grouped into two classes[15]: (1) intergenic miRNAs (miRNA-coding genes located in between protein-coding genes); and (2) intragenic miRNAs (miRNA-coding genes located within their host protein-coding genes). The intragenic miRNAs can be divided into the following subclasses: (a) intronic miRNAs (miRNA-coding genes located within introns of their host protein-coding genes); (b) exonic miRNAs (miRNA-coding genes located within exons of host protein-coding genes); (c) 3’ untranslated region (3’UTR) miRNAs (miRNA-coding genes located within 3’UTR of host protein-coding genes); and (d) 5’UTR miRNAs (miRNA-coding genes located within 5’UTR of host protein-coding genes).
According to our analysis, for the human miRNAs identified thus far, a majority of miRNAs belong to intergenic (42%) and intronic miRNAs (44%), and the other three categories are rare with the exonic miRNAs (7%), 3’UTR miRNAs (1.5%) and 5’UTR miRNAs (1%).
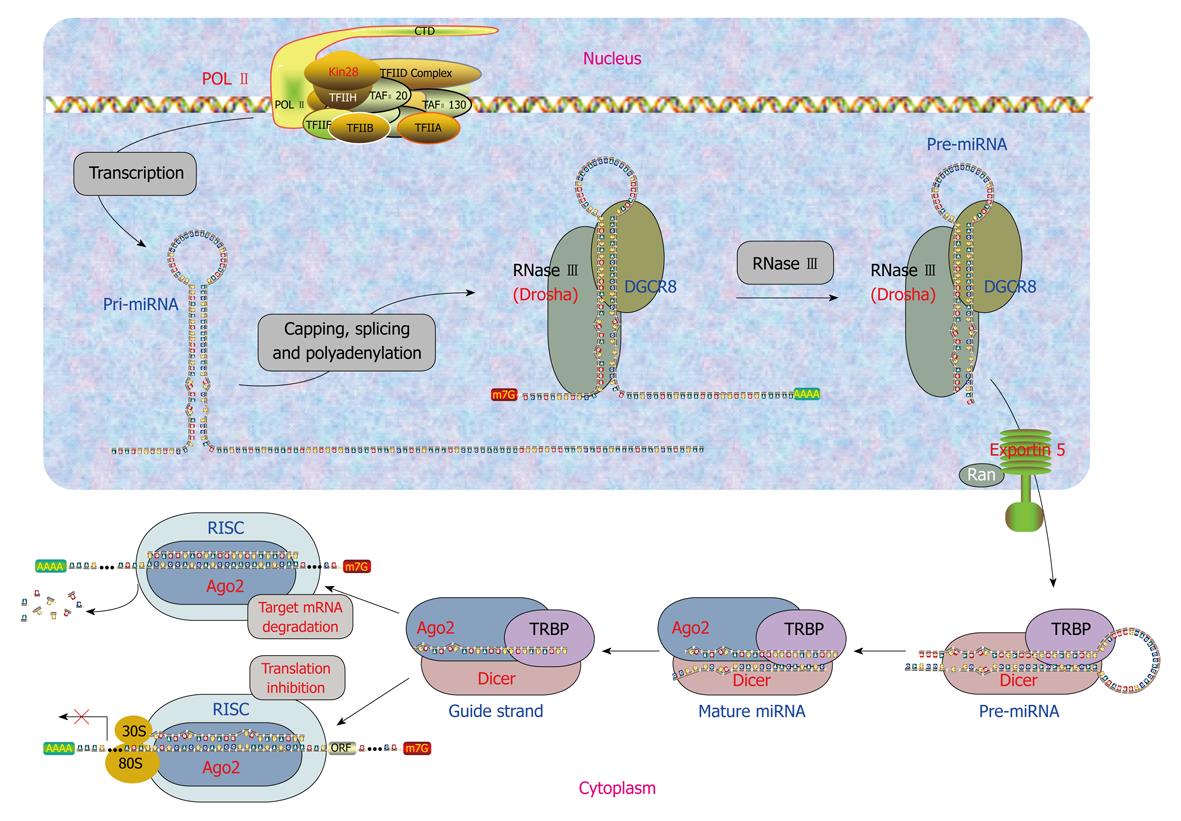
Clearly, miRNAs either have their own genes or are associated with their host genes. Accordingly, miRNAs are generated by two different mechanisms. Biogenesis of miRNAs can be summarized as a five-step process (Figure 1) as follows.
Generation of primary miRNAs: transcription of miRNA genes: The intergenic miRNA genes are first transcribed as long transcripts, called primary miRNAs (pri-miRNAs) mostly by RNA polymerase II or RNA polymerase III[17]. The pri-miRNAs are capped and polyadenylated, and can reach several kilobases in length[18,19]. The clustered miRNA genes in polycistronic transcripts are likely to be coordinately regulated[20]. The intronic miRNAs are processed by sharing the same promoter and other regulatory elements of the host genes. They are first transcribed along with their host genes by RNA polymerase II and then processed by Drosha independent pathway from excised introns by the RNA splicing machinery for their biogenesis in Drosophila, C elegans and mammals[21-23].
Generation of precursor miRNAs: endonuclease processing of pri-miRNAs: The pri-miRNAs are processed to precursor miRNAs (pre-miRNAs) by the RNase endonuclease-III Drosha and its partner DGCR8/Pasha in the nucleus[24-27]. These pre-miRNAs are about 60-100 nucleotides (nts) with a stem-loop or hairpin secondary structure. Specific RNA cleavage by Drosha predetermines the mature miRNA sequence and provides the substrates for subsequent processing steps. Cleavage of a pri-miRNA by microprocessor begins with DGCR8 recognizing the junction between single-stranded RNA and double-stranded RNA typical of a pri-miRNA[28]. Then, Drosha is brought close to its substrate through interaction with DGCR8 and cleaves the stem of a pri-miRNA 11 nts away from the two single-stranded segments.
miRNA precursor-containing introns have recently been designated as “mirtrons”[29]. Mirtrons are derived from certain debranched introns that fold into hairpin structures with 5′ monophosphates and 3′ 2-nt hydroxyl overhangs, which mimic the structural hallmarks of pre-miRNAs and enter the miRNA-processing pathway[22,23]. The discovery of mirtrons suggests that any RNA, with a size comparable to a pre-miRNA and all the structural features of a pre-miRNA, can be utilized by the miRNA processing machinery, and potentially give rise to a functional miRNA.
Nucleus to cytoplasm translocation of pre-miRNAs: Pre-miRNAs then get exported to the cytoplasm from the nucleus through nuclear pores by RanGTP and exportin-5[30-32]. After a pre-miRNA is exported to the cytoplasm, RanGTP is hydrolyzed by RanGAP to RanGDP, and the pre-miRNA is released from Exp-5.
Generation of mature miRNAs: endonuclease processing of pre-miRNAs: In the cytoplasm, pre-miRNAs are further processed by Dicer in animals, which is a highly conserved, cytoplasmic RNase III ribonuclease that chops pre-miRNAs into the duplex form of mature miRNAs of around 22 nts containing a guide strand and a passenger strand (miRNA/miRNA*), with 2-nt overhangs at the 3’ termini[19]. Like other RNase III family proteins, Dicer interacts with double-stranded RNA-binding protein partners. In mammalian cells, Dicer associates with transactivation-response element RNA-binding protein (TRBP) and protein activator of the interferon-induced protein kinase (PACT)[33,34]. In plants, miRNAs are cleaved into miRNA:miRNA* duplex possibly by Dicer-like enzyme 1 in the nucleus rather than in the cytoplasm[20,24], then the duplex is translocated into the cytoplasm by HASTY, the plant ortholog of exportin 5[20]. The strands of this duplex separate and release mature miRNA of 19-25 nts in length[20,24]. Plant miRNAs undergo further modification by methylation at the 3’ end by HEN1[35].
