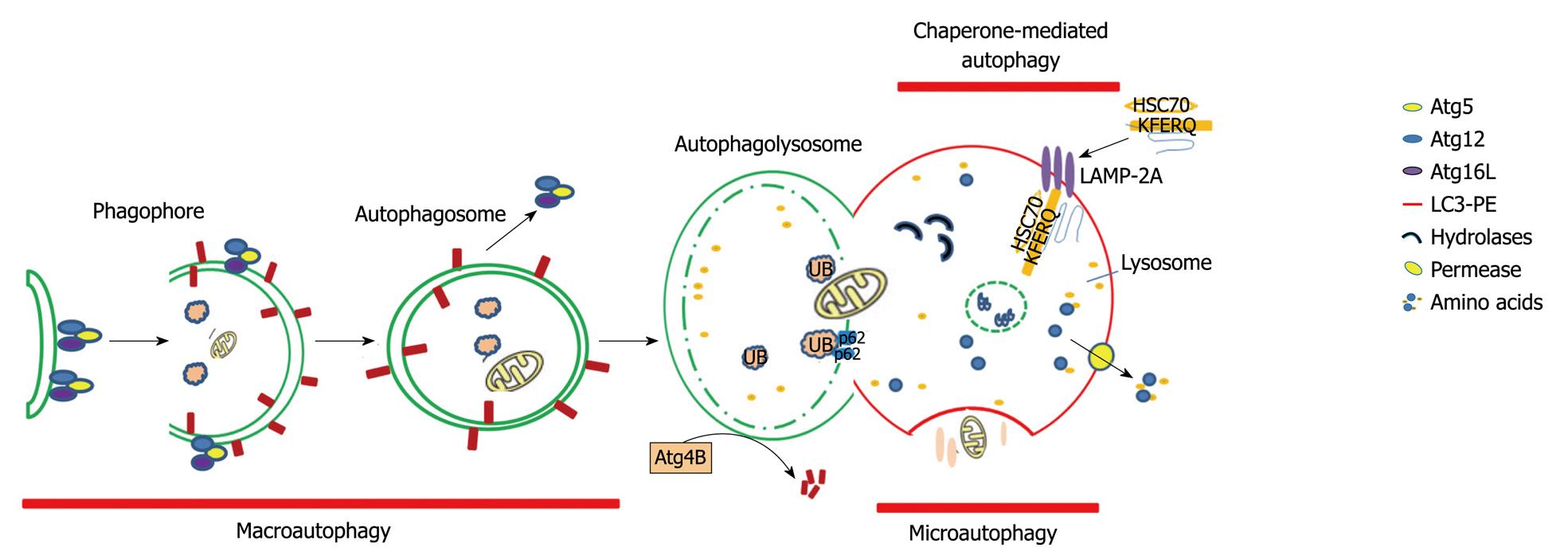
AUTOPHAGY IN LIVER PATHOPHYSIOLOGY
Removal of intracellular protein aggregates
Autophagy has now been recognized to be able to help clear up protein aggregates. The two degradation systems, the ubiquitin-proteasome system and autophagy, are both activated by protein aggregates, but they can differentially degrade different forms of the substrates[20]. Autophagy seems to be able to degrade all forms of misfolded proteins whereas proteasomal degradation is likely limited to soluble proteins[21]. Increased accumulation of ubiquitin positive protein aggregates has been observed in Atg7 liver specific knockout mice, suggesting that autophagy is constitutively acting on the turnover of cytoplasmic proteins, a process that has been classified as “basal autophagy”[22,23].
