Copyright
©The Author(s) 2016.
World J Biol Chem. May 26, 2016; 7(2): 206-222
Published online May 26, 2016. doi: 10.4331/wjbc.v7.i2.206
Published online May 26, 2016. doi: 10.4331/wjbc.v7.i2.206
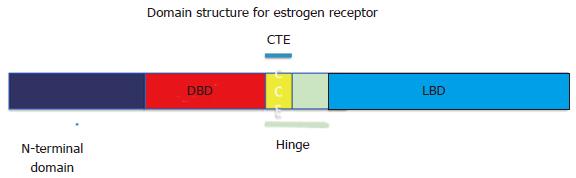
Figure 1 Domain structure for estrogen receptor.
The structural and functional domains include the N-terminal domain, DBD, CTE, the flexible hinge region and LBD. DBD: DNA binding domain; CTE: C-terminal extension; LBD: Ligand binding domain.
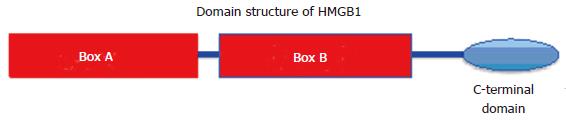
Figure 2 The structural and functional domains of high mobility group protein 1 protein.
The tripartite structure includes the A and B boxes which are basic and the acidic C-terminal domain made up entirely of acidic (asp/glu) residues. HMGB1: High mobility group protein 1.
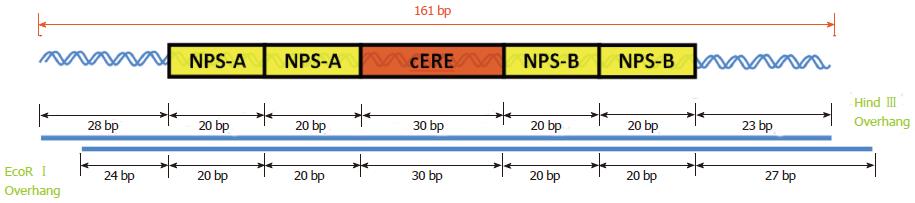
Figure 3 A schematic drawing of the 161 bp DNA containing four nucleosome positioning sequences (yellow boxes), that straddle the 30 bp consensus estrogen response element (red box) at the dyad axis.
The 161 bp DNA was excised by EcoRI and HindIII digestion of the pGEM-Q2-2E2 plasmid. cERE: Consensus estrogen response element.
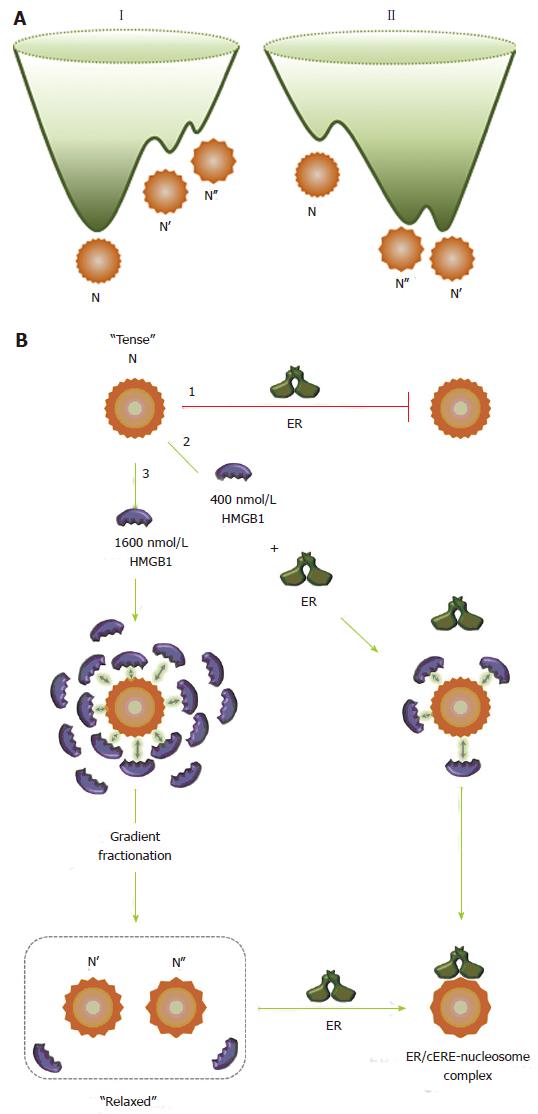
Figure 4 High mobility group protein 1 relaxes the canonical nucleosome structure and facilitates estrogen receptor binding.
A: Energy landscapes for canonical (I) and HMGB1-restructured nucleosomes (II). A hypothetical representation for the energy landscape of the canonical nucleosome, N, and the HMGB1-restructured nucleosomes, N’ and N’’. Using conventional isolation protocols, the canonical nucleosome, N, is the predominant and thermodynamically most stable conformation. N’ and N’’ are in low abundance, higher energy conformational isomers that are kinetically trapped near the bottom of energy landscape I. HMGB1 interaction with N reduces intranucleosomal constraints, which resets the energy landscape (II), resulting in a population shift in which the N population significantly decreases and the population of the more “relaxed” and accessible N’ and N’’ states increases. The more unstable form, N’’, sets in a shallower potential well than that for N’. Although interactions with HMGB1 provide the driving force to restructure N into these states, these forms remain stable and although in equilibrium with the canonical state under many solution conditions, can revert to the canonical nucleosome on challenge with increasing concentrations of NaCl and DNA; B: Interaction of the nucleosome with HMGB1 and ER. The canonical nucleosome (N) represents a “tense” and relatively inaccessible conformational isomer (pathway 1) and ER does not bind to the canonical nucleosome state. In the presence of 400 nmol/L HMGB1 (pathway 2), due to the transient and dynamic “hit and run” interaction of HMGB1 with the nucleosome, represented by arrows, the intranucleosomal constraints are relaxed, which facilitates ER binding. ER binds to the nucleosome to form ER/cERE nucleosome complex. In the presence of 1600 nmol/L (pathway 3), the intranucleosomal constraints are relaxed due to increased “hit and run” interaction of HMGB1. After gradient fractionation, the restructured nucleosomes (N’ and N’’) are isolated and contain only low nmol/L levels of HMGB1, which maintain the more accessible and “relaxed” conformational isomers (N’ and N’’) that permit ER binding. HMGB1: High mobility group protein 1; ER: Estrogen receptor.
- Citation: Scovell WM. High mobility group protein 1: A collaborator in nucleosome dynamics and estrogen-responsive gene expression. World J Biol Chem 2016; 7(2): 206-222
- URL: https://www.wjgnet.com/1949-8454/full/v7/i2/206.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v7.i2.206