Formation of miRISC: Mature miRNAs gets integrated into a RNA-induced silencing complex (RISC) to form the miRNA:RISC complex (miRISC). Only one strand of miRNA/miRNA*, the guide strand, is successfully incorporated into RISC, while the other strand, the passenger strand, is eliminated. Strand selection may be determined by the relative thermodynamic stability of two ends of miRNA duplexes[36,37]. The strand with less stability at the 5’ end is favorably loaded onto RISC, whereas the passenger strand is released or destroyed. miRISC contains several proteins such as Dicer, TRBP, PACT, and Gemin3, but the components directly associated with miRNAs are Argonaute proteins (Ago). These proteins contain four domains: the N-terminal, PAZ, middle, and Piwi domains. The PAZ domain binds to the 3’ end of guide miRNA, while the other three domains form a unique structure, creating grooves for target mRNA and guide miRNA interactions[38-41]. In mammalian cells, four Ago proteins have been identified, all of which can bind to endogenous miRNAs[42]. Despite the sequence similarity among these Ago proteins, only Ago2 exhibits endonuclease activity to slice complementary mRNA sequences between positions 10 and 11 from the 5’ end of guide strand miRNA. Therefore, human Ago2 is a component of both miRISC and siRISC (siRNA-induced silencing complex), a RISC assembled with exogenously introduced siRNA. The roles of various Ago proteins in mammalian RISC are ambiguous, but the division of labor among Ago proteins in Drosophila is well defined. Drosophila Ago1 and Ago2 have been shown by biochemical and genetic evidence to participate in two separate pathways: Ago1 interacts with miRNA in translational repression, whereas Ago2 associates with siRNA for target cleavage[15,43,44].
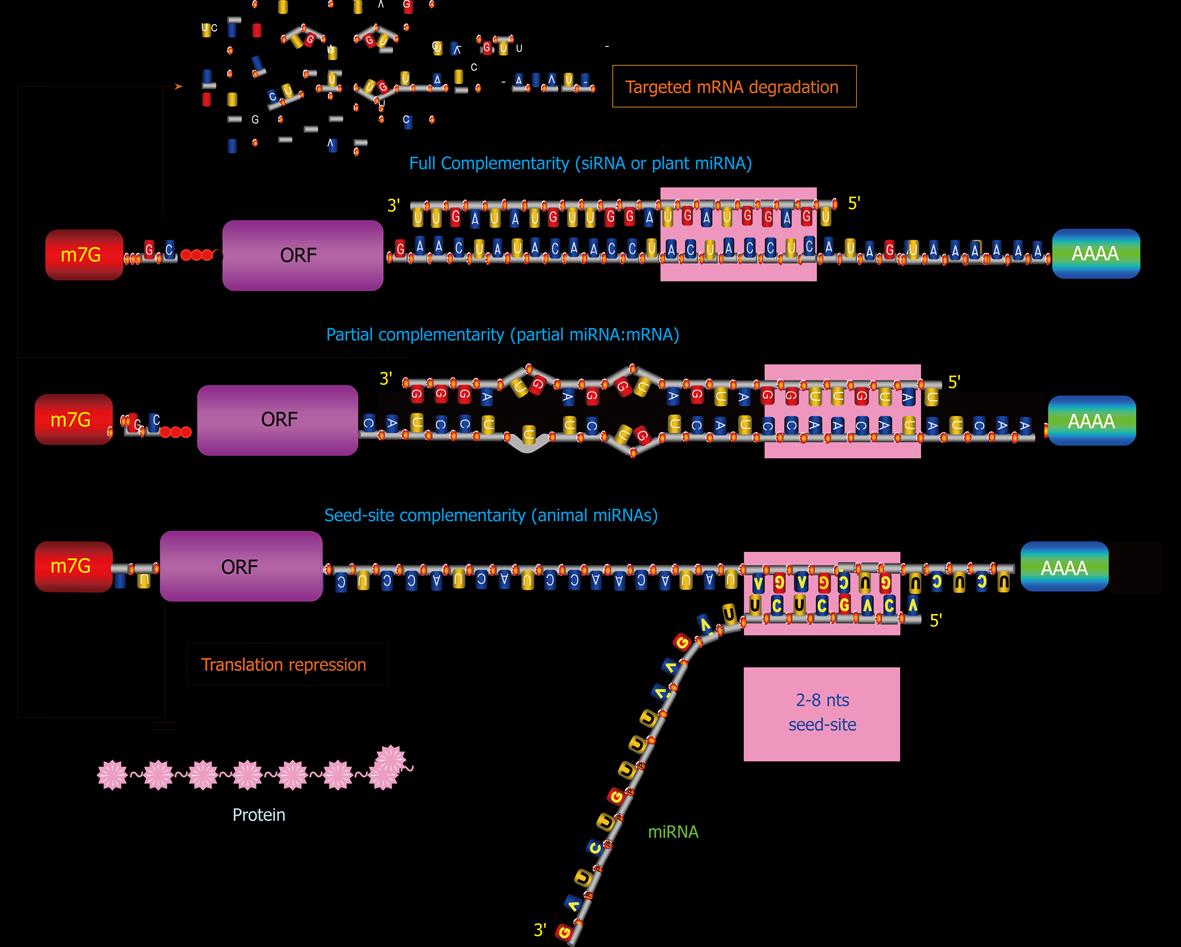
miRNAs exist in double-stranded form (duplex), activate in single-stranded form (simplex), and act in complex form miRISC. Mature miRNAs confer sequence specificities to the RISC complex. Subsequent binding of a miRNA in the miRISC to the 3’UTR of its target mRNA through a Watson-Crick basepairing mechanism with its 5’-end 2-8 nts exactly complementary to recognition motif within the target (taking into account that an RNA-RNA hybrid can also contain G-U matches). This 5’-end 2-8 nt region is termed “seed sequence” or “seed site”, for it is critical for miRNA actions[45,46]. Partial complementarity with the rest of the sequence of a miRNA also plays a role in producing post-transcriptional regulation of gene expression, presumably by stabilizing the miRNA:mRNA interaction. Moreover, the mid and 3’-end regions of a miRNA may also be important for forming miRISC. Studies have shown that in addition to 3’UTR, coding region and 5’UTR can also interact with miRNAs to induce gene silencing[47-49].
In mammalian species, the assembly of the miRNA/RISC on a 3′UTR can potentially influence protein production by enhancing de-adenylation with subsequent degradation of the mRNA or by repressing translation initiation or both[50-53], depending upon at least the following factors (Figure 2): (1) The overall degree of complementarity of the binding site; (2) The number of recognition motif corresponding to 5’-end 2-8 nts of the miRNA; and (3) The accessibility of the bindings sites (as determined by free energy states)[54-56].
The greater the degree of complementarity of accessible binding sites, the more likely a miRNA degrades its targeted mRNA. The loose binding constraints allow one miRNA to bind to several sites within one 3′UTR. Perfectly complementary targets (full miRNA:mRNA interaction) are efficiently silenced by the endonucleolytic cleavage activity of some Ago proteins[50,57,58], but the vast majority of predicted targets in animals are only partially paired (partial miRNA:mRNA interaction)[45,46,59-62] and can hardly be cleaved[63]. Some miRNAs have only seed-site complementarity (seed-site miRNA:mRNA) and this interaction primarily leads to translation inhibition. And those miRNAs that display imperfect sequence complementarities with target mRNAs primarily result in translational inhibition[45,46,54-56]. The mechanisms for translational inhibition remain largely unknown, although inhibition of translation initiation has been identified as one such mechanism by several studies[51,64]. Greater actions may be elicited by a miRNA if it has more than one accessible binding sites in its targeted miRNA, presumably by the cooperative miRNA:mRNA interactions from different sites. mRNA degradation by miRISC is initiated by deadenylation and decapping of the targeted mRNAs[56]. A recent study demonstrated, however, that miRNAs can also act to enhance translation when AU-rich elements and miRNA target sites coexist at proximity in the target mRNA and when the cells are in the state of cell-cycle arrest[65].