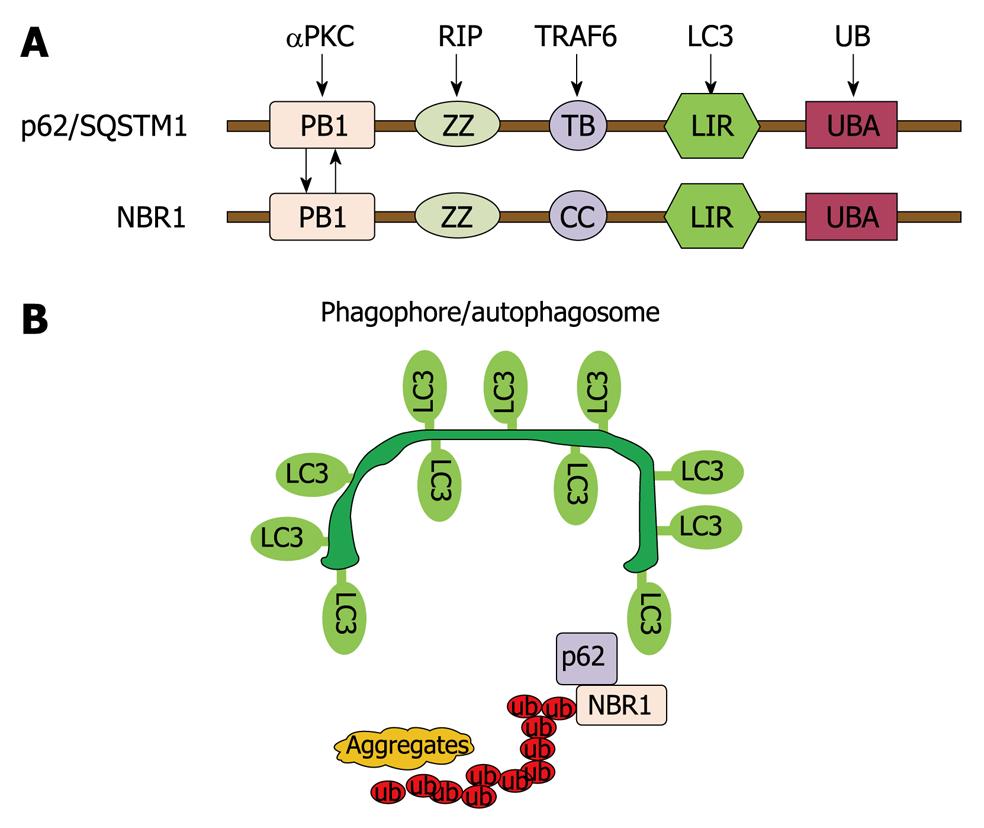
Although autophagy is generally thought to be a non-selective lysosomal degradation pathway, there are many examples showing that autophagy can be selective. In addition to providing nutrition during starvation, selective autophagic degradation of intracellular misfolded proteins plays an important homeostatic function. Insufficient removal of these misfolded proteins may cause protein aggregate-related pathogenesis (discussed below). Accumulating evidence now supports ubiquitination is a candidate signal for autophagic degradation of misfolded and aggregated proteins. Recent studies suggest that this degradative process is mediated through the mammalian protein p62/SQSTM1. p62 directly binds to poly- or mono-ubiquitin through its C-terminal ubiquitin binding domain (UBA) and also binds directly with autophagy proteins light chain-3 (LC3) and GABARAP, and thus acts as a cargo adapter for ubiquitinated proteins and links them to autophagy degradation[24-26]. Using Atg8 as the bait, neighbor of BRCA1 gene 1 (NBR1) is identified as an additional LC3- and Ub-binding protein, which is structurally and functionally like p62 (Figure 2). Inhibition of autophagy leads to the accumulation of protein aggregates that are both p62 and NBR1 positive[27]. Therefore, it is suggested that p62 together with NBR1 promotes autophagic degradation of ubiquitinated proteins. p62 has been found to be localized in Mallory bodies in alcoholic liver disease and p62 may be required for their formations[28].
Figure 2 Autophagy regulates protein homeostasis through interaction with p62 and NBR1.
A: A schematic diagram showing the domain organization of p62 and NBR1 proteins. PB1: Phox and Bem1p domain; ZZ: Zinc finger domain; TB: TRAF6-binding domain; CC: Coiled-coil domain; LIR: LC3-interacting region; UBA: Ub-associated domain; B: p62 and NBR1 are autophagy receptors that interact with both ubiquitin-positive protein aggregates through their UBA domains and target them to autophagosomes through their LIR regions with LC3 on the autophagosomal membranes, thereby promoting autophagy of ubiquitinated targets.
Disruption of basal autophagy in the liver, by generating the liver specific Atg7 in knockout mice, leads to the accumulation of inclusion bodies, abnormal membrane structures, accumulation of peroxisomes and deformed mitochondria, resulting in hepatomegaly and liver injury. Interestingly, deletion of p62 markedly attenuates liver injury induced by the autophagy deficiency due to the deficiency of Atg7[23,29]. The protective effects of the loss of p62 were thought to be due to the suppression of inclusion bodies formation in the liver; however, recent studies suggest that p62 may have multiple functions. Accumulated p62, due to autophagy defects, can promote oxidative stress, alter nuclear factor-κB (NF-κB) regulation and gene expression, and promote tumorigenesis[30]. In addition, p62 can also promote caspase-8 activation by inducing caspase-8 aggregation[31]. Therefore, homeostasis of p62 via autophagy is vital for many cellular functions.