The loose binding constraints also allow one miRNA to bind multiple mRNA targets within the transcriptome. This multiplicity endows miRNAs in principle with the ability to inhibit several genes at once, leading to a much stronger biological response due to multiple effects on one pathway or coordinated effects on several pathways. It has been predicted that each single miRNA can have > 1000 target genes and each single protein-coding gene can be regulated by multiple miRNAs[45,46,54-56,66,67]. This is at least partially a result of a lax requirement of complementarity for miRNA:mRNA interaction[52]. This implies that actions of miRNAs are sequence- or motif-specific, but not gene-specific; different genes can have same binding motifs for a given miRNA and a given gene can have multiple binding motifs for distinct miRNAs. On the downside, the relaxed stringency of miRNAs, with regard to their potential targets, enhances the possibility of disadvantageous off-target effects on inappropriate mRNAs. Another disadvantage is the difficulty for researchers trying to interpret the biological significance of altered expression of a given miRNA by determination of its relevant downstream targets. Based on the characteristics of miRNA actions, I have postulated that a miRNA should be viewed as a regulator of a cellular function or a cellular program, not of a single gene[14].
miRNAs are an abundant RNA species constituting > 3% of the predicted human genes, which regulates about 30% of protein-coding genes[52]. The high sequence conservation across metazoan species and the ability of individual miRNAs to regulate the expression of multiple genes confer strong evolutionary pressure and participation of miRNAs in essential biologic processes such as cell proliferation, differentiation, apoptosis, metabolism, stress-response, etc.[66,67].
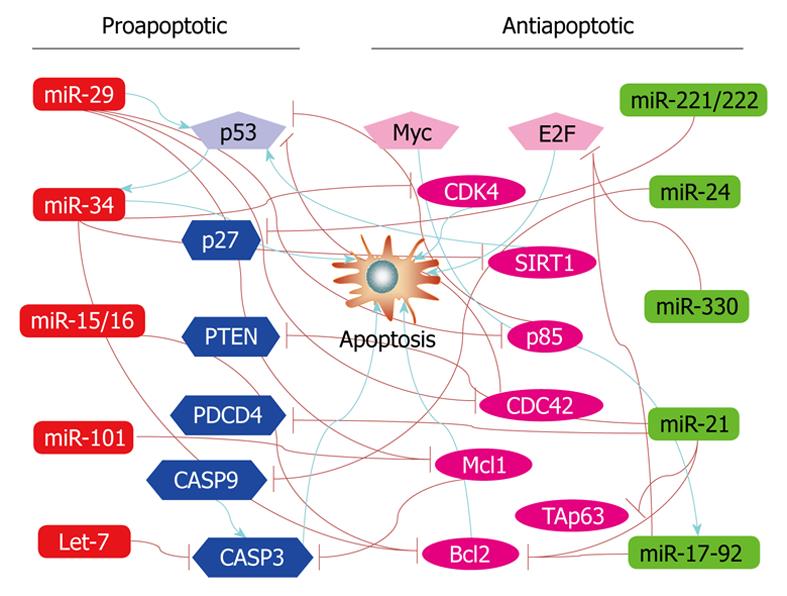
To date, no less than 30 individual miRNAs have been known to play a role in regulating apoptosis. These include the let-7 family, miR-1, miR-1d, miR-7, miR-14, miR-15a, miR-16-1, miR-17 cluster (miR-17-5p, miR-18, miR-19a, miR-19b, miR-20 and miR-92), miR-21, miR-29, miR-34a, miR-133, miR-146a, miR-146b, miR-148, miR-191, miR-204, miR-210, miR-214, miR-216, miR-278, miR-296, miR-335, miR-Lat, and bantam. The list is expected to expand quickly with more studies. Indeed, we have performed a bioinformatics prediction of the vertebrate miRNAs available to date in miRBase using a target scan computational analysis with miRBase and miRanda and surprisingly found that nearly all known vertebrate miRNAs (approximately 70%) have at least one target gene related to cell death or survival. This suggests that a majority of, if not all, vertebrate miRNAs can regulate apoptosis in at least some cell types.
Among the known apoptosis-regulating miRNAs, some are designated as anti-apoptotic and others as proapoptotic miRNAs. This distinction is primarily based on experimental results from a particular cell type.
In general, the following miRNAs are considered anti-apoptotic: miR-17-5p, miR-20a, miR-21, miR-133, miR-146a, miR-146b, miR-191, miR-14, bantam, miR-1d, miR-7, miR-148, miR-204, miR-210, miR-216, miR-296, and miR-Lat. For example, inhibition of miR-17-5p and miR-20a with their antisense oligonucleotides, induces apoptosis in lung cancer cells, indicating that miR-17-5p and miR-20a can protect these cells against apoptosis[68]. Knockdown of miR-21 in cultured glioblastoma cells resulted in a significant drop in cell number. This reduction was accompanied by increases in caspase-3 and -7 enzymatic activities and TUNEL staining[69,70]. Similarly, in MCF-7 human breast cancer cells, miR-21 also elicits anti-apoptotic effects[71,72]. miR-Lat was reported to protect neuroblastoma cells against apoptotic death[73].
On the other hand, several miRNAs have been referred to proapoptotic ones, which include let-7 family, miR-15a, miR-16-1, miR-29, miR-34a, miR-34b, miR-34c, miR-1, and miR-214. The best example of proapoptotic miRNAs is probably miR-15a and miR-16-1 that when forced to express induces apoptosis in chronic lymphocytic leukemia cells[74]. Equally interesting is the finding that when Wi38 human diploid fibroblasts transduced with an AMO against miR-34 were treated with the apoptosis-inducing agent staurosporine, fewer early apoptotic cells and more viable cells were observed. It has been proposed that miR-34 can mediate key effects associated with p53 function for p53 transactivates gene expression of miR-34[75]. Similar relationships between miR-34a and p53 have also been confirmed in other cells, such as H1299 human lung cancer cells, MCF-7 human breast cancer cells, U-2OS osteosarcoma cells[76], and in p53 wild-type HCT116 colon cancer cells[77]. Expression of miR-34a causes dramatic reprogramming of gene expression and promotes apoptosis[78]. A most recent study further revealed that overexpression of miR-34a causes a dramatic reduction in cell proliferation through the induction of a caspase-dependent apoptotic pathway in three neuroblastoma cell lines, Kelly, NGP and SK-N-AS[79]. On the other hand, Bommer et al[80] demonstrated that the expression of miR-34b and miR-34c was dramatically reduced in 6 of 14 (43%) non-small cell lung cancers (NSCLCs) and that the restoration of miR-34 expression inhibited growth of NSCLC cells.
It is noticed that the previous work on miRNAs and apoptosis has been mostly limited to the context of cancer, while studies on apoptosis regulation by miRNAs in non-cancer cells have been sparse. We have recently found that miR-133, one of the muscle-specific miRNAs, produced antiapoptotic actions in neonatal rat ventricular myocytes[81]. Noticeably, this cytoprotective effect may be weakened in hypertrophic heart since miR-133 level has been found significantly reduced under such conditions and this may contribute to increased tendency of apoptosis induction in hypertrophic myocytes[82-84]. Our study demonstrated that miR-1, another muscle-specific miRNAs, promotes the apoptosis induced by oxidative stress in cardiac cells counteracting with miR-133[81]. Intriguingly, our earlier study revealed that miR-1 level was elevated by 2-3 folds in ischemic myocardium[85]. Whether this altered miR-1 expression is linked to increased apoptotic cell death in myocardial infarction[86,87] merits further studies to verify.
Nonetheless, precaution must be taken when attempting to categorize a miRNA by its role in apoptosis. Each miRNA has the potential to regulate > 1000 protein-coding genes that could well be a mixture of some antiapoptotic and proapoptotic genes[29]. Whether a miRNA is antiapoptotic or proapoptotic may largely depend upon the cell-specific expression of genes involving in apoptosis and survival. There indeed have been a few instances reinforcing the needs to consider cell context as an important index for the role of miRNAs in apoptosis. As mentioned above, miR-21 has been considered as antiapoptotic in glioblastoma[69] and MCF-7 cells[71]. In HeLa cells, however, miR-21 does the opposite; inhibition of miR-21 increased the number of surviving cells[88]. Moreover, inhibition of miR-21 in A549 human lung cancer cells fails to alter cell death or growth[88]. Evidently, a same miRNA can have three different actions, antiapoptotic, proapoptotic or neutral, in different cell types. Similarly, miR-24 promotes growth in A549 cells, but inhibits growth in HeLa cells[88].
Many human diseases are related to cell proliferation and cell death or a loss of balance between cell proliferation and cell death. The regenerative types of disease (such as cancer) are associated with increased cell proliferation and/or decreased cell death (particularly apoptosis). On the contrary, the degenerative types of disease (such as heart failure) are in general linked to increased apoptotic cell death. From this point of view, it is quite plausible that the miRNAs able to regulate apoptosis are importantly involved in the human diseases associated with cell proliferation and apoptosis, and are vivid targets for therapeutic interventions of the disease.
Mounting evidence from both basic and clinical studies indicates that miRNAs are aberrantly expressed in cancer, and different types of cancer have different expression profiles of miRNAs, which can lead to a novel cancer-specific and cancer type-selective treatment strategy. The implication of miRNAs in cancer therapy is obvious and there have been several excellent review articles detailing miRNAs and cancer[89-105].
miRNAs involved in cancer cell apoptosis: Around 30 miRNAs have been characterized for their pro- or anti-apoptotic properties. This editorial does not intend to do an exhausted description of every single miRNAs; instead it aims to highlight a few important points. (1) Deregulated expression of miRNAs has been characterized in a variety of human cancers including chronic lymphocytic leukemia[106], breast[107], lung cancers[108], pancreatic cancer, and six solid tumors including breast, colon, lung, pancreas, prostate and stomach[109]. The miRNAs that have attracted tremendous attention from both scientists and clinicians mainly include miR-21, miR-34a, miR-15a, miR-16-1, miR-17-5p, miR-20a, and let-7. The findings could aid in the diagnosis and prognosis of cancer[108]; (2) Some of the miRNAs are conserved and the others are distinct among various types of human cancers in terms of their expression deregulation. For example, miR-21, miR-191 and miR-17-5p are abnormally overexpressed in six solid tumors studied, including breast, colon, lung, pancreas, prostate and stomach[109]. By comparison, other miRNAs such as miR-155, miR-146 and miR-20a are more restricted to certain types of tumors. The differential alterations of miRNAs provide opportunities for interventions aiming at particular types of cancer; (3) Studies have well demonstrated the feasibility of targeting the relevant miRNAs to facilitate cancer cell death and/or to inhibit cancer cell growth under in vitro conditions. Use of the xenograft mouse model to confirm the anti-tumor efficacy under in vivo conditions has also been documented. Si et al[71] showed that one transient transfection with knockdown of miR-21 by AMO is sufficient to cause substantial inhibition of tumor growth in MCF-7 human breast cancer cells and in the xenograft tumors generated by MCF-7 cells, by effectuating an increase in apoptosis associated with downregulation of Bcl-2 expression. This finding with miR-21 and MCF-7 in nude mice was confirmed by an independent group[72]. In another study on miR-21, the intracranial graft survival of miR-21-antagonized (by AMO) gliomas was shown to be diminished, as indicated by a sharp reduction of tumor volumes in vivo compared with control-treated gliomas[70]. Meng et al[111] described a detailed investigation on the anticancer potential of let-7 depletion by AMO and the underlying molecular mechanisms. The authors revealed an increase in basal expression of phosphor-Stat3 and decrease in neurofibromatosis 2 (NF2) in homogenates from xenograft tumors generated by Mz-IL-6 human malignant cholangiocytes injected subcutaneously. Intratumoral administration of let-7a AMO increased NF2 and decreased phosphor-Stat3 expression in Mz-IL-6 xenografts in vivo. Moreover, they observed a decrease in tumor growth consistent with increased gemcitabine toxicity in response to let-7a AMO when compared with the tumors that were untreated. Subcutaneous administration of exogenous miR-34a considerably suppressed in vivo growth of HCT116 and RKO human colon cancer cells in tumors in nude mice[77]. The c13orf25/miR-17 cluster, which is responsible for 13q31-q32 amplification in malignant lymphoma, contains the miRNA-17-18-19-20-92 polycistron. Tazawa et al[77] demonstrated that the nude mice injected with rat fibroblasts pretreated with both miR-17 polycistron and Myc by transfection showed accelerated tumor growth as compared with those injected with only Myc transfectant cells. The finding suggests that an anti-miR-17 antisense should be able to suppress tumor growth; and (4) Also important is that some of the signaling molecules that can mediate the apoptosis-regulating actions of miRNAs have been identified. Understanding the molecular mechanisms should allow for more rational considerations of miRNAs as a target for cancer therapy.
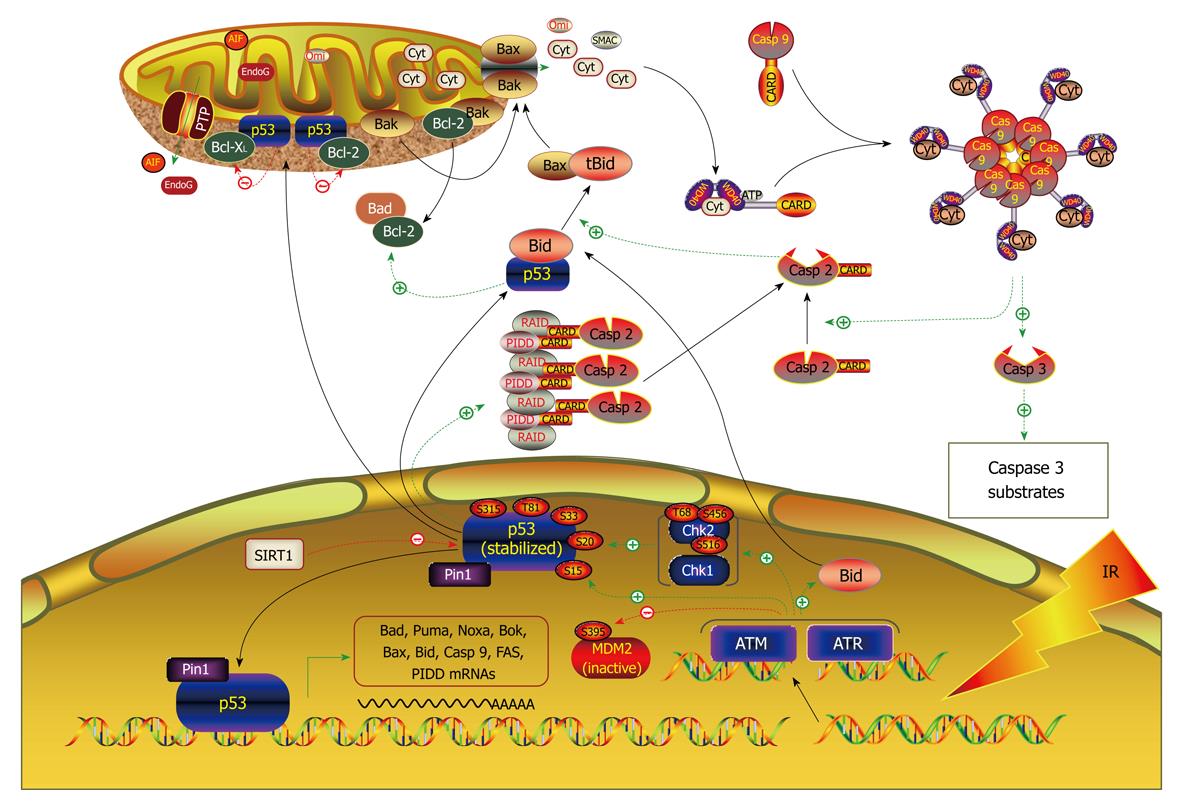
Signaling pathways mediating the effect of miRNAs on cancer cell apoptosis: The miRNAs known to be involved in regulation of apoptosis seem to be associated with most of the signaling pathways (Figure 3). Cimmino et al[74] demonstrated that miR-15a and miR-16-1 expression is inversely correlated to Bcl-2 expression in chronic lymphocytic leukemia and that both miRNAs negatively regulate Bcl-2 at a post-transcriptional level. The regulation of Bcl-2 protein levels by these two miRNAs depends on the potential binding site in the 3’UTR of Bcl-2. Bcl-2 repression by these miR-15a and miR-16-1 induces apoptosis in the leukemic cell line model. Johnson et al[112] found that the 3′UTRs of the human RAS oncogenes contain multiple complementary sites for let-7, allowing let-7 to regulate RAS expression through post-transcriptional repression. Lung tumor tissues display significantly reduced levels of let-7 and significantly increased levels of RAS protein relative to normal lung tissue, suggesting let-7 regulation of RAS as a mechanism for let-7 in lung oncogenesis. RAS as target for let-7 repression was verified by their