α-1-antitrypsin (AT) deficiency
Study of AT deficiency, which causes liver inflammation and carcinogenesis, was one of the first lines of evidence suggesting a role of autophagy in diseases associated with aggregate-prone proteins[32]. AT, the archetype of the Serpin supergene family, is the principal blood-borne inhibitor of destructive neutrophil proteases including elastase, cathepsin G, and proteinase[33,34]. The classical form of α1AT deficiency affects 1 in 1800 live births in Northern European and North American populations[34]. The normal AT protein is secreted from hepatic cells into the bloodstream, where it inhibits the neutrophil proteases. However, a mutation in the AT gene results in misfolding of the mutant protein, which cannot transport from the endoplasmic reticulum (ER) and becomes stuck in the ER as an aggregated form[20]. In the liver cells of AT deficiency patients, an increased number of autophagosome is readily observed[35]. Autophagy mainly serves to degrade the mutant AT aggregates in the ER, whereas the soluble mutant proteins are subjected to ER-associated degradation (ERAD) by proteasomes[20,36].
Hypofibrinogenemia
Hypofibrinogenemia is another liver ER storage disease. A mutant form of fibrinogen, named Aguadilla γD, forms protein aggregates in the hepatic ER, causing similar pathological alterations to AT deficiency[37]. Although most of the mutant forms can be degraded via the ERAD pathway, autophagy helps to degrade excess aberrant polypeptide formed aggregates within the ER[37]. These studies suggest a protective role of autophagy in relieving the cytotoxicity associated with abnormal protein aggregates in the ER. When the unfolded protein response and ERAD is saturated or impaired, the accumulated abnormal proteins in the ER cause ER stress. The ER stress signaling pathways, such as Ire1/, PERK/eIF2α and JNK, may be involved in the ER-accumulated aggregates induced-autophagy[38,39]. Autophagy may help to remove part of the abnormal ER, presumably together with the accumulated protein aggregates to maintain organelle homeostasis. In this context, autophagy serves as an “ERAD-like” mechanism and contributes to ER quality control[21].
Alcoholic Mallory body
Autophagy may also play a role in alcohol-induced liver pathogenesis. Earlier studies have shown that alcohol fed to rats produced hepatomegaly, associated with enlargement of the hepatocytes and protein accumulation[40,41]. The mechanisms for the alcohol-induced protein accumulation in hepatocytes are not completely known. It is suggested that alcohol exposure can alter the proteolytic activity of hepatic lysosomes[42], alter the trafficking of lysosomal enzymes[43], and probably alter microtubule structures and vesicle protein trafficking in hepatocytes[44]. Moreover, there is also evidence that ethanol administration can inhibit proteasome activity, likely due to the ethanol metabolism and generation of reactive oxygen species (ROS)[45]. Suppression of proteasome activity and induction of ROS have been shown to be able to induce autophagy in other cell types and systems. We recently found that binge drinking of alcohol indeed could induce autophagy in the mouse liver (Ding et al manuscript in press).
Besides the general protein accumulation, alcohol exposure also leads to the formation of inclusion bodies known as Mallory bodies in hepatocytes, which are frequently observed in alcoholic hepatitis and cirrhosis[46]. Mallory bodies are filaments of intermediate diameters with intermediate filament components[47]. These structures contain cytokeratin 8 and cytokeratin 18 and ubiquitin positive protein aggregates and share many similar characteristics with other inclusion bodies found in neuronal degenerative diseases, such as Lewy bodies in Alzheimer’s disease and Huntington inclusions bodies in Huntington’s disease[47]. Although Mallory bodies have no longer been considered just as a marker of alcoholic disease, the biological significance of Mallory bodies in alcohol-induced liver injury is still unclear. Moreover, the mechanisms for the induction of Mallory bodies are also not completely known. Inhibition of proteasome activity by proteasome inhibitor can induce Mallory body-like structures in cultured cells and in mouse liver. Interestingly, induction of autophagy by rapamycin, a mTOR inhibitor, significantly suppresses Mallory body formation both in vitro and in vivo, suggesting autophagy plays an important role in alcoholic Mallory body formation and induction of autophagy may help to attenuate Mallory body formation[48,49].
Degradation of organelles via autophagy
Unlike proteasomal degradation, autophagy can degrade not only cytosolic proteins but also organelles such as mitochondria, peroxisome and ER.
Degradation of peroxisome by autophagy (pexophagy)
Autophagy selectively removes peroxisomes (pexophagy), which was first discovered in yeast when the culture medium was switched and peroxisomal function was no longer required for growth[50]. It was further found that both macroautophagy and microautophagy could be used to degrade peroxisomes in yeast. For example, in P. pastoris, glucose-induced peroxisome degradation mainly occurs through microautophagy, whereas ethanol-induced degradation utilizes macroautophagy[51].
In mammalian cells, autophagic degradation of peroxisomes has been observed in hepatocytes treated with clofibrate or dioctyl phthalate[52], two drugs that activate the peroxisome proliferators-activated receptors to induce accumulation of peroxisome in mammalian cells[52,53]. The requirement of autophagy to degrade peroxisomes in hepatocytes is further proved in a recent study using the liver specific Atg7-deficient mouse challenged with phthalate esters, an agent that can induce marked increase of peroxisome numbers and size in the liver[54].
Pexophagy has been well known to be a selective process, and its mechanisms have been best studied in yeast. In P. pastoris, PAtg30 functions as an adapter molecule to interact with peroxisomal membrane proteins PpPex14 and PpPex3 and autophagy proteins PpAtg11 and PpAtg17, and in turn, links peroxisomes to autophagy degradation[55]. In mammalian cells, it seems that peroxisomes can be removed by autophagy similar to the ubiquitinated protein aggregates. A recent study reveals that fusion of a single ubiquitin moiety to a peroxisome integral membrane protein, PMP34, is sufficient to trigger selective autophagy degradation of peroxisomes. Interestingly, this kind of selective pexophagy is also mediated by p62, similar to the role of p62 in the autophagic degradation of protein aggregates as discussed above[56].
Mitophagy
Enveloped mitochondria in autophagosomes have been observed by De Duve et al[1] as early as 1966 in drug injected rat liver cells. This process is now termed “mitophagy”[1,57]. Increasing evidence now supports mitophagy as a selective process. In yeast, Atg32, a mitochondria-anchored protein, has recently been found to be essential for selective mitophagy, although a mammalian homologue of Atg32 has not been found[58,59]. Except for Atg32, Uth1p and Aup1 have also been found to be involved in mitochondrial autophagy although it seems that they only play roles in certain models[60,61]. The mechanisms of mitophagy are more complicated in mammalian cells and the following mechanisms discussed below have been implicated in mammalian cell mitophagy.
Mitochondrial permeability transition (MPT)
MPT, an event that has long been proposed to regulate apoptosis and necrosis in mammalian cells, may also play a role in regulating mitophagy. When cultured hepatocytes were deprived of nutrition, mitochondria became depolarized and moved into acidic vesicles[62]. Cyclosporin A, a MPT inhibitor, significantly inhibited mitochondria depolarization and mitophagy during nutrient deprivation in hepatocytes[62]. Besides the nutrient deprivation, mitophagy in hepatocytes was also induced when selected mitochondria inside living hepatocytes were subjected to laser-induced photodamage[63]. Mitophagy after nutrition deprivation was further confirmed by using cultured hepatocytes from the GFP-LC3 transgenic mouse, in which some mitochondria were enveloped by the green GFP-LC3 signals[63]. As mitophagy could selectively remove those damaged mitochondria, it has been proposed that mitophagy could be protective against cell death, as these mitochondria produce toxic free radicals and release mitochondria apoptotic factors[57]. Indeed, in drug induced pathogenesis of Reye syndrome, salicylate induced mitochondria damage by inducing MPT in hepatocyte[64]. Interestingly, autophagic degradation of damaged mitochondria was found in liver biopsies of Reye syndrome patients[65], and also in an influenza B virus model of Reye’s syndrome in mice[66].