experimental results. In a separate study by Meng et al[111], however, let-7a was shown to act by a different mechanism; it enhances IL-6 production and subsequent constitutive Stat-3 phosphorylation and activation via direct post-transcriptional repression of NF2 in human malignant cholangiocytes Mz-1 and Mz-IL-6 cells. Intratumoral administration of AMO against let-7a increased NF2 and decreased Stat-3 phosphorylation in Mz-IL-6 xenografts in vivo[110]. Overexpression of miR-21 paradoxically upregulates Bcl-2 in MCF-7 breast cancer cells, possibly via an indirect pathway[71]. Regulation of PTEN/Akt and Bcl-2 by miR-21 is in agreement with its cytoprotective ability against apoptosis. Two most recent studies revealed that the tumor suppressor protein programmed cell death 4 (PDCD4) is a functionally important target for miR-21 in breast cancer cells[113] and colorectal cancer cells[114]. E2F1, a direct downstream target for c-myc, is negatively regulated by miR-17-5p and miR-20a, which may account partially for the antiapoptotic effects of these miRNAs[115,116]. miR-29 was found to be an endogenous regulator of Mcl-1 protein expression, and thereby, apoptosis[117]. Enforced miR-29b expression reduced expression of Mcl-1 protein, an anti-apoptotic Bcl-2 family member, in KMCH malignant cholangiocarcinoma cells, and sensitized the cancer cells to tumor necrosis factor-related apoptosis-inducing ligand cytotoxicity. This effect was direct, as miR-29b negatively regulated the expression of a Mcl-1 3’UTR-based reporter construct. miR-34a has recently attracted tremendous attention owing its ability to mediate p53-induced apoptosis[75,76,78,80]. Data have now been available indicating that miR-34a directly targets the E2F3 mRNA and significantly reduces the levels of E2F3 protein in neuroblastoma[79]. Similar results were obtained by an independent group in two colon cancer cell lines, HCT 116 and RKO. The authors showed that introduction of miR-34a into the cells elicits a profound inhibition of cell proliferation accompanying the down-regulation of the E2F family[77]. In addition, the same group also found that miR-34a causes upregulation of the HBP1 gene, which is associated with oncogenic RAS-induced premature senescence. miR-27a reportedly produces antiapoptotic effect in SKBr3 human breast cancer cells[118]. Reverse transcriptase polymerase chain reaction (RT-PCR) study showed that miR-27a AMO significantly upregulates ZBTB10/RINZF, a Sp1 repressor, and RYBP/DEDAF, an apoptotic facilitator, in SKBr3 cells. Northern blots confirmed the up-regulation. Repression of these two target genes by miR-27a may underlie its antiapoptotic efficacy. The latency-associated transcript (LAT) of herpes simplex virus-1 (HSV-1) inhibits apoptosis and maintains latency by promoting the survival of infected neurons. Gupta and colleagues[73] found that the antiapoptotic action of LAT depends on a miRNA. The group first identified a new miRNA from the exon 1 region of the HSV-1 LAT gene and they named it miR-LAT. Subsequently, they confirmed that miR-LAT exerts its antiapoptotic effect by downregulating the transforming growth factor (TGF)-1 and SMAD3 expression, both of which are functionally linked in the TGF-pathway (Figure 4).
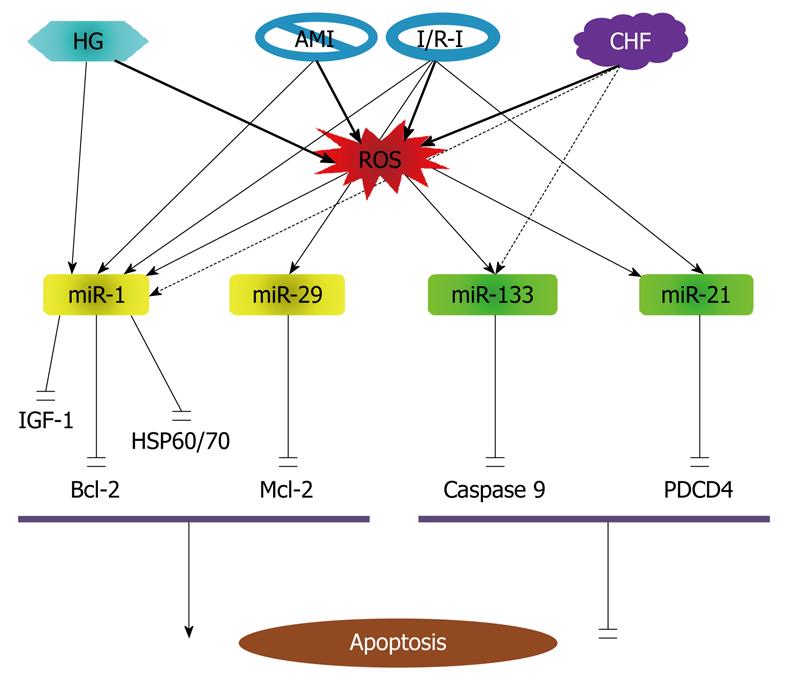
It is noticed that the previous work on miRNAs and apoptosis has been mostly limited to the context of cancer, while studies on apoptosis regulation by miRNAs in non-cancer cells have been sparse. The first evidence for the role of miRNAs in cardiomyocyte apoptosis was obtained in 2007 from my laboratory demonstrating the proapoptotic effect of miR-1 and anti-apoptotic effect of miR-133 in response to oxidative stress[81]. Subsequent studies in 2010 and 2008 revealed the involvement of other miRNAs such as miR-21, miR-24 and miR-29 in ischemic myocardial injury[119,120]. The currently known apoptosis-regulating miRNAs in cardiomyocytes are illustrated in Figure 5.
Role of miR-1 and miR-133: Oxidative stress constitutes one of the major threats to the living system and accumulative oxidative damage has been implicated in many degenerative conditions including the aforementioned diseases such as heart failure, atherosclerosis and Alzheimer’s disease[121-125], and even aging process, all of which are associated with progressive decrease in cell density. Indeed, oxidative stress is one of the most crucial factors causing neurodegenerative disorders. There is also a growing body of evidence that oxidative stress increases in myocardial failure and may contribute to the structural and functional changes that lead to disease progression[123]. Moreover, oxidative stress is a well-known factor promoting apoptosis[126-129].
We found that miR-1 level is significantly increased in response to oxidative stress in cardiomyocytes[81]. This overexpression is involved in regulation of apoptotic cell death. Overexpression of miR-1 in H9c2 rat myoblasts provoked apoptotic cell death, which was partially rescued by treatment with miR-133[81]. Similar effects on oxidative stress-induced apoptosis were observed in hydrogen peroxide (H2O2)-treated H9c2 cells, wherein cotransfection with miR-1 led to 60% reduction in the IC50 value necessary for oxidative stress-induced DNA fragmentation, and cotransfection with miR-133 resulted in a 40% increase in the IC50 value required for oxidative stress-induced DNA fragmentation. Qualitatively similar results were obtained when rat neonatal ventricular myocytes were exposed to H2O2. Subsequent computational analyses predicted that heat shock protein HSP60 and HSP70 were targets for miR-1 and that caspase-9 was a target for miR-133. This was verified experimentally by showing that miR-1 suppressed HSP60 and HSP70 protein levels by about 70% and 60%, respectively, whereas miR-133 decreases protein levels of caspase-9 and caspase-9 activity levels in rat H9c2 cells. The post-transcriptional repression of HSP60 and HSP70 and caspase-9 was confirmed by luciferase reporter experiments. We postulated that the relative levels of miR-1 and miR-133 may determine cell fate.