Mitochondrial fragmentation
Mitochondria are dynamic organelles undergoing fusion and fission constantly. It is tempting to speculate that fragmented mitochondria are more readily taken up by autophagosomes due to their size. In a nitric oxide (NO)-induced neuron damage model, it was found that Fiss1, a protein that regulates mitochondria fission, was involved in mitophagy[67]. We and others found that inhibition of mitochondria fragmentation such as by overexpression of a mutant form of mitochondrial fission molecular, Drp1K38A, can also suppress mitophagy[67]. Moreover, using the Mfn1 deficient mouse embryonic fibroblasts, in which mitochondria are already fragmented due to the lack of mitochondrial fusion protein Mfn1 in these cells, we found a much higher rate of mitophagy in Mfn1-deficient cells than that of wild type cells (Ding et al, unpublished observations). Interestingly, in the nutrition deprivation-induced mitophagy in hepatocytes, it is found that only a portion of individual mitochondria becomes sequestered, in some cases sequestered from both the ends and middle parts of mitochondria[63]. These data tend to support that the mitochondrial fission process may also be coordinated with autophagosome formation[63].
Nix and BNIP3
How the damaged mitochondria are recognized by the autophagy machinery in mammalian cells is not clear. BNIP3 (Bcl-2/E1B-19kDa interacting protein 3) was first identified in a yeast two-hybrid screen for proteins that interact with adenovirus E1B 19 kDa[68]. BNIP3 is a pro-apoptotic mitochondrial protein that contains a Bcl-2 homology 3 (BH3) domain and a carboxyl terminal transmembrane (TM) domain[69,70]. BNIP3 is inserted into the outer mitochondrial membrane through its C-terminus transmembrane domain while its N-terminus is exposed in the cytoplasm. Unlike other BH3-only pro-apoptotic proteins, the TM domain of BNIP3, but not its BH3 domain, is required for mitochondria targeting and pro-apoptotic function[70,71]. Nix/BNIP3L is a homolog of BNIP3 and they share 53%-56% amino acid sequence identity[72]. In addition to apoptosis, BNIP3 has been implicated in necrosis and autophagic cell death[73-75]. However, BNIP3 is not ubiquitously expressed under normal conditions. It is only expressed in skeletal muscle and brain at a low level under physiological conditions. It is markedly expressed in regions of solid tumors or normal tissue in response to hypoxia and appears to be regulated by hypoxia-inducible factor (HIF), which binds to a site on the BNIP3 promoter[76,77].
BNIP3 has been found to be important for ceramide or arsenic trioxide induced autophagy in malignant glioma cells[75,78]. Using cultured mouse embryonic fibroblast (MEF) cells, it is demonstrated that mitophagy is induced by hypoxia. This mitophagy requires the HIF-1-inducible expression of BNIP3. Mitophagy serves as an adaptive metabolic response to prevent increased levels of ROS via removal of damaged mitochondria, and in turn to mitigate cell death[79]. The critical role of BNIP3 in mitophagy has further been supported by an elegant genetic model. During the maturation, reticulocytes completely eliminate their mitochondria partly through autophagy, a process that provides a physiological model to study mitophagy. In Nix-deficient mice, mitochondrial clearance in reticulocytes is significantly inhibited or retarded, suggesting that Nix is required for the selective elimination of mitochondria[80]. Later on, it was discovered by another group that the role of Nix for mitophagy is likely due to the loss of mitochondria membrane potential (MMP) induced by Nix, because treatment with a mitochondria uncoupler or a BH3 mimetic, induces the loss of MMP and restores the sequestration of mitochondria into autophagosomes in Nix-deficient erythroid cells[81]. Their results thus suggest that Nix-dependent loss of MMP is important for targeting damaged mitochondria to autophagosomes. This notion may also help to explain why mitochondrial permeability transition is involved in hepatocytes mitophagy, because in most cases, the onset of mitochondria permeability transition can lead to the loss of MMP.
Parkin and ubiquitin
As discussed above, ubiquitin plays an important role for the autophagic removal of not only protein aggregates but also organelles such as peroxisomes. This is mainly achieved through several adapter molecules, such as p62 and NBR1, which can directly interact with poly- and mono-ubiquitin and LC3. Therefore, it is very tempting to hypothesize that ubiquitin may also play a role in mitophagy. Indeed, it is recently found that Parkin, an ubiquitin E3 ligase, could be recruited selectively to impaired mitochondria and to promote their degradation via autophagy[82]. Interestingly, although Parkin was first identified as a gene implicated in autosomal recessive Parkinsonism, Parkin knockout mice have enhanced hepatocyte proliferation and hepatocellular carcinoma (HCC)[83]. It is not known whether the lack of Parkin in the hepatocytes would affect the hepatocyte mitochondrial turnover resulting in an increased number of damaged mitochondria, increased levels of oxidative stress and genome instability, which contribute to tumorigenesis. Although direct experimental evidence is not yet available to show the role of ubiquitination of mitochondria in mitophagy, it has been noted that sperm-derived mitochondria are completely eliminated after fertilization to ensure that only maternal mitochondrial DNAs are inherited. Interestingly, sperm mitochondria have been found to be tagged with ubiquitin, although whether autophagy was involved in this process has not been determined.
ER-phagy
The ER was first identified as being selectively sequestrated by autophagic vacuoles as early as 1973, when hepatocytes were previously treated with phenobarbital followed by cessation of the treatment[84]. In this case, based on morphological study, the elimination is mainly of smooth ER. This was later confirmed by a study using a biochemical approach, in which two typical ER membrane proteins, phenobarbital (PB)-inducible cytochrome P-450 and NADPH-cytochrome P-450 reductase, were selectively degraded by autophagy in rat liver when rats were treated with phenobarbital followed by removal[85]. It will be interesting if this model can also be applied in the liver specific Atg-7 knock out mouse.
Currently, how ER is selectively removed by autophagy is not known. The ER is a major intracellular site for proper protein folding and posttranslational modifications. Disrupting the oxidized environment of ER by dithiothreitol (DTT), calcium homeostasis by thapsigargin, or inhibition of glycosylation by tunicamycin, can all lead to the accumulation of misfolded proteins in the ER and causes the so called unfolded protein response (UPR)[21]. We and others have demonstrated that ER stress can induce autophagy, likely through the UPR components such as Ire1, perk and eif2α or the ER calcium leakage[38,39,86,87].
Lipohagy
Lipid droplets (LDs) are intracellular storage depots for neutral lipid that are found in all kinds of cells, ranging from bacteria to human. The LD has been considered as an organelle with a polar lipid monolayer membrane that envelops the hydrophobic core of triglycerides (TGs), diacyglycerol (DG), cholesterol ester (CE), and other esters in various proportions[88]. The phospholipid composition of the LD is very similar to the ER membrane, which includes phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI)[89]. There are also a variety of proteins associated with the LD membrane. For example, more than 10 Rab proteins, including Rab5, -7, -11 and -33, have been detected in isolated LDs. However, among them, only Rab18 has been confirmed by microscopic co-localization studies[90]. In addition to the Rab proteins, PAT proteins are perhaps the most characterized LD associated proteins. PAT proteins, named after perilipin, ADRP, and the tail-interacting protein of 47 kDa (TIP47), mainly regulate cytosolic lipase mediated lipolysis, which has been thought to be a major pathway for the regulation of lipid homeostasis[88]. However, recent work by the Czaja and Cuervo groups clearly demonstrates that autophagy also plays an important role in lipid homoeostasis in hepatocytes by autophagic lipolysis[91]. Suppression of the autophagic pathway, either by a genetic or pharmacological approach, leads to the accumulation of LDs in hepatocytes and other cells. The autophagic marker LC3-II is highly enriched in the LD fractions, and LDs are found to be enveloped by GFP-LC3 positive vesicles. More importantly, it seems that autophagy plays an important role in the clearance of the accumulated LDs in hepatocytes, in particular, in response to the methionine- and choline-deficient (MCD) diet or oleate addition-induced lipid load[91]. However, in starvation-induced hepatic lipid accumulation, it is found that knockout of Atg7 actually leads to less lipid accumulation in the liver, suggesting that different stress-induced lipid accumulation or a different source of lipids maybe differentially regulated by autophagy or some of the Atg proteins may have non-autophagic functions such as to regulate the LD formation[92]. Nevertheless, these findings open a new possible therapeutic approach for treating liver steatosis induced by a high fat diet or obesity via induction of autophagy.
Xenophagy for hepatitis virus
There is an increasing body of evidence now supporting autophagy and/or the autophagy genes as having both anti-viral and pro-viral capacities against various viruses. Autophagy can directly recognize and enwrap virions and/or viral components and target them for degradation in lysosomes, a process termed as “xenophagy”[11,93,94]. Autophagy may also regulate the innate and adaptive immune system to protect against viral infections. In order to counteract autophagy to survive, it is not surprising that some viruses can use some mechanisms to either inhibit autophagy or escape from autophagy recognition. In support of this concept, it has been shown that herpes viruses and lentiviruses can use some viral proteins to inhibit autophagy. For example, ICP34.5, a neurovirulence protein from Herpes simplex virus type 1 (HSV-1), binds protein phosphatase 1α to counter PKR-mediated phosphorylation of eIF2α and in turn suppresses autophagy. In addition, ICP34.5 may also suppress autophagy by binding to the autophagy-promoting protein Beclin 1[93]. Some other intracellular pathogens can escape from autophagic degradation by either suppressing the fusion of autophagosomes with lysosomes or escaping autophagy recognition[95,96].