It is noteworthy that expression of miR-1 is upregulated in coronary disease, acute myocardium ischemia (AMI) and ischemia/reperfusion injury (I/R-I)[85,130], but is downregulated in cardiac hypertrophy/heart failure[121,131,132], wherein apoptosis is an important mode of cell death[86,87]. In this regard, it seems that increase in miR-1 during AMI and I/R-I contributes to apoptosis in these settings, and decrease in miR-1 in cardiac hypertrophy/heart failure is an adaptive change to minimize its deleterious effect. On the other hand, the cytoprotective effect of miR-133 may be weakened in cardiac hypertrophy/heart failure since miR-133 level has been found significantly reduced under such conditions and this may contribute to increased tendency of apoptosis induction in hypertrophic myocytes[82-84]. These notes merits future studies to verify.
The proapoptotic action of miR-1 in cardiomyocytes has been subsequently reproduced by other laboratories. A 2008 study by Yu et al[133] investigated the possible miRNA mechanism for glucose cardiotoxicity. Glucose toxicity is an important initiator of cardiovascular disease, contributing to the development of cardiomyocyte death and diabetic complications. Yu et al[133] showed that H9C2 cells exposed to high glucose have increased miR-1 expression level, decreased mitochondrial membrane potential, increased cytochrome-c release, and increased apoptosis. Glucose induced mitochondrial dysfunction, cytochrome-c release and apoptosis were blocked by IGF-1. miR-1 mimics, but not mutant miR-1, blocked the capacity of IGF-1 to prevent glucose-induced mitochondrial dysfunction, cytochrome-c release and apoptosis. The finding indicates that IGF-1 inhibits glucose-induced mitochondrial dysfunction, cytochrome-c release and apoptosis via downregulating miR-1 expression. The finding, according to the authors, provides a miRNA mechanism for the deleterious effects of glucose in the development of cardiomyocyte death and diabetic complications.
In a most recent study in 2009, Tang et al[130] studied the role of miR-1 in a rat model of I/R-I. The level of miR-1 was found inversely correlated with Bcl-2 protein expression in cardiomyocytes with I/R-I. The in vitro level of miR-1 was dramatically increased in response to H2O2. Overexpression of miR-1 facilitated H2O2-induced apoptosis in cardiomyocytes. Inhibition of miR-1 by antisense inhibitory oligonucleotides caused marked resistance to H2O2. Through bioinformatics, the authors identified the potential target sites for miR-1 on the 3’UTR of Bcl-2. miR-1 significantly reduced the expression of Bcl-2 at both mRNA and protein levels. The post-transcriptional repression of Bcl-2 was further confirmed by luciferase reporter experiments. The data indicate that miR-1 is not only proarrhythmic in AMI[85] but also proapoptptic in I/R-I. The findings from these above studies also indicate that the proapoptotic action of miR-1 involves multiple death signaling pathways including HSP60/70, IGF-1 and Bcl-1, in a coordinated manner or separately under different situations.
Role of miR-21: In addition to miR-133, it appears that miR-21 is also an anti-apoptotic miRNA. miR-21 has been found upregulated in various cancers (malignant glioblastoma, colorectal carcinoma, cervical adenocarcinoma) and, based on its in silico-predicted proapoptotic gene targets, has been proposed as a potential antiapoptotic miRNA[71,114,115]. Knockdown of miR-21 in glioblastoma cells leads to caspase activation and apoptotic cell death, and depletion of miR-21 in vascular smooth muscle cells (VSMCs) results in a dose-dependent increase in apoptosis and decrease in cell proliferation[68]. As noted above, several independent groups have shown that the expression levels of miR-21 are increased by 2-4 folds in hypertrophied heart[134-137]. Although miR-21 levels were not examined in relation to myocyte apoptosis in these studies, the upregulation of miR-21 may represent a prosurvival stress response in response to hemodynamic pressure overload.
Yin et al[138] investigated the role of miRNA in protection against I/R-I in heart. Mice subjected to cytoprotective heat-shock (HS) showed a significant increase of miR-1 by 80%, miR-21 by 100% and miR-24 by 60% in the heart, as determined by qPCR. miRNAs isolated from HS mice and injected into non-HS mice significantly reduced infarct size after I/R-I, which was associated with the inhibition of pro-apoptotic genes and increase in anti-apoptotic genes. Chemically synthesized miR-21 also reduced infarct size, whereas a miR-21 inhibitor abolished this effect. These studies provide evidence for the potential role of endogenous miRNA in cardioprotection following I/R-I. miRNA treatment caused profound changes in several apoptotic related genes as determined by gene microarray analysis. The caspase family members 1, 2, 8 and 14 were suppressed in the hearts of HS mice treated with miRNA as compared with the controls. Except for BNIP-3, most of the pro-apoptotic genes, including Bid (BH3 interacting domain death agonist), Bcl-10 (B-cell leukemia/lymphoma 10), Cidea (cell death-inducing DNA fragmentation factor, alpha subunit-like effector A), Ltbr (lymphotoxin B receptor), Trp53 (transformation related protein 53), Fas (TNF receptor superfamily member) and Fasl (Fas ligand, TNF superfamily, member 6), were also repressed. On the other hand, the anti-apoptotic genes, Bag-3 (Bcl-2-associated athanogene and Prdx2 (Peroxiredoxin 2) were increased. According to previous studies, miR-1 has apoptosis-promoting effect[81,130,133], miR-21 is anti-apoptotic in cardiac cells, and miR-24 is anti-apoptotic in cancer cells[139]. In particular, miR-1 was found to induce cardiomyocytes apoptosis in a rat model of I/R-I[130]. It is possible that the anti-apoptotic effect of these miRNAs in I/R-I is a net outcome of the counteracting effects of these miRNAs. Moreover, most of the apoptosis-related proteins reported in this study are predicted not to be the targets for these miRNAs and whether the observed changes in their expression were ascribed to the upregulation of these miRNAs are not certain; by logic, the upregulation of the anti-apoptotic proteins could not be the direct consequence of the upregulation of the miRNAs.
In a subsequent study, Cheng et al[139] confirmed the anti-apoptoptic effect of miR-21 in cardiac cells. Using quantitative real-time RT-PCR (qRT-PCR), the authors demonstrated that miR-21 was upregulated in cardiac myocytes after treatment with H2O2. H2O2-induced cardiac cell death and apoptosis were increased by miR-21 inhibitor and was decreased by pre-miR-21. PDCD4 was established as a direct target of miR-21 in cardiac myocytes. Pre-miR-21-mediated protective effect on cardiac myocyte injury was inhibited in H2O2-treated cardiac cells via adenovirus-mediated overexpression of PDCD4 without miR-21 binding site. Moreover, activator protein 1 was a downstream signaling molecule of PDCD4 that was involved in miR-21-mediated effect on cardiac myocytes. The results suggest that miR-21 is sensitive to H2O2 stimulation. miR-21 participates in H2O2-mediated gene regulation and functional modulation in cardiac myocytes. miR-21 might play an essential role in heart diseases related to oxidative stress such as cardiac hypertrophy, heart failure, myocardial infarction, and myocardial I/R-I.