In the liver, both hepatitis B and C viruses have been shown to be involved in the regulation of autophagy. Beclin-1, an essential autophagy protein, is found to be upregulated in hepatitis B virus-infected cancerous liver tissues. Enforced expression of HBV X protein induces Beclin-1 upregulation in cultured hepatoma cells and, more importantly, enhanced starvation-induced autophagy[97]. In contrast to hepatitis B, hepatitis C virus replication is more complicated. Transfection of HCV viral RNA into Huh7.5 cells leads to the accumulation of autophagosomes and this induction seems to depend on HCV virus-induced ER stress and an unfolded protein response (UPR)[98]. However, this autophagic response is not complete because the long lived protein degradation is not changed, suggesting accumulated autophagosomes are either due to a defect of fusion with lysosomes or alterations of the lysosomes due to the infection of HCV. Interestingly, siRNA knockdown of some essential autophagy genes, such as Atg7, LC3, Beclin-1, Atg5 and Atg12 all suppress HCV replication[98,99]. Moreover, chloroquine, an autophagy inhibitor by increasing lysosomal pH, also significantly suppresses HCV replication in hepatocytes[100]. However, it is found that HCV proteins failed to co-localize with autophagy proteins in infected cells, suggesting the HCV replication complex does not assemble on autophagic vesicles[101]. It remains unknown exactly how autophagy proteins affect HCV replication, and it is possible that the autophagy pathway may provide an initial membranous support for translation of incoming RNA before the accumulation of viral proteins or some autophagy proteins may have non-autophagic effects for viral replication.
AUTOPHAGY IN LIVER TUMORIGENESIS AND TUMOR METASTASIS
As one protein degradation and recycling pathway, autophagy has been generally believed to be a pro-survival pathway. In nutrient starvation conditions, the pro-survival function of autophagy has been very well characterized[102]. Under the conditions of nutrient starvation, autophagy can recycle the macromolecules and thus help to overcome the moment of stress[102]. This hypothesis is clearly supported by the fact that deletion of autophagy genes leads to increased cell death under nutrient deprivation. Autophagy’s role in organism survival has been observed in yeast, plants, worms, flies and mice. Atg5, Atg3 or Atg7 knockout mice die during the neonatal period when the placental blood is no longer supplied. Atg5 knockout mice exhibit reduced amino acid concentrations in plasma and tissues and show signs of energy depletion. This situation can be considered a form of starvation, during which autophagy is critical for survival[23,103]. Autophagy also acts in a protective role during other cell stress, and in this setting, autophagy is used as a strategy to remove either toxic protein aggregates or damaged mitochondria and mitochondrial-generated ROS that could activate apoptosis[30,38,39]. However, autophagy can also contribute to cell death if the process is over activated and deregulated, resulting in excessive catabolism and/or hijacking of the apoptosis machinery[104,105]. When hepatocytes are under starvation conditions, it was reported that ferritin could be degraded in the autophagosomes. The subsequent generated pool of free iron sensitized hepatocytes to be killed by oxidative stress, likely through the iron-mediated Fenton-reaction and, in turn, enhanced oxidative stress[106].
Although constitutive autophagy is important for cellular homeostasis and cell survival, paradoxically, loss of autophagy has been found to promote tumorigenesis. An essential autophagy gene, Beclin 1, was frequently found monoallelically deleted in many human cancers, such as breast, prostate and ovarian cancers[107]. Mice with allelic loss of Beclin 1 are prone to HCC, lung adenocarcinoma, mammary hyperplasia, and lymphoma. Loss of heterozygosity of UVRAG, a Beclin 1 interacting protein, is frequently observed in colon cancers[108,109]. Moreover, loss of other autophagy regulatory genes, such as bif-1 and atg4C, also increased tumorigenesis in mice[109,110]. To further support the concept that autophagy may suppress tumorigenesis, many other known tumor suppressor genes, such as Lkt, Ampk, Pten, are positive regulators of autophagy[111-114]. In contrast, many oncogenes products, including phosphatidylinosital 3-kinase, Akt and anti-apoptotic Bcl-2 family proteins, suppress autophagy[115].
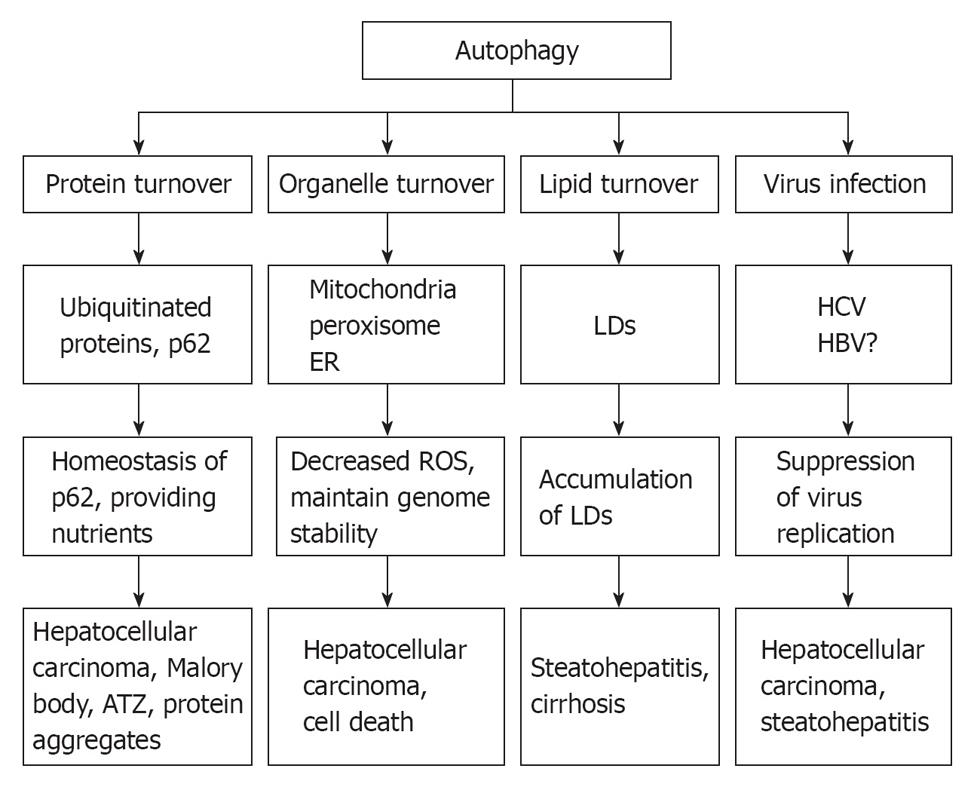
Mice that have autophagy defects develop liver injury, steatohepatitis and HCC[16,29,91,115]. Autophagy defects can lead to an increased level of oxidative stress, accumulation of damaged mitochondria and intracellular p62, an adaptor protein that functions to direct polyubiquitinated proteins to autophagosomes for degradation. Sustained p62 expression resulting from autophagy defects is sufficient to alter NF-κB regulation and gene expression and to promote tumorigenesis. In contrast, suppression of ROS production and p62 expression inhibit tumorigenesis[30]. Increased levels of p62 have been documented in alcoholic liver disease as a major component of Mallory body and alcoholic liver injury and have been implicated to promote HCC[28,116]. However, whether p62 contributes to alcoholic related HCC is not known. Steatohepatitis has been implicated to promote HCC but also could result from autophagy suppression. Moreover, hepatitis C virus can also inhibit autophagy and thus may provide an additional mechanism to promote HCC[98]. Taken together, autophagy plays multiple essential roles in liver pathophysiology by removing misfolded proteins, regulating hepatocellular organelle turn over, maintaining hepatic lipid homeostasis, and influencing hepatitis virus infection (Figure 3). Therefore stimulation of autophagy in liver may thus have therapeutic effects to mitigate steatohepatitis, mitochondria damage, accumulation of 62 and virus infection and may provide a novel means to suppress HCC.
Figure 3 Role of autophagy in liver pathophysiology.
At least 4 different roles that autophagy may play in liver physiology and liver diseases: remove misfolded proteins, regulate hepatocellular organelle turn over, maintain hepatic lipid homeostasis, and influence hepatitis virus infection. As a result, defects in autophagy may lead to accumulation of alcoholic Mallory bodies, α-antitrypsin deficiency-induced liver injury, increased hepatocyte cell death, steatohepatitis and hepatocellular carcinoma. ER: Endoplasmic reticulum; ROS: Reactive oxygen species; LDs: Lipid droplets.