Role of miR-29: Pioglitazone, a peroxisome proliferator-activated receptor (PPAR)-γ agonist, has been documented by numerous studies to be able to limit myocardial infarct size in experimental animals[115,116]. Ye et al[119] assessed the effects of PPAR-γ activation on myocardial miRNA expression and the role of miRNAs in I/R-I in the rat heart after pioglitazone administration using miRNA microarray methods, followed by Northern blot verification. They found that miR-29a and miR-29c levels were decreased after 7-d treatment with pioglitazone. In H9c2 cells, the effects of pioglitazone and rosiglitazone on miR-29 expression levels were blocked by a selective PPAR-γ inhibitor GW9662. Down-regulation of miR-29 by antisense inhibitor or by pioglitazone protected H9c2 cells from simulated IR injury, with increased cell survival and decreased caspase-3 activity. In contrast, overexpressing miR-29 promoted apoptosis and completely blocked the protective effect of pioglitazone. Antagomirs against miR-29a or -29c significantly reduced myocardial infarct size and apoptosis in hearts subjected to I/R-I. Western blot analyses demonstrated that Mcl-2, an anti-apoptotic Bcl-2 family member, was increased by miR-29 inhibition, similar to the finding in cancer cells[117]. Clearly, down-regulation of miR-29 protected hearts against I/R-I through its anti-apoptotic activity. miR-29 thus represents another proapoptotic miRNA in cardiac cells, in addition to miR-1[81].
It is known that proliferative vascular diseases share similar cellular events and molecular mechanisms with cancer, and neointimal lesion formation is the pathological basis of proliferative vascular diseases. Neointimal growth is the balance between proliferation and apoptosis of VSMCs. The increased VSMC proliferation or the relative decreased VSMC apoptosis are responsible for neointimal lesion formation. In a recent study of the potential roles of miRNAs in VSMCs proliferation and apoptosis[68], the authors found that miR-21 level was up-regulated in proliferative VSMCs and depletion of miR-21 resulted in decreased cell proliferation and increased cell apoptosis in a dose-dependent manner in cultured rat aortic VSMCs. This suggests that miR-21 has a proproliferative and anti-apoptotic effect on VSMCs. miR-21 inhibition upregulated, whereas miR-21 overexpression downregulated, expression of PTEN protein in VSMCs, using both “loss-of-function” and “gain-of-function” approaches. They further demonstrated that inhibition of miR-21 downregulated, whilst overexpression of miR-21 upregulated, the level of Akt protein that mediates survival signal in a cell. These results are consistent with the expression changes of PTEN. In contrast to PTEN, miR-21 knockdown decreased and overexpression increased expression of antiapoptotic Bcl-2 protein. The authors suggested that Bcl-2 might be an indirect target of miR-21 in VSMCs by suppressing expression of a gene that negatively regulates Bcl-2 expression or that miR-21 might be able to directly affect Bcl-2 expression via binding to the sequence outside the 3’UTR. Thus, Bcl-2 might be involved in the anti-apoptotic action of miR-21 in VAMCs. The authors believe that miR-21 may be a new therapeutic target for proliferative vascular diseases such as atherosclerosis, postangioplasty restenosis, transplantation arteriopathy, and stroke[68].
One of the major roles miRNAs play is the ability of these molecules to control cell death that bears a wide range of biological and pathological implications. While this issue has been firmly established and well advanced in the field of oncology with a realistic hope in developing novel diagnostic/prognostic biomarkers and therapeutic agents as well, relatively little is yet known about the regulation of cardiac and vascular apoptosis by miRNAs and the role of the regulation in cardiovascular pathogenesis. Nonetheless, there are many reasons to believe that miRNAs are much more importantly involved in apoptosis in the cardiovascular system than we currently know; future studies will prove it. We are now just beginning to understand their role as gatekeepers of cell death.
Though a great advancement in elucidating the role and mechanisms of miRNAs in apoptosis and their implications has been made over the past years, many questions remain unanswered. Whether aberrant expression of apoptosis-responsive miRNAs is a consequence or a cause or both or merely a bystander? How do miRNAs in a pool, which are associated with a pathological condition with some being antiapoptotic and others proapoptotic, interplay to produce a final net outcome? How does a single miRNA which can target both pro- and anti-apoptotic protein-coding genes avoid producing skewed effect? The apoptosis-regulating miRNAs may be antiapoptotic or proapoptotic or even neutral, depending upon the cell context (cell types and cellular environment), or specifically, upon whether a cell contains relevant genes for the miRNAs. Intricate fine-tuning of gene expression by miRNAs is presumably under differential influence of degrees of homology and numbers of target sites as well as the context of expression profiles of various genes in the cells of interest. The apoptosis-regulatory effects of miRNAs are likely mediated by multiple signaling pathways in a cell. It is also possible that different signaling pathways underlie the actions of miRNAs in different cells. Moreover, a particular effect of a given miRNA on apoptosis may also be a net outcome of the balance between apoptotic and survival signaling pathways. The apoptosis-regulatory effects of miRNAs are also related to tissue-specific expression of miRNAs. Different tissues/cells have distinct miRNA expression profiles, though the expression of an individual miRNA may not be tissue/cell-specific. The expression patterns of miRNAs in a given tissue/cell may be different under different pathological conditions[16]. These facts make miRNAs amenable for therapeutic intervention of human disease. The obvious implications of miRNAs related to their ability to regulate apoptosis are the potential applications of miRNA-interfering approaches to the treatment of human cancer and cardiovascular disease. Studies on miRNAs related to cancer have been much more advanced compared with those to heart problems. At present, several strategies are available for miRNA-interfering; they include AMO techniques, exogenous miRNA, miR-Mask techniques, and miRNA Mimic techniques[15]. It is therefore plausible to anticipate the promise of unitizing miRNA-interfering as a vivid approach for gene therapy of human disease. Yet we have merely made a first step towards the application of miRNA-interfering technologies. Not until we will have had thorough answers to these questions after rigorous fundamental and clinical studies, will we have better ideas about miRNAs as targets for the development of therapeutic agents for human disease.
Peer reviewers: Dr. Guenther Witzany, Telos-Philosophische Praxis, Vogelsangstrasse 18c, A-5111-Buermoos, Austria; Yiider Tseng, PhD, Associate Professor, Department of Chemical Engineering, University of Florida, Room 223, Museum Road, Chemical Engineering Building, Gainesville, FL 32611-6005, United States
S- Editor Cheng JX L- Editor Ma JY E- Editor Zheng XM
| 1. | Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239-257. |
| 2. | Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1:111-121. |
| 3. | Jaffe R, Flugelman MY, Halon DA, Lewis BS. Ventricular remodeling: from bedside to molecule. Adv Exp Med Biol. 1997;430:257-266. |
| 4. | Sabbah HN, Sharov VG, Goldstein S. Programmed cell death in the progression of heart failure. Ann Med. 1998;30 Suppl 1:33-38. |
| 5. | Hamet P, deBlois D, Dam TV, Richard L, Teiger E, Tea BS, Orlov SN, Tremblay J. Apoptosis and vascular wall remodeling in hypertension. Can J Physiol Pharmacol. 1996;74:850-861. |
| 6. | Behl C. Apoptosis and Alzheimer's disease. J Neural Transm. 2000;107:1325-1344. |
| 7. | Offen D, Elkon H, Melamed E. Apoptosis as a general cell death pathway in neurodegenerative diseases. J Neural Transm Suppl. 2000;153-166. |
| 8. | Intengan HD, Schiffrin EL. Vascular remodeling in hypertension: roles of apoptosis, inflammation, and fibrosis. Hypertension. 2001;38:581-587. |
| 9. | Palojoki E, Saraste A, Eriksson A, Pulkki K, Kallajoki M, Voipio-Pulkki LM, Tikkanen I. Cardiomyocyte apoptosis and ventricular remodeling after myocardial infarction in rats. Am J Physiol Heart Circ Physiol. 2001;280:H2726-H2731. |
| 10. | Grimm S, Noteborn M. Anticancer genes: inducers of tumour-specific cell death signalling. Trends Mol Med. 2010;16:88-96. |
| 11. | Mahmood Z, Shukla Y. Death receptors: targets for cancer therapy. Exp Cell Res. 2010;316:887-899. |
| 12. | Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27 Suppl 2:S52-S57. |
| 13. | Yang B, Lu Y, Wang Z. Control of cardiac excitability by microRNAs. Cardiovasc Res. 2008;79:571-580. |
| 15. | Wang Z. MicroRNA interference technologies. New York: Springer-Verlag 2009; . |
| 16. | Wang Z, Yang B. MicroRNA expression detection methods. New York: Springer-Verlag 2010; . |
| 17. | Ying SY, Lin SL. Intronic microRNAs. Biochem Biophys Res Commun. 2005;326:515-520. |
| 18. | Cuellar TL, McManus MT. MicroRNAs and endocrine biology. J Endocrinol. 2005;187:327-332. |
| 19. | Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376-385. |
| 21. | Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328-336. |
| 22. | Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89-100. |
| 23. | Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83-86. |
| 24. | Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663-4670. |
| 25. | Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231-235. |
| 26. | Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235-240. |
| 27. | Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162-2167. |
| 28. | Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887-901. |
| 29. | Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203-1217. |
| 30. | Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-191. |
| 31. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. |
| 32. | Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011-3016. |
| 33. | Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740-744. |
| 34. | Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522-532. |
| 35. | Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932-935. |
| 36. | Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209-216. |
| 37. | Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199-208. |
| 38. | Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004;101:1679-1684. |
| 39. | Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666-670. |
| 40. | Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434-1437. |
| 41. | Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663-666. |
| 42. | Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343-349. |
| 43. | Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733-2742. |
| 44. | Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655-1666. |
| 45. | Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-798. |
| 46. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. |
| 47. | Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008;283:20045-20052. |
| 48. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. |
| 49. | Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124-1128. |
| 50. | Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-596. |
| 51. | Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573-1576. |
| 52. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. |
| 53. | Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833-838. |
| 55. | Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243-249. |
| 56. | Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118-126. |
| 57. | Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056-2060. |
| 58. | Davis E, Caiment F, Tordoir X, Cavaillé J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743-749. |
| 59. | Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. |
| 60. | Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500. |
| 61. | Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. |
| 62. | Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529-535. |
| 63. | Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599-606. |
| 64. | Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961-16966. |
| 65. | Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-1934. |
| 66. | Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653-4662. |
| 68. | Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579-1588. |
| 69. | Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-6033. |
| 70. | Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994-9000. |
| 71. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. |
| 72. | Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328-14336. |
| 73. | Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82-85. |
| 74. | Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944-13949. |
| 75. | Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731-743. |
| 76. | Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586-1593. |
| 77. | Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472-15477. |
| 78. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. |
| 79. | Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017-5022. |
| 80. | Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298-1307. |
| 81. | Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045-3052. |
| 82. | Tea BS, Dam TV, Moreau P, Hamet P, deBlois D. Apoptosis during regression of cardiac hypertrophy in spontaneously hypertensive rats. Temporal regulation and spatial heterogeneity. Hypertension. 1999;34:229-235. |
| 83. | Teiger E, Than VD, Richard L, Wisnewsky C, Tea BS, Gaboury L, Tremblay J, Schwartz K, Hamet P. Apoptosis in pressure overload-induced heart hypertrophy in the rat. J Clin Invest. 1996;97:2891-2897. |
| 84. | Izumiya Y, Kim S, Izumi Y, Yoshida K, Yoshiyama M, Matsuzawa A, Ichijo H, Iwao H. Apoptosis signal-regulating kinase 1 plays a pivotal role in angiotensin II-induced cardiac hypertrophy and remodeling. Circ Res. 2003;93:874-883. |
| 85. | Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486-491. |
| 86. | Rodríguez M, Lucchesi BR, Schaper J. Apoptosis in myocardial infarction. Ann Med. 2002;34:470-479. |
| 87. | Zidar N, Jera J, Maja J, Dusan S. Caspases in myocardial infarction. Adv Clin Chem. 2007;44:1-33. |
| 88. | Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-1297. |
| 89. | Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16 Spec No 1:R106-R113. |
| 90. | Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293-300. |
| 92. | Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220-6227. |
| 94. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. |
| 95. | Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4-9. |
| 96. | Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776-780. |
| 97. | Slack FJ, Weidhaas JB. MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2006;2:73-82. |
| 98. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. |
| 99. | Yue J, Tigyi G. MicroRNA trafficking and human cancer. Cancer Biol Ther. 2006;5:573-578. |
| 100. | Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7:560-564. |
| 101. | Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert Opin Ther Targets. 2007;11:1119-1129. |
| 102. | Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther. 2007;7:1009-1019. |
| 103. | Giannakakis A, Coukos G, Hatzigeorgiou A, Sandaltzopoulos R, Zhang L. miRNA genetic alterations in human cancers. Expert Opin Biol Ther. 2007;7:1375-1386. |
| 104. | McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253-258. |
| 105. | Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509-3512. |
| 106. | Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. |
| 107. | Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755-11760. |
| 108. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. |
| 109. | Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189-198. |
| 110. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. |
| 111. | Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256-64. |
| 112. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. |
| 113. | Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026-1033. |
| 114. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. |
| 115. | Wynne AM, Mocanu MM, Yellon DM. Pioglitazone mimics preconditioning in the isolated perfused rat heart: a role for the prosurvival kinases PI3K and P42/44MAPK. J Cardiovasc Pharmacol. 2005;46:817-822. |
| 116. | Yasuda S, Kobayashi H, Iwasa M, Kawamura I, Sumi S, Narentuoya B, Yamaki T, Ushikoshi H, Nishigaki K, Nagashima K. Antidiabetic drug pioglitazone protects the heart via activation of PPAR-gamma receptors, PI3-kinase, Akt, and eNOS pathway in a rabbit model of myocardial infarction. Am J Physiol Heart Circ Physiol. 2009;296:H1558-H1565. |
| 117. | Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. |
| 118. | Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133-6140. |
| 119. | Ye Y, Hu Z, Lin Y, Zhang C, Perez-Polo JR. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-{gamma} agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2010;Epub ahead of print. |
| 120. | Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia-reperfusion in mice. FEBS Lett. 2008;582:4137-4142. |
| 121. | Ferrari R, Agnoletti L, Comini L, Gaia G, Bachetti T, Cargnoni A, Ceconi C, Curello S, Visioli O. Oxidative stress during myocardial ischaemia and heart failure. Eur Heart J. 1998;19 Suppl B:B2-B11. |
| 122. | Ide T, Tsutsui H, Kinugawa S, Suematsu N, Hayashidani S, Ichikawa K, Utsumi H, Machida Y, Egashira K, Takeshita A. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ Res. 2000;86:152-157. |
| 123. | Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, Jones PK, Ghanbari H, Wataya T, Shimohama S. Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:759-767. |
| 124. | Patel VA, Zhang QJ, Siddle K, Soos MA, Goddard M, Weissberg PL, Bennett MR. Defect in insulin-like growth factor-1 survival mechanism in atherosclerotic plaque-derived vascular smooth muscle cells is mediated by reduced surface binding and signaling. Circ Res. 2001;88:895-902. |
| 125. | Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134-147. |
| 126. | Antunes F, Cadenas E, Brunk UT. Apoptosis induced by exposure to a low steady-state concentration of H2O2 is a consequence of lysosomal rupture. Biochem J. 2001;356:549-555. |
| 127. | Ren JG, Xia HL, Just T, Dai YR. Hydroxyl radical-induced apoptosis in human tumor cells is associated with telomere shortening but not telomerase inhibition and caspase activation. FEBS Lett. 2001;488:123-132. |
| 128. | Han H, Wang H, Long H, Nattel S, Wang Z. Oxidative preconditioning and apoptosis in L-cells. Roles of protein kinase B and mitogen-activated protein kinases. J Biol Chem. 2001;276:26357-26364. |
| 129. | Wang H, Zhang Y, Cao L, Han H, Wang J, Yang B, Nattel S, Wang Z. HERG K+ channel, a regulator of tumor cell apoptosis and proliferation. Cancer Res. 2002;62:4843-4848. |
| 130. | Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J. 2009;50:377-387. |
| 131. | Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613-618. |
| 132. | Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416-424. |
| 133. | Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, Li Y. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun. 2008;376:548-552. |
| 134. | Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137-1141. |
| 135. | Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831-1840. |
| 136. | Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514-29525. |
| 137. | Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;Epub ahead of print. |
| 138. | Yin C, Salloum FN, Kukreja RC. A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res. 2009;104:572-575. |
| 139. | Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5-